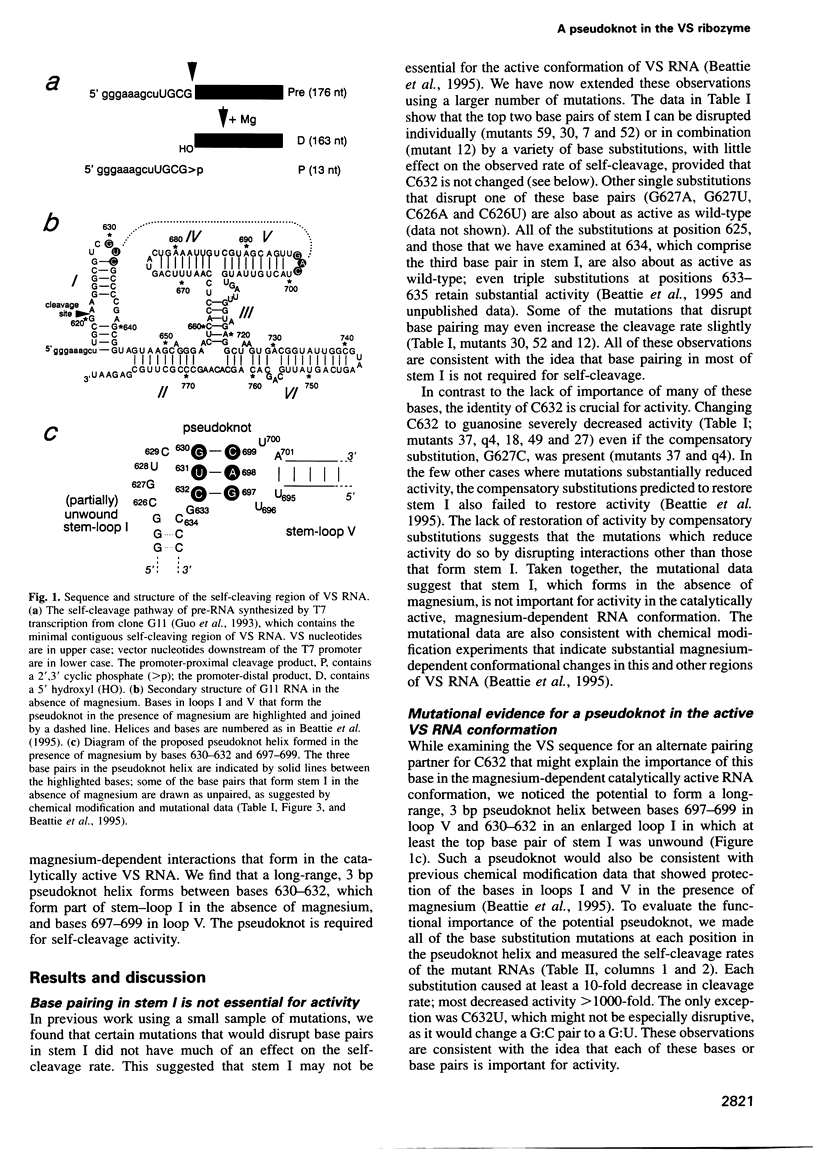
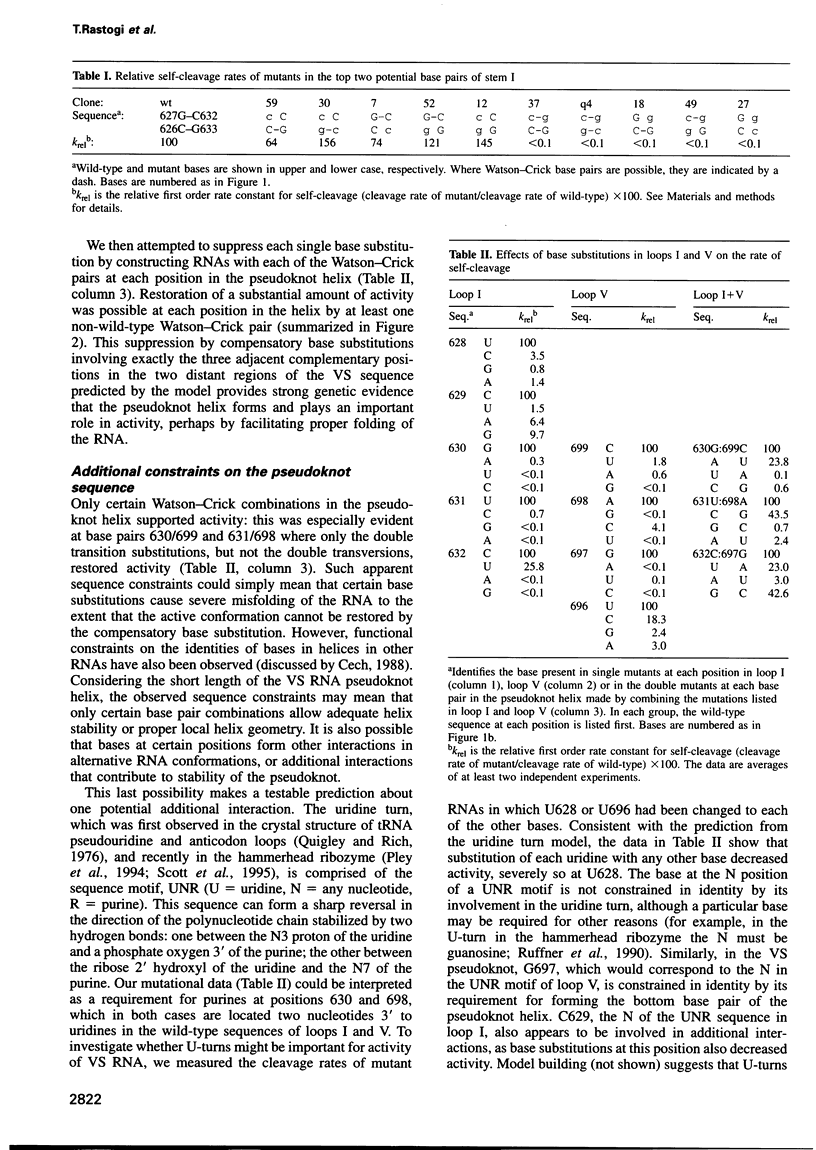
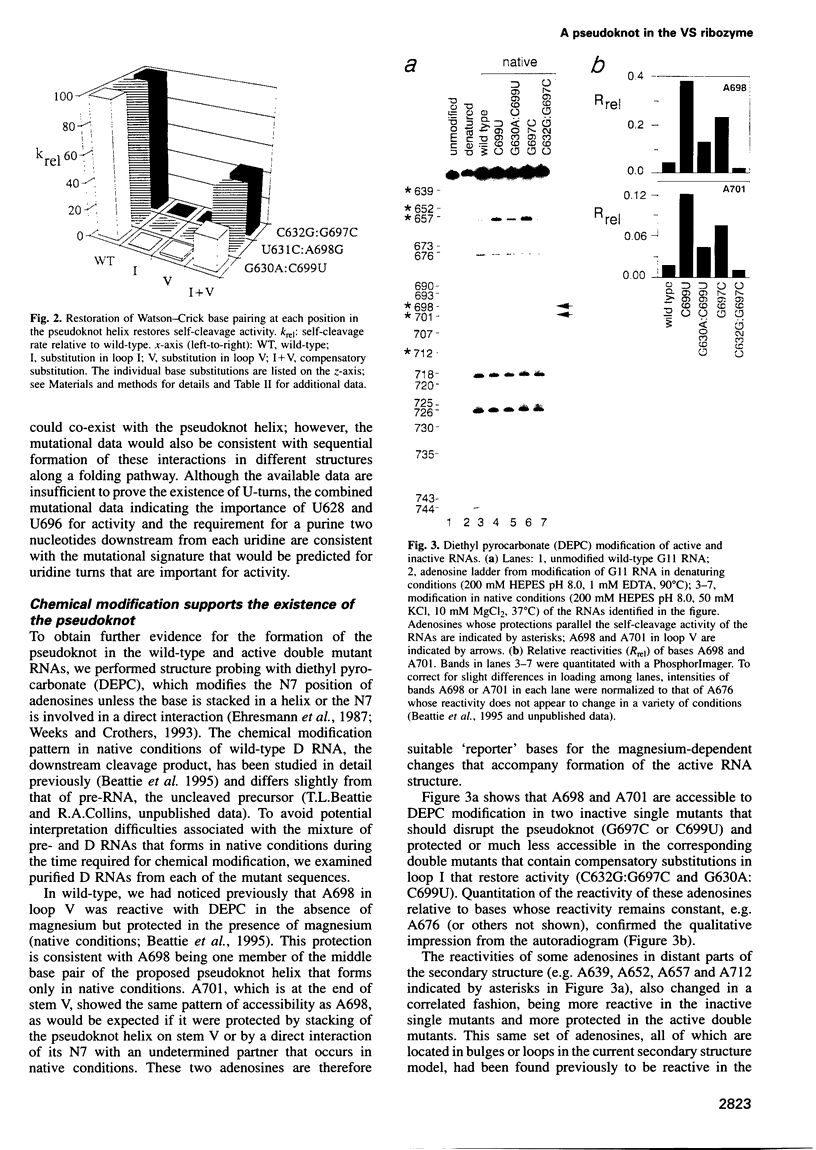
Abstract
Four small RNA self-cleaving domains, the hammerhead, hairpin, hepatitis delta virus and Neurospora VS ribozymes, have been identified previously in naturally occurring RNAs. The secondary structures of these ribozymes are reasonably well understood, but little is known about long-range interactions that form the catalytically active tertiary conformations. Our previous work, which identified several secondary structure elements of the VS ribozyme, also showed that many additional bases were protected by magnesium-dependent interactions, implying that several tertiary contacts remained to be identified. Here we have used site-directed mutagenesis and chemical modification to characterize the first long-range interaction identified in VS RNA. This interaction contains a 3 bp pseudoknot helix that is required for tertiary folding and self-cleavage activity of the VS ribozyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beattie T. L., Olive J. E., Collins R. A. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Chang K. Y., Tinoco I., Jr Characterization of a "kissing" hairpin complex derived from the human immunodeficiency virus genome. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8705–8709. doi: 10.1073/pnas.91.18.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. A., Olive J. E. Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry. 1993 Mar 23;32(11):2795–2799. doi: 10.1021/bi00062a009. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Saville B. J. Independent transfer of mitochondrial chromosomes and plasmids during unstable vegetative fusion in Neurospora. Nature. 1990 May 10;345(6271):177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992 Dec 1;31(47):11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- Eguchi Y., Tomizawa J. Complexes formed by complementary RNA stem-loops. Their formations, structures and interaction with ColE1 Rom protein. J Mol Biol. 1991 Aug 20;220(4):831–842. doi: 10.1016/0022-2836(91)90356-b. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluick T. C., Draper D. E. Thermodynamics of folding a pseudoknotted mRNA fragment. J Mol Biol. 1994 Aug 12;241(2):246–262. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- Guo H. C., Collins R. A. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J. 1995 Jan 16;14(2):368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. C., De Abreu D. M., Tillier E. R., Saville B. J., Olive J. E., Collins R. A. Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J Mol Biol. 1993 Jul 20;232(2):351–361. doi: 10.1006/jmbi.1993.1395. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Nolan J. M., Malhotra A., Brown J. W., Harvey S. C., Pace N. R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994 Sep 1;13(17):3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S., Berzal-Herranz A., Chowrira B. M., Butcher S. E., Burke J. M. Substrate selection rules for the hairpin ribozyme determined by in vitro selection, mutation, and analysis of mismatched substrates. Genes Dev. 1993 Jan;7(1):130–138. doi: 10.1101/gad.7.1.130. [DOI] [PubMed] [Google Scholar]
- Kennell J. C., Saville B. J., Mohr S., Kuiper M. T., Sabourin J. R., Collins R. A., Lambowitz A. M. The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev. 1995 Feb 1;9(3):294–303. doi: 10.1101/gad.9.3.294. [DOI] [PubMed] [Google Scholar]
- Krol A., Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Marino J. P., Gregorian R. S., Jr, Csankovszki G., Crothers D. M. Bent helix formation between RNA hairpins with complementary loops. Science. 1995 Jun 9;268(5216):1448–1454. doi: 10.1126/science.7539549. [DOI] [PubMed] [Google Scholar]
- Michel F., Ferat J. L. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Paillart J. C., Marquet R., Skripkin E., Ehresmann B., Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994 Nov 4;269(44):27486–27493. [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. Assessment of disparate structural features in three models of the hepatitis delta virus ribozyme. Nucleic Acids Res. 1993 Aug 25;21(17):3959–3965. doi: 10.1093/nar/21.17.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Saville B. J., Collins R. A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990 May 18;61(4):685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- Saville B. J., Collins R. A. RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8826–8830. doi: 10.1073/pnas.88.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. G., Finch J. T., Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995 Jun 30;81(7):991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S. T., Tuschl T., Eckstein F. Probing RNA tertiary structure: interhelical crosslinking of the hammerhead ribozyme. RNA. 1995 Aug;1(6):575–583. [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A. M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994 Jun 1;4(6):488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. Major groove accessibility of RNA. Science. 1993 Sep 17;261(5128):1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]