Abstract
The best understood system for the transport of macromolecules between the cytoplasm and the nucleus is the classical nuclear import pathway. In this pathway, a protein containing a classical basic nuclear localization signal (NLS) is imported by a heterodimeric import receptor consisting of the β-karyopherin importin β, which mediates interactions with the nuclear pore complex, and the adaptor protein importin α, which directly binds the classical NLS. Here we review recent studies that have advanced our understanding of this pathway and also take a bioinformatics approach to analyze the likely prevalence of this system in vivo. Finally, we describe how a predicted NLS within a protein of interest can be confirmed experimentally to be functionally important.
In eukaryotic cells, the genetic material and transcriptional machinery of the nucleus are separated from the translational machinery and metabolic systems of the cytoplasm by the nuclear envelope. This segregation facilitates the precise regulation of cellular processes such as gene expression (1), signal transduction (2), and cell cycle progression (3) through selective regulation of bidirectional transport between the nucleus and the cytoplasm. However, this physical separation also necessitates the existence of molecular machinery that specifically recognizes cargo in one compartment, translocates it through the nuclear pore, and releases it in the other compartment. Nuclear transport systems of this kind were first proposed when a nuclear targeting signal in the simian virus 40 (SV40)2 large T antigen was characterized more than 20 years ago (4, 5). Since then, several pathways for nucleocytoplasmic transport have been described, of which the classical nuclear import pathway is the best characterized. The integration of detailed structural information on the components of the pathway, whole genome surveys, and extensive molecular analysis has generated powerful insight into the crucial interactions that underlie this pathway. Here we review recent studies that have defined key aspects of the cargo/import receptor interaction in the classical nuclear import cycle and present results of a bioinformatics-based assessment of the likely prevalence of this system within the model eukaryotic organism, Saccharomyces cerevisiae.
Overview of Nucleocytoplasmic Transport
Transport of macromolecules into and out of the nucleus occurs through large, proteinaceous structures called nuclear pore complexes (NPCs) (6–9). Nuclear pore complexes allow passive diffusion of ions and small proteins (<40 kDa) but restrict passage of larger molecules to those containing an appropriate targeting signal (10, 11). The pores are constructed from a class of proteins called “nucleoporins,” a subset of which contains a tandem series of phenylalanine-glycine (FG) repeats that line the central transport channel of the pore (8, 12–14).
The active transport of macromolecular cargo between the cytoplasmic and nuclear compartments is facilitated by specific soluble carrier proteins. These carriers are collectively referred to as “karyopherins” (15), with those involved in import and export termed “importins” (16) and “exportins” (17), respectively. This distinction may be artificial though, because in an ideal system, a transport factor would import one molecule, switch to export mode, and return to the cytoplasm carrying a different cargo. For example, both import and export cargos for the S. cerevisiae Msn5 receptor have been identified (18–22). Many transport receptors are members of the importin β superfamily and are called collectively “β-karyopherins.” Cargo proteins can bind directly to β-karyopherins; however, in classical nuclear import, the interaction between the β-karyopherin and the cargo is mediated by the adaptor molecule importin α. Recent modeling studies have shown that addition of an adaptor to the system results in lower transport efficiency, but this feature also may facilitate the formation of a higher nuclear cytoplasmic gradient (23).
The energy for nuclear transport is provided by the small Ras family GTPase, Ran (24). Like other GTPases (25), Ran cycles between a GTP- and a GDP-bound state. The nucleotide state of Ran is modulated by regulatory proteins, primarily the Ran guanine nucleotide exchange factor (RanGEF) in the nucleus and the Ran GTPase-activating protein (RanGAP) in the cytoplasm (26–29). Because these key regulatory factors are compartmentalized, the different forms of Ran are asymmetrically distributed in the cell, with RanGTP enriched in the nucleus and RanGDP enriched in the cytoplasm (30, 31). This compartmentalization allows Ran to impart directionality to nuclear transport by acting as a molecular switch that controls the binding and release of cargo. Therefore, import receptors bind cargo in the cytoplasm in the absence of RanGTP and release cargo in the nucleus upon RanGTP binding to the complex. In contrast, export receptors bind cargo in the nucleus in complex with RanGTP with hydrolysis to GDP in the cytoplasm triggering dissociation.
Classical Nuclear Import Cycle
The best understood pathway of nucleocytoplasmic transport is the classical nuclear import pathway (Fig. 1). Here, importin α recognizes and binds cargo in the cytoplasm, linking it to the β-karyopherin, importin β (32). Importin β then mediates interaction of the trimeric complex with the nuclear pore as it translocates into the nucleus. Once the import complex reaches the nucleus, it is dissociated by RanGTP. Binding of RanGTP to importin β causes a conformational change that results in the release of the importin α-cargo complex (33). An autoinhibitory region on the importin β-binding (IBB) domain of importin α (34, 35), the nucleoporin Nup2 (Nup50 or Npap60 in vertebrates) (36–38), and the export receptor for importin α, Cse1/RanGTP (CAS/RanGTP in vertebrates) (39) then work together to deliver the cargo into the nucleus. Finally, Cse1/RanGTP recycles importin α back to the cytoplasm in preparation for another round of import (40, 41).
FIGURE 1. The classical nuclear import cycle.
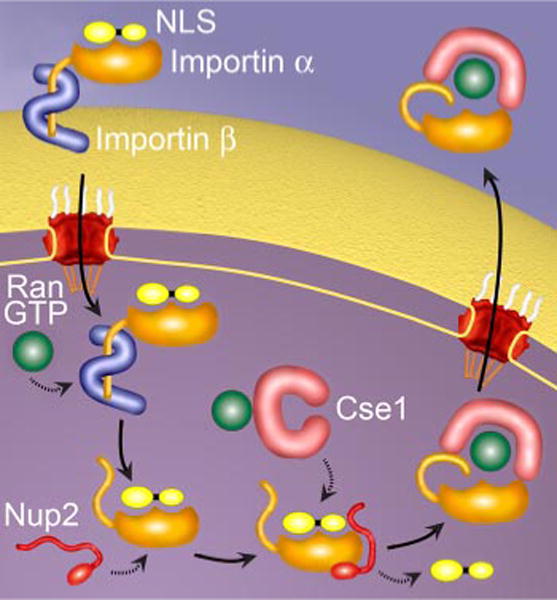
In the cytoplasm, cargo containing a cNLS is bound by the heterodimeric import receptor, importin α/importin (32). Importin α recognizes the cNLS, and importin β mediates interactions with the nuclear pore during translocation. Once inside the nucleus, RanGTP binding causes a conformational change in importin β, which releases the IBB region of importin α. This autoinhibitory domain, together with Nup2 and Cse1, facilitates cNLS dissociation and delivery of the cNLS cargo in the nucleus (35, 36). Finally, importin α is recycled back to the cytoplasm by the export receptor, Cse1, in complex with RanGTP (40, 41).
Classical Nuclear Localization Signal
The first step of nuclear import occurs when an importin discriminates between its cargo and other cellular proteins. Proteins destined for transport into the nucleus contain amino acid targeting sequences called nuclear localization signals (NLSs). The best characterized transport signal is the classical NLS (cNLS) for nuclear protein import, which consists of either one (monopartite) or two (bipartite) stretches of basic amino acids (4, 42, 43). Monopartite cNLSs are exemplified by the SV40 large T antigen NLS (126PKKKRRV132) and bipartite cNLSs are exemplified by the nucleoplasmin NLS (155KRPAATKKAGQAKKKK170). Consecutive residues from the N-terminal lysine of the monopartite NLS are referred to as P1, P2, etc. Structural (44, 45) and thermodynamic (46) studies have identified many of the key requirements for a cNLS. These studies show that a monopartite cNLS requires a lysine in the P1 position, followed by basic residues in positions P2 and P4 to yield a loose consensus sequence of K(K/R)X(K/R). Additionally, through alanine scanning of the Myc, monopartite SV40, and artificial bipartite SV40 cNLSs, it was found that the binding affinity of a cNLS for importin α measured in vitro correlates with the steady state nuclear accumulation and import rate of the corresponding cNLS cargo in vivo (46, 47). These data establish boundaries for the binding capacity of a cNLS, where a functional cNLS has a binding constant of ~10 nM for importin α. Non-functional near-cNLS sequences either bind importin α too weakly to be effectively imported or too tightly to be efficiently released from the receptor in the nucleus, although karyopherin release factors such as Nup2 and Cse1 can aid in this cargo release step (36–39, 48). Recent computer simulations also indicate that the import rate of cNLS cargo depends strongly on the rate of formation of the import complex (23). Therefore, the import and accumulation of cNLS cargo in the nucleus is affected by both the affinity of the cNLS cargo for importin α and by the concentration of the importin α receptor itself (23, 47). This relationship is also characteristic of other NLS cargo/receptor interactions (49).
Importin α/cNLS Interaction
The molecular basis for recognition of a cNLS by importin α has been defined using x-ray crystallography techniques (44, 45, 50, 51). These structural studies reveal that importin α is composed of a large curved domain consisting of ten armadillo (ARM) motifs, each of which is constructed from three α-helices (Fig. 2A), and a flexible N-terminal domain (the IBB domain) required for both binding to importin β (32) and cargo dissociation (34, 52, 53). The regular sequence of ARM repeats generates a gently curving, elongated molecule where the major and minor cNLS-binding sites are located within a shallow groove on the concave face. Both pockets are formed by solvent-exposed, conserved tryptophans together with a set of invariant asparagines four residues downstream (34, 50). The major pocket, which lies nearer the N terminus, binds monopartite cNLSs and the larger stretch of basic residues in bipartite cNLSs. The minor pocket is more C-terminal and binds the smaller stretch of basic residues in bipartite cNLSs. cNLS motifs bind to the NLS-binding pockets in an extended conformation with their main chains running anti-parallel to the direction of the importin α chain. The key cNLS lysine side chains then lie between the stacked hydrophobic indole side chains of the conserved tryptophans and form salt bridges with negatively charged residues lining the binding groove, whereas the asparagines make key main chain contacts (34, 50).
FIGURE 2.
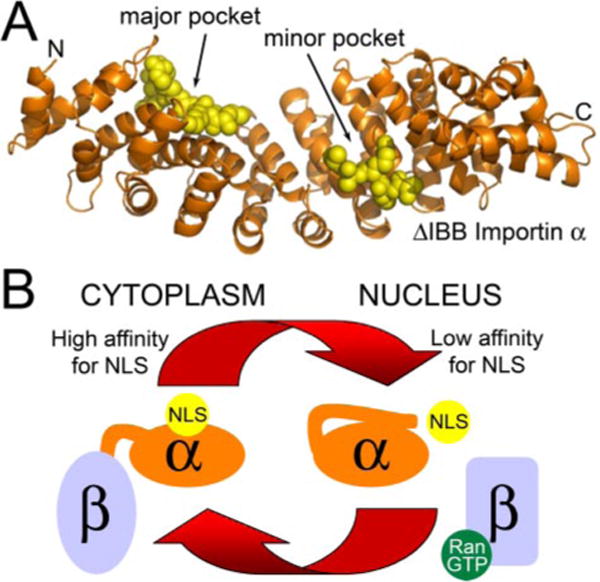
A, the importin α/cNLS interaction. cNLS peptides bound to the major and minor binding pockets of S. cerevisiae importin α lacking the IBB domain (Protein Data Bank entry 1BK6) (50). Importin β(amino acids 88–530) is shown in orange. Two SV40 peptides bound to the major and minor binding pockets are shown in yellow. B, regulation of cNLS cargo binding to importin α. In the cytoplasm, importin β is bound to the flexible autoinhibitory domain of importin α, sequestering it from the NLS-binding pocket and allowing cNLSs to bind to importin α with high affinity. Once in the nucleus, RanGTP binding to importin β causes a conformational change that releases the autoinhibitory domain, which can then compete for binding to the NLS-binding pocket. This competition contributes to a low affinity of the cNLS for importin α in the nucleus, facilitating cNLS cargo delivery.
This characteristic binding mechanism, which is found for importin α with each of its cNLS-bearing cargos (34, 50), allows an autoinhibitory region within importin α to specifically compete for binding with all cNLSs, contributing to the delivery of cNLS cargo in the nucleus (Fig. 2B). This autoinhibitory sequence consists of KRR residues within the N-terminal domain of importin α (35, 53) that mimic the basic cNLS. Upon entry of the trimeric import complex into the nucleus, the N-terminal domain of importin α is released by importin β, freeing it to fold over and interact with the major cNLS-binding pocket on the body of importin α (34, 50, 51). Because both monopartite and bipartite cNLSs bind to the major pocket, the autoinhibitory region affects the binding affinity of both types of cNLS, preventing re-binding and, therefore, facilitating the release of cNLS cargo within the nucleus.
How Common Is cNLS Import?
The classical NLS is often thought of as the prototypical NLS. It was the first NLS to be characterized, and as such, many examples of proteins using the classical import pathway have been characterized. Because of the surfeit of known cNLS-containing proteins, many have assumed that this pathway is the most prevalent in the cell; however, no studies have established empirically the proportion of cargos imported via this mechanism. It is possible that other pathways account for a large amount of nuclear traffic and that examples are lacking simply because the methods for quick and easy identification of non-classical NLSs are lacking. To address this issue, we have taken advantage of the well annotated S. cerevisiae genome and known consensus sequences for monopartite and bipartite cNLSs to estimate the prevalence of classical NLSs.
For this analysis, the search algorithm from PSORT II (54) for monopartite cNLSs and bipartite cNLSs was used to query three sets of data. First, we examined the known yeast proteome represented by the 5850 proteins in the S. cerevisiae GenBank™ (55) to identify the fraction of yeast proteins predicted to contain a cNLS and, hence, which have the potential to enter the nucleus via the classical import pathway. Second, we analyzed the 1515 proteins localized to either the nucleus or the nucleolus in a comprehensive subcellular localization study that was performed with a global yeast GFP-fusion library (56). We reasoned that these proteins were targeted to the nucleus by some nuclear import pathway and that a fraction probably used classical nuclear import. Third, we analyzed the 224 proteins that interact with importin α according to the BioGRID data base (57), because proteins that contain functional cNLSs should interact with importin α. Given that the bipartite consensus sequence consists of the monopartite consensus sequence plus a linker and upstream basic residues, all proteins that contain a putative bipartite sequence also, by definition, contain a monopartite consensus sequence. To eliminate overlap, proteins containing a bipartite cNLS were removed from the monopartite tally.
The complete results of this analysis are available in Table 1 of the supplementary data and are summarized in Fig. 3. In the GenBank™ set of 5850 proteins, 968 (16.5%) proteins contain a predicted bipartite cNLS, and 1671 (28.6%) contain a predicted monopartite cNLS. This result suggests that classical nuclear import may indeed be as important as the name implies because about 45% of the proteins in the cell have the potential to enter the nucleus via the classical nuclear import pathway. This is a sizeable proportion, especially because only 25.8% of proteins localize to the nucleus at steady state when chromosomally tagged with GFP (56). Of the 1515 proteins that have been localized to the nucleus in the global GFP screen, 391 (25.8%) contain a putative bipartite cNLS and 468 (30.9%) contain a putative monopartite cNLS. Therefore, about 57% of steady-state nuclear proteins are predicted to use classical nuclear import, whereas about 43% may use other mechanisms to enter the nucleus. In the set of 224 proteins that interact with importin α, 49 (21.9%) contain a predicted bipartite cNLS and 61 (27.2%) contain a predicted monopartite cNLS, meaning that about one-half do not contain predicted cNLSs. At first, this number seems surprisingly high because cNLSs are conventionally thought to mediate interactions with importin α; however, many of the interactions in the BioGRID data base are genetic in nature and do not require or even necessarily imply a direct physical interaction. In addition, many proteins that interact with importin α in non-cargo roles are included, such as importin β (the import partner for importin α) and Cse1 (the export factor for importin α), which interact with importin α through non-NLS-mediated interfaces (39, 58). The current definition of a cNLS also may need refining, because some of these proteins may contain cNLSs and utilize traditional importin α-mediated import but escape recognition by the standard algorithm. For example, the primary sequence of the STAT1 (signal transducers and activators of transcription 1) protein does not contain a functional classical targeting sequence; however, upon dimerization, each subunit contributes basic residues that form a cNLS recognized by importin α (59). Although a large fraction of the proteins examined in these three data bases show evidence for import by the classical system, our analysis collectively indicates that alternative nuclear localization mechanisms also are likely to be prevalent. Thus, further studies and novel experimental approaches will be necessary to fully explore these additional systems.
FIGURE 3. The prevalence of classical nuclear import.
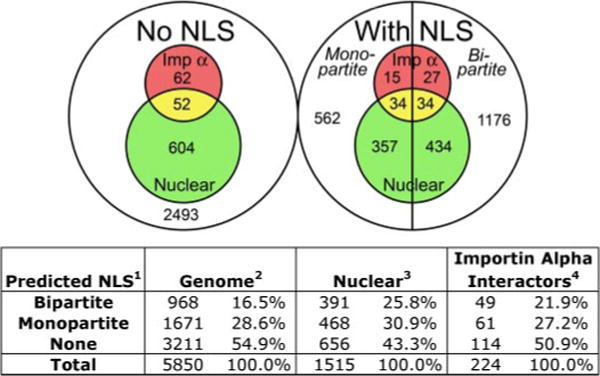
The PSORT II algorithm for monopartite and bipartite cNLSs was used to query three data sets: the 5850 proteins in the S. cerevisiae GenBank™ (Genome) (55), the 1515 proteins localized to either the nucleus or the nucleolus in the global yeast GFP-fusion library (Nuclear, green) (56), and the 224 proteins that interact with importin α according to the BioGRID data base (Importin Alpha Interactors, red) (57). The Venn diagram indicates the number of proteins from each data set with no predicted cNLS (left circle) or with a monopartite or bipartite cNLS (right circle). The corresponding percentages are listed in the table below.
Demonstrating That an NLS Is Functional in Vivo
These consensus sequence algorithms and bioinformatics approaches are valuable for predicting putative cNLS motifs because they can suggest potential pathways of import and provide a starting point for establishing how a protein of interest enters the nucleus. Importantly, they provide a quick portal into the world of nuclear transport by facilitating the inquiries of researchers interested in the import mechanism of their favorite protein. However, they can only suggest probable residues that may constitute a cNLS. Before a putative cNLS can be deemed a functional targeting sequence, it must meet four criteria (60). First, the sequence must be necessary for import, meaning that transport of the protein of interest into the nucleus is dramatically hindered when the sequence is deleted or, preferably, altered. The most common approach to test whether a sequence is necessary is to mutate one or often more of the consensus residues to alanine and determine whether the nuclear localization of the protein decreases. Second, the sequence must be sufficient to target an unrelated protein to the nucleus. Typically, the suspected sequence is fused to the terminus of GFP that most closely mimics the native location of the cNLS in the protein of interest and the location of the reporter protein is assessed visually. Third, the protein of interest must directly interact with its putative import receptor, and this interaction must be mediated by the identified sequence. The best way to address this particular question is to perform direct in vitro binding experiments with purified proteins in the presence of either RanGDP or RanGTP. Import complexes are dissociated by RanGTP; therefore, in these binding assays, the protein of interest should interact with its import receptor in the presence of RanGDP but not in the presence of RanGTP. Fourth, to determine which import pathway the protein of interest utilizes in vivo, it should be demonstrated that disabling the nuclear transport machinery in question disrupts import of the protein. For example, in S. cerevisiae, the localization of the protein of interest could be assessed at the restrictive temperature in a strain containing a temperature-sensitive allele of the essential gene encoding either importin α or importin β. Unfortunately, testing the import pathway is much more complicated in non-yeast systems because specific inhibitors of nuclear import have not yet been developed. After a putative sequence has met these requirements, then it may be deemed a functional NLS for the protein of interest. However, it is important to note that a specific protein may have more than one nuclear targeting sequence that may be used by different pathways or respond to different types of signals, highlighting the complexity and intricacy of nuclear transport in vivo.
Conclusions
In this review, we have dissected the recognition of classical NLS cargo in detail by analyzing the consensus sequence of a cNLS and the details of how it interacts with its import receptor, importin α. We have also assessed the prevalence of the classical nuclear import system in the cell by determining the fraction of proteins that contain a predicted cNLS. However, binding cargo is only the first step in an extremely complex and complicated cycle. Several recent studies have shed light on many of the other steps of classical import such as the cargo release step, where the structure of importin α with the karyopherin release factor Nup2 has been solved (37, 38), and importin α recycling, where the structure of importin α with its export receptor Cse1 has been deciphered (39). Future studies will focus on illuminating one of the enduring mysteries of transport, the translocation of cargo through the nuclear pore. Additionally, although a great deal is understood about the classical nuclear import pathway, very little is known about the details of other β-karyopherin-mediated transport pathways. Lee et al. (61) have begun to address this question and have recently proposed rules for NLS recognition by karyopherin β2 (also known as transportin). Thorough analogous structural studies complemented with bioinformatics analyses similar to the one performed here should facilitate the identification of the mechanism of interaction between other non-classical NLSs and their receptors, allowing development of a more comprehensive understanding of nucleocytoplasmic transport in vivo.
Supplementary Material
Acknowledgments
We thank past and present members of the Corbett laboratory, especially Dr. Michelle T. Harreman.
Footnotes
This minireview will be reprinted in the 2007 Minireview Compendium, which will be available in January, 2008.
This work is dedicated to the memory of our friend and colleague, Dr. Alec E. Hodel.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
The abbreviations used are: SV40, simian virus 40; NLS, nuclear localization signal; cNLS, classical nuclear localization signal; IBB, importin β-binding; GFP, green fluorescent protein.
References
- 1.Kaffman A, O’Shea EK. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 2.Johnson HM, Subramaniam PS, Olsnes S, Jans DA. Bioessays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- 3.Cyert MS. J Biol Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 4.Kalderon D, Richardson WD, Markham AF, Smith AE. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 5.Kalderon D, Roberts BL, Richardson WD, Smith AE. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 6.Fahrenkrog B, Aebi U. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 7.Stoffler D, Fahrenkrog B, Aebi U. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 8.Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 9.Fahrenkrog B, Aebi U. Results Probl Cell Differ. 2002;35:25–48. doi: 10.1007/978-3-540-44603-3_2. [DOI] [PubMed] [Google Scholar]
- 10.Paine PL, Moore LC, Horowitz SB. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 11.Bonner WM. Protein Migration and Accumulation in Nuclei. Academic Press; New York: 1978. [Google Scholar]
- 12.Stewart M, Baker RP, Bayliss R, Clayton L, Grant RP, Littlewood T, Matsuura Y. FEBS Lett. 2001;498:145–149. doi: 10.1016/s0014-5793(01)02489-9. [DOI] [PubMed] [Google Scholar]
- 13.Suntharalingam M, Wente SR. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 14.Tran EJ, Wente SR. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 15.Radu A, Blobel G, Moore MS. Proc Natl Acad Sci U S A. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorlich D, Prehn S, Laskey RA, Hartmann E. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Stade K, Ford CS, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K, Blobel G. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaffman A, Rank NM, O’Neill EM, Huang LS, O’Shea EK. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 20.Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M. Genes Dev. 1999;13:2284–2300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeVit MJ, Johnston M. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 22.Mahanty SK, Wang Y, Farley FW, Elion EA. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 23.Riddick G, Macara IG. J Cell Biol. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quimby BB, Dasso M. Curr Opin Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- 25.Bourne HR, Sanders DA, McCormick F. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 26.Corbett AH, Koepp DM, Lee MS, Schlenstedt G, Hopper AK, Silver PA. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 28.Klebe C, Prinz H, Wittinghofer A, Goody RS. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 29.Bischoff FR, Ponstingl H. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 30.Kalab P, Weis K, Heald R. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 31.Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Science. 2002;295:488–491. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 32.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Matsuura Y, Liu SM, Stewart M. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 34.Kobe B. Nat Struct Biol. 1999;6:301–304. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 35.Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. J Biol Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- 36.Gilchrist D, Mykytka B, Rexach M. J Biol Chem. 2002;277:18161–18172. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. EMBO J. 2003;22:5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuura Y, Stewart M. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuura Y, Stewart M. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 40.Hood JK, Silver PA. J Biol Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 41.Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 42.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 43.Dingwall C, Laskey RA. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 44.Conti E, Kuriyan J. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 45.Fontes MR, Teh T, Kobe B. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 46.Hodel MR, Corbett AH, Hodel AE. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 47.Hodel AE, Harreman MT, Pulliam KF, Harben ME, Holmes JS, Hodel MR, Berland KM, Corbett AH. J Biol Chem. 2006;281:23545–23556. doi: 10.1074/jbc.M601718200. [DOI] [PubMed] [Google Scholar]
- 48.Gilchrist D, Rexach M. J Biol Chem. 2003;278:51937–51949. doi: 10.1074/jbc.M307371200. [DOI] [PubMed] [Google Scholar]
- 49.Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. J Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 51.Fontes MR, Teh T, Toth G, John A, Pavo I, Jans DA, Kobe B. Biochem J. 2003;375:339–349. doi: 10.1042/BJ20030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fanara P, Hodel MR, Corbett AH, Hodel AE. J Biol Chem. 2000;275:21218–21223. doi: 10.1074/jbc.M002217200. [DOI] [PubMed] [Google Scholar]
- 53.Harreman MT, Cohen PE, Hodel MR, Truscott GJ, Corbett AH, Hodel AE. J Biol Chem. 2003;278:21361–21369. doi: 10.1074/jbc.M301114200. [DOI] [PubMed] [Google Scholar]
- 54.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 55.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 57.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cingolani G, Petosa C, Weis K, Müller CW. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 59.Fagerlund R, Melen K, Kinnunen L, Julkunen I. J Biol Chem. 2002;277:30072–30078. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 60.Damelin M, Silver PA, Corbett AH. Methods Enzymol. 2002;351:587–607. doi: 10.1016/s0076-6879(02)51870-x. [DOI] [PubMed] [Google Scholar]
- 61.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


