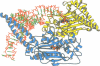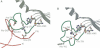Abstract
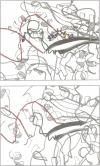
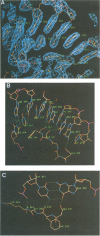
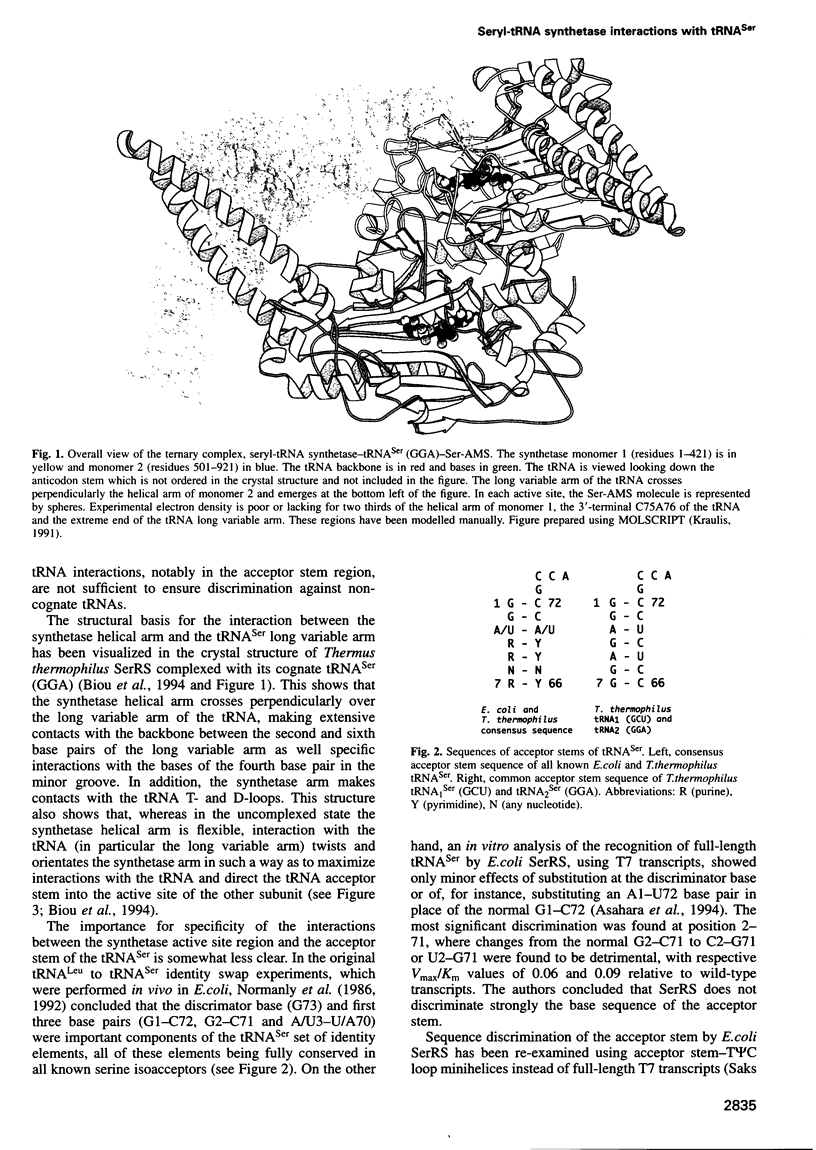
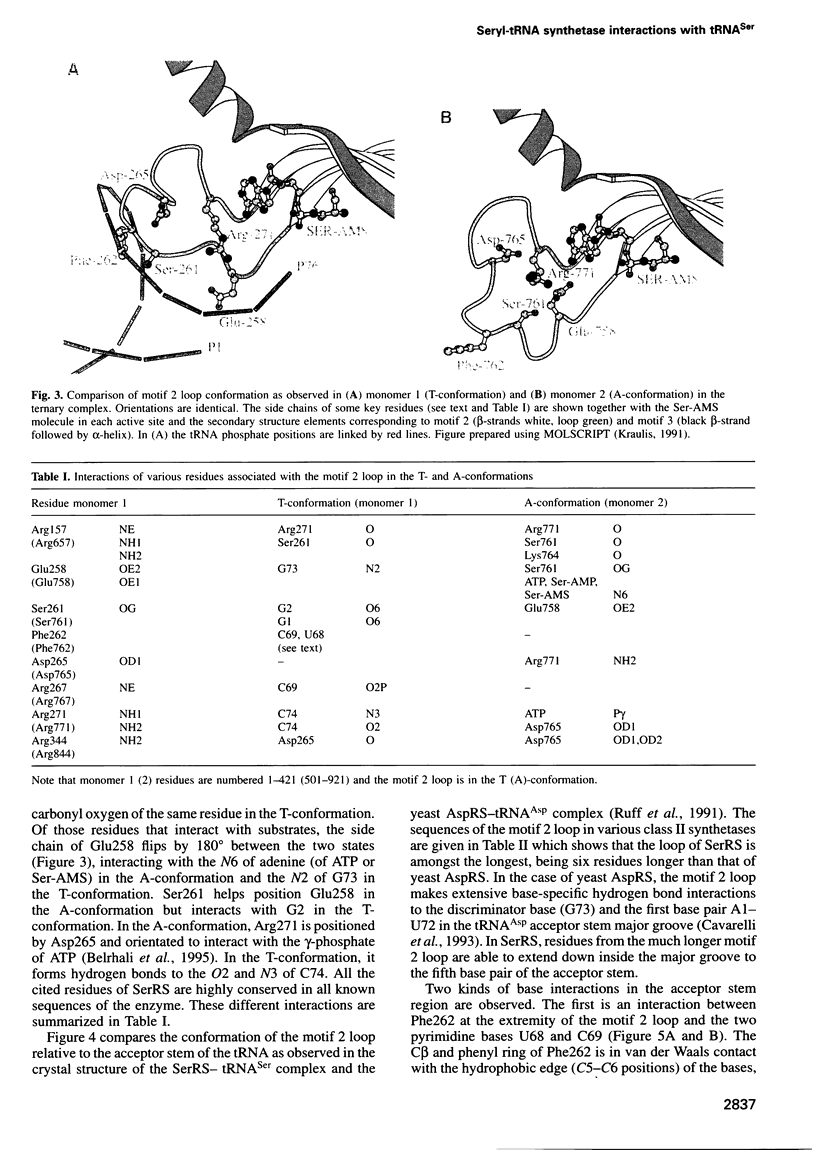
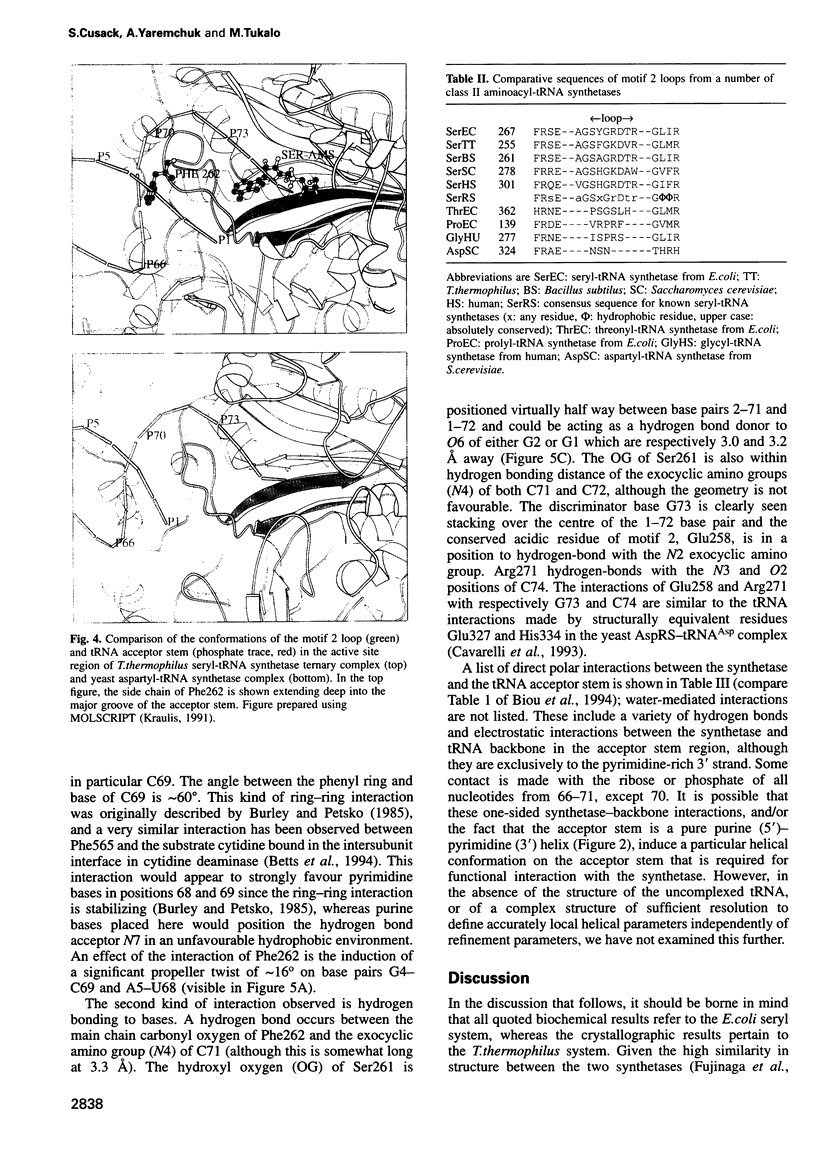
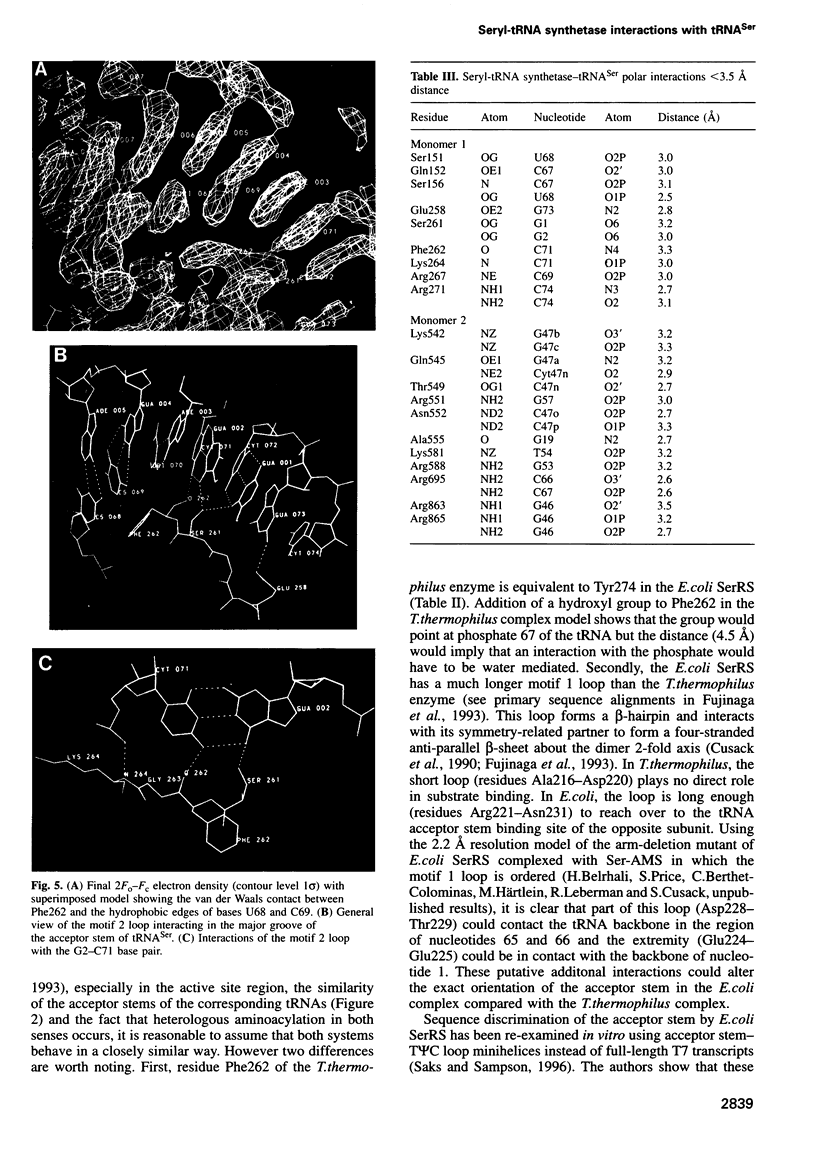
The low temperature crystal structure of the ternary complex of Thermus thermophilus seryl-tRNA synthetase with tRNA(Ser) (GGA) and a non-hydrolysable seryl-adenylate analogue has been refined at 2.7 angstrom resolution. The analogue is found in both active sites of the synthetase dimer but there is only one tRNA bound across the two subunits. The motif 2 loop of the active site into which the single tRNA enters interacts within the major groove of the acceptor stem. In particular, a novel ring-ring interaction between Phe262 on the extremity of this loop and the edges of bases U68 and C69 explains the conservation of pyrimidine bases at these positions in serine isoaccepting tRNAs. This active site takes on a significantly different ordered conformation from that observed in the other subunit, which lacks tRNA. Upon tRNA binding, a number of active site residues previously found interacting with the ATP or adenylate now switch to participate in tRNA recognition. These results shed further light on the structural dynamics of the overall aminoacylation reaction in class II synthetases by revealing a mechanism which may promote an ordered passage through the activation and transfer steps.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asahara H., Himeno H., Tamura K., Hasegawa T., Watanabe K., Shimizu M. Recognition nucleotides of Escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr). J Mol Biol. 1993 May 20;231(2):219–229. doi: 10.1006/jmbi.1993.1277. [DOI] [PubMed] [Google Scholar]
- Asahara H., Himeno H., Tamura K., Nameki N., Hasegawa T., Shimizu M. Escherichia coli seryl-tRNA synthetase recognizes tRNA(Ser) by its characteristic tertiary structure. J Mol Biol. 1994 Feb 25;236(3):738–748. doi: 10.1006/jmbi.1994.1186. [DOI] [PubMed] [Google Scholar]
- Belrhali H., Yaremchuk A., Tukalo M., Berthet-Colominas C., Rasmussen B., Bösecke P., Diat O., Cusack S. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure. 1995 Apr 15;3(4):341–352. doi: 10.1016/s0969-2126(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Belrhali H., Yaremchuk A., Tukalo M., Larsen K., Berthet-Colominas C., Leberman R., Beijer B., Sproat B., Als-Nielsen J., Grübel G. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science. 1994 Mar 11;263(5152):1432–1436. doi: 10.1126/science.8128224. [DOI] [PubMed] [Google Scholar]
- Betts L., Xiang S., Short S. A., Wolfenden R., Carter C. W., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994 Jan 14;235(2):635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994 Mar 11;263(5152):1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Borel F., Vincent C., Leberman R., Härtlein M. Seryl-tRNA synthetase from Escherichia coli: implication of its N-terminal domain in aminoacylation activity and specificity. Nucleic Acids Res. 1994 Aug 11;22(15):2963–2969. doi: 10.1093/nar/22.15.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Eriani G., Rees B., Ruff M., Boeglin M., Mitschler A., Martin F., Gangloff J., Thierry J. C., Moras D. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994 Jan 15;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Cusack S. Eleven down and nine to go. Nat Struct Biol. 1995 Oct;2(10):824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fujinaga M., Berthet-Colominas C., Yaremchuk A. D., Tukalo M. A., Cusack S. Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 A resolution. J Mol Biol. 1993 Nov 5;234(1):222–233. doi: 10.1006/jmbi.1993.1576. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Miyano M., Himeno H., Sano Y., Kimura K., Shimizu M. Identity determinants of E. coli threonine tRNA. Biochem Biophys Res Commun. 1992 Apr 15;184(1):478–484. doi: 10.1016/0006-291x(92)91219-g. [DOI] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. Conversion of aminoacylation specificity from tRNA(Tyr) to tRNA(Ser) in vitro. Nucleic Acids Res. 1990 Dec 11;18(23):6815–6819. doi: 10.1093/nar/18.23.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlein M., Cusack S. Structure, function and evolution of seryl-tRNA synthetases: implications for the evolution of aminoacyl-tRNA synthetases and the genetic code. J Mol Evol. 1995 May;40(5):519–530. doi: 10.1007/BF00166620. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Nagai K. Recruiting proteins to the RNA world. Nat Struct Biol. 1995 Jul;2(7):518–522. doi: 10.1038/nsb0795-518. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ollick T., Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G., Vojtechovsky J., Clowney L., Brünger A. T., Berman H. M. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr D Biol Crystallogr. 1996 Jan 1;52(Pt 1):57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Saks M. E., Sampson J. R., Abelson J. N. The transfer RNA identity problem: a search for rules. Science. 1994 Jan 14;263(5144):191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- Saks M. E., Sampson J. R. Variant minihelix RNAs reveal sequence-specific recognition of the helical tRNA(Ser) acceptor stem by E.coli seryl-tRNA synthetase. EMBO J. 1996 Jun 3;15(11):2843–2849. [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., Saks M. E. Contributions of discrete tRNA(Ser) domains to aminoacylation by E.coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993 Sep 25;21(19):4467–4475. doi: 10.1093/nar/21.19.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C., Borel F., Willison J. C., Leberman R., Härtlein M. Seryl-tRNA synthetase from Escherichia coli: functional evidence for cross-dimer tRNA binding during aminoacylation. Nucleic Acids Res. 1995 Apr 11;23(7):1113–1118. doi: 10.1093/nar/23.7.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Q., Gross H. J. The long extra arms of human tRNA((Ser)Sec) and tRNA(Ser) function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993 Dec 11;21(24):5589–5594. doi: 10.1093/nar/21.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk A. D., Tukalo M. A., Krikliviy I., Malchenko N., Biou V., Berthet-Colominas C., Cusack S. A new crystal form of the complex between seryl-tRNA synthetase and tRNA(Ser) from Thermus thermophilus that diffracts to 2.8 A resolution. FEBS Lett. 1992 Sep 28;310(2):157–161. doi: 10.1016/0014-5793(92)81319-h. [DOI] [PubMed] [Google Scholar]