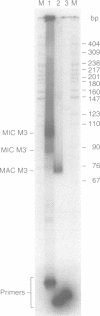
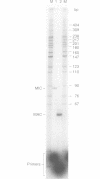
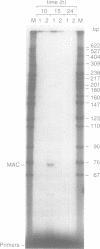
Abstract
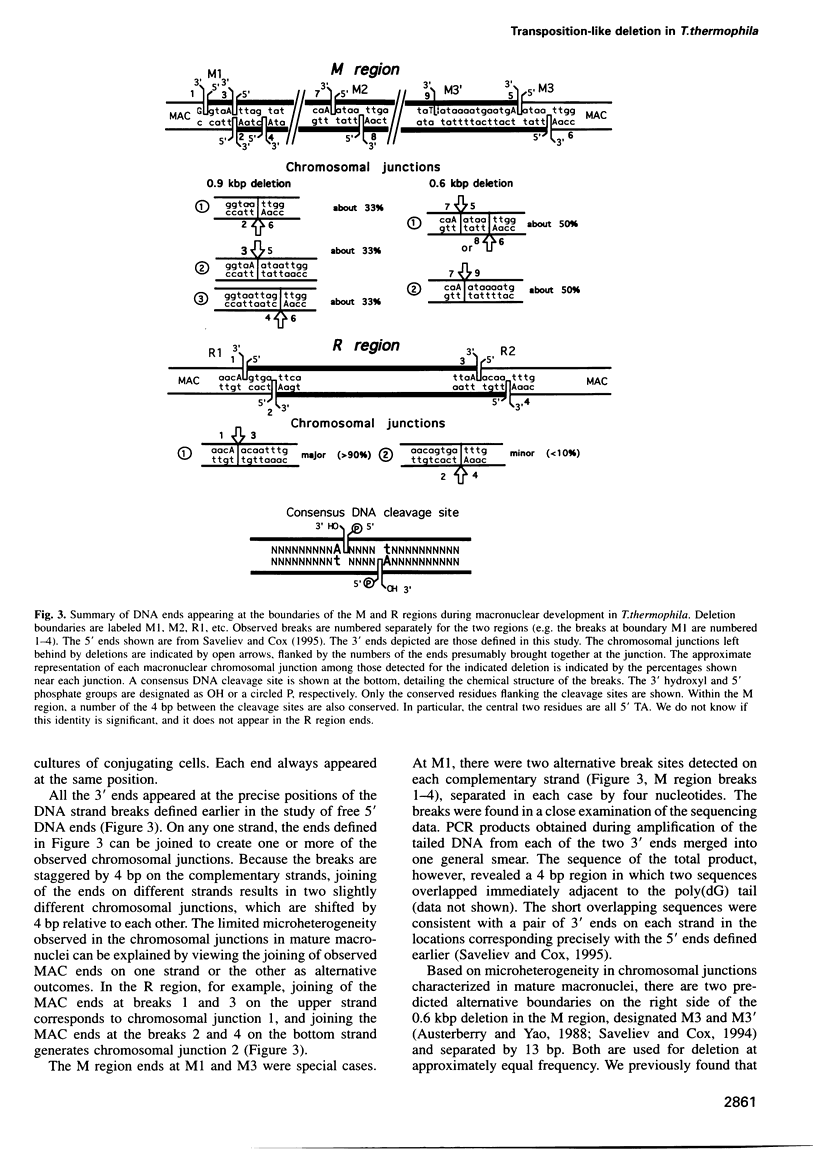
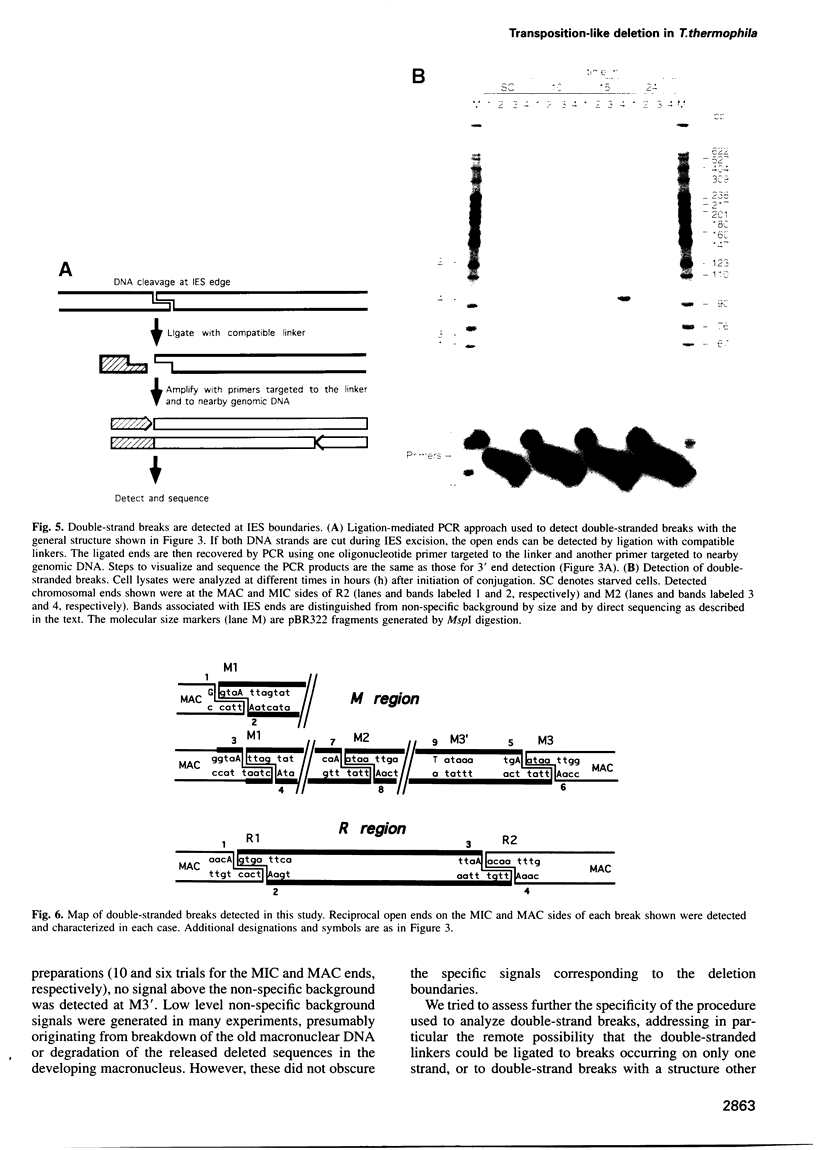
We provide a molecular description of key intermediates in the deletion of two internal eliminated sequences (IES elements), the M and R regions, during macronuclear development in Tetrahymena thermophila. Using a variety of PCR-based methods in vivo, double-strand breaks are detected that are generated by hydrolytic cleavage and correspond closely to the observed chromosomal junctions left behind in the macronuclei. The breaks exhibit a temporal and structural relationship to the deletion reaction that provides strong evidence that they are intermediates in the deletion pathway. Breaks in the individual strands are staggered by 4 bp, producing a four nucleotide 5' extension. Evidence is presented that breaks do not occur simultaneously at both ends. The results are most consistent with a deletion mechanism featuring initiation by double-strand cleavage at one end of the deleted element, followed by transesterification to generate the macronuclear junction on one DNA strand. An adenosine residue is found at all the nucleophilic 3' ends used in the postulated transesterification step. Evidence for the transesterification step is provided by detection of a 3' hydroxyl that would be liberated by such a step at a deletion boundary where no other DNA strand ends are detected.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austerberry C. F., Allis C. D., Yao M. C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C. F., Snyder R. O., Yao M. C. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res. 1989 Sep 25;17(18):7263–7272. doi: 10.1093/nar/17.18.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C. F., Yao M. C. Nucleotide sequence structure and consistency of a developmentally regulated DNA deletion in Tetrahymena thermophila. Mol Cell Biol. 1987 Jan;7(1):435–443. doi: 10.1128/mcb.7.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C. F., Yao M. C. Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol Cell Biol. 1988 Sep;8(9):3947–3950. doi: 10.1128/mcb.8.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. B. Pair formation in tetrahymena pyriformis, an inducible developmental system. J Exp Zool. 1974 Jun;188(3):337–344. doi: 10.1002/jez.1401880309. [DOI] [PubMed] [Google Scholar]
- Cherry J. M., Blackburn E. H. The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell. 1985 Dec;43(3 Pt 2):747–758. doi: 10.1016/0092-8674(85)90248-x. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Deng G., Wu R. Terminal transferase: use of the tailing of DNA and for in vitro mutagenesis. Methods Enzymol. 1983;100:96–116. doi: 10.1016/0076-6879(83)00047-6. [DOI] [PubMed] [Google Scholar]
- Doak T. G., Doerder F. P., Jahn C. L., Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common "D35E" motif. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R., James C., Yao M. C. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 1993 Dec;7(12A):2357–2365. doi: 10.1101/gad.7.12a.2357. [DOI] [PubMed] [Google Scholar]
- Godiska R., Yao M. C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990 Jun 29;61(7):1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- Heinonen T. Y., Pearlman R. E. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J Biol Chem. 1994 Jul 1;269(26):17428–17433. [PubMed] [Google Scholar]
- Jaraczewski J. W., Jahn C. L. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993 Jan;7(1):95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- Katoh M., Hirono M., Takemasa T., Kimura M., Watanabe Y. A micronucleus-specific sequence exists in the 5'-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 1993 May 25;21(10):2409–2414. doi: 10.1093/nar/21.10.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L. A., Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995 Jun 11;23(11):2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L. A., Turner L. R., LaPlante J. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 1993 Jan;7(1):84–94. doi: 10.1101/gad.7.1.84. [DOI] [PubMed] [Google Scholar]
- Loh E. Y., Elliott J. F., Cwirla S., Lanier L. L., Davis M. M. Polymerase chain reaction with single-sided specificity: analysis of T cell receptor delta chain. Science. 1989 Jan 13;243(4888):217–220. doi: 10.1126/science.2463672. [DOI] [PubMed] [Google Scholar]
- Madireddi M. T., Davis M. C., Allis C. D. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994 Oct;165(2):418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982 Jul;140(1):227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- Saveliev S. V., Cox M. M. The fate of deleted DNA produced during programmed genomic deletion events in Tetrahymena thermophila. Nucleic Acids Res. 1994 Dec 25;22(25):5695–5701. doi: 10.1093/nar/22.25.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev S. V., Cox M. M. Transient DNA breaks associated with programmed genomic deletion events in conjugating cells of Tetrahymena thermophila. Genes Dev. 1995 Jan 15;9(2):248–255. doi: 10.1101/gad.9.2.248. [DOI] [PubMed] [Google Scholar]
- Wells J. M., Ellingson J. L., Catt D. M., Berger P. J., Karrer K. M. A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol Cell Biol. 1994 Sep;14(9):5939–5949. doi: 10.1128/mcb.14.9.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Doak T. G., Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993 Dec;12(12):4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 1996 Jan;12(1):26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Detection of circular excised DNA deletion elements in Tetrahymena thermophila during development. Nucleic Acids Res. 1994 Dec 25;22(25):5702–5708. doi: 10.1093/nar/22.25.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]