Abstract
A chemoenzymatic approach for synthesizing heparan sulfate oligosaccharides with a reactive diazoacetyl saccharide residue is reported. The resultant oligosaccharides were demonstrated to serve as specific inhibitors for heparan sulfate sulfotransferases, offering a new set of tools to probe the structural selectivity for heparan sulfate-binding proteins.
Heparan sulfate (HS) is a highly sulfated polysaccharide abundantly present on the cell surface and in the extracellular matrix. It participates in a wide range of physiological and pathophysiological functions, including embryonic development,1 inflammatory responses,2 blood coagulation,3 and viral/bacterial infections.4 HS consists of a disaccharide repeating unit with glucuronic acid (GlcA) or iduronic acid (IdoA) and glucosamine (GlcN), each of which is capable of carrying sulfo groups. HS achieves its functions by interacting with a variety of proteins.5 The positions of sulfo groups and the locations of the IdoA residues are critically important for their binding specificities.6 There is strong demand for methods to decipher the interactions of specific HS structures with their protein targets.
Here, we report a new approach for synthesizing active heparan sulfate based probes (AHSBP). The probe consists of an HS oligosaccharide functionalized with an N-diazoacetyl moiety (Fig 1). It utilizes the saccharide motif as a guiding system to find the protein target site. The diazoacetyl group,7 while stable under neutral pH, upon binding and acidification, could be activated to covalently couple the oligosaccharide with the protein and inhibit protein activity. The effects of AHSBP on two HS biosynthetic enzymes, including 2-O-sulfotransferase (2-OST) and 3-O-sulfotransferase (3-OST), were examined to demonstrate the utility of these compounds.
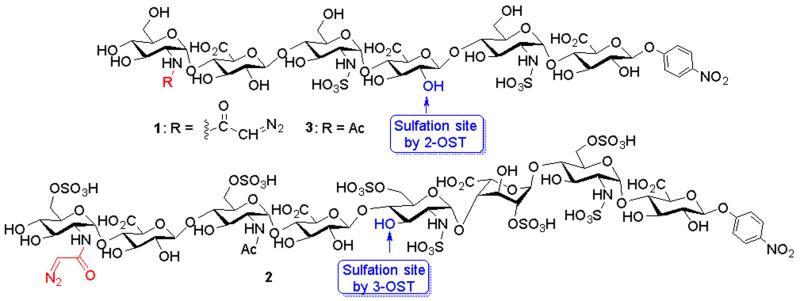
Fig 1.
Structures of the AHSBP synthesized. 1 and 2 are the substrates for 2-OST and 3-OST respectively. The hydroxyl groups that can be modified by 2-OST and 3-OST are indicated.
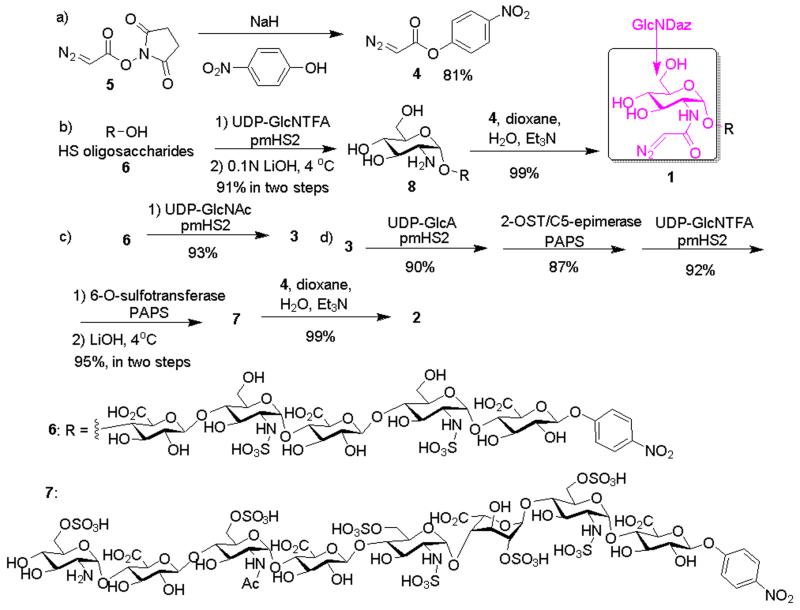
Synthesis of AHSBP started from the preparation of diazo acylating agent p-nitrophenyl diazoacetate 4, which was obtained from the N-hydroxysuccinimide diazoacetate 58 in 81% yield (Scheme 1a).8 The oligosaccharides bearing diazoacetyl functionalized glucosamine (GlcNDaz) residues were synthesized through a chemoenzymatic approach (Scheme 1b). Various reaction conditions to introduce the diazoacetyl moiety to 8 were investigated. Near quantitative yield was obtained by mixing compound 4 and the HS oligosaccharide 8 at a 10:1 ratio in a mixture of 1,4-dioxane/H2O (2:1) using trimethylamine as the base. Attempts to use compound 5 to directly react with the N-unsubstituted glucosamine 8 failed (Supplementary Table 1), probably due to the hydrolytic instability of 5 compared to the p-nitrophenyl ester 4. 2 was synthesized from 7 that was prepared from 3 through four enzymatic steps (Scheme 1c and 1d). A control compound, 3, was also synthesized (Scheme 1c). 3 has the identical number of saccharide residue and sulfo groups as 1, except that a GlcNAc residue was used to substitute the GlcNDaz at the non-reducing end (Supplementary Figs S10-S12).
Scheme 1.
Synthesis of AHSBP
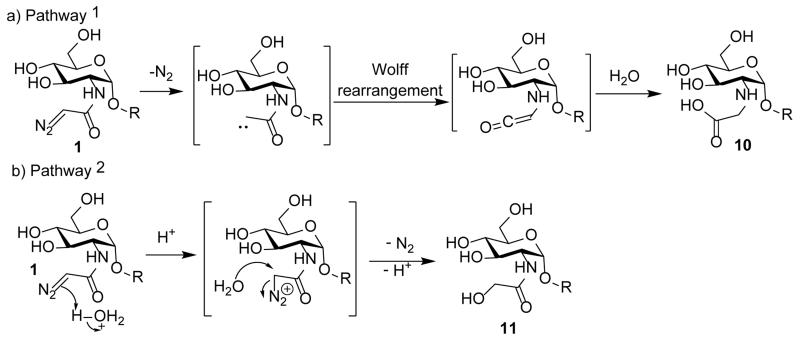
While diazo compounds can be activated by light, we explored the alternative of their activation by acid. After 1 was incubated in water at pH 5.5 for one hour, one major product was obtained from the solution. Electrospray ionization mass spectrometry (ESI-MS) analysis demonstrated the product had the molar mass of 1369.3 indicating the addition of a water molecule and the loss of nitrogen gas (Supplementary Figs S15 and S16). There are two possible mechanisms for product formation. The first is that the diazoacetamide underwent Wolff rearrangement with subsequent hydrolysis of the isocyanate leading to a carboxylic acid 10 (Scheme 2a).9, 10 The other pathway is that protonation of the diazoacetamide triggers the release of nitrogen, which is followed by nucleophilic attack by water generating the glycolamide 11 (Scheme 2b), an isomer of 10.
Scheme 2.
Two potential pathways for activation of the GlcNDaz residue.
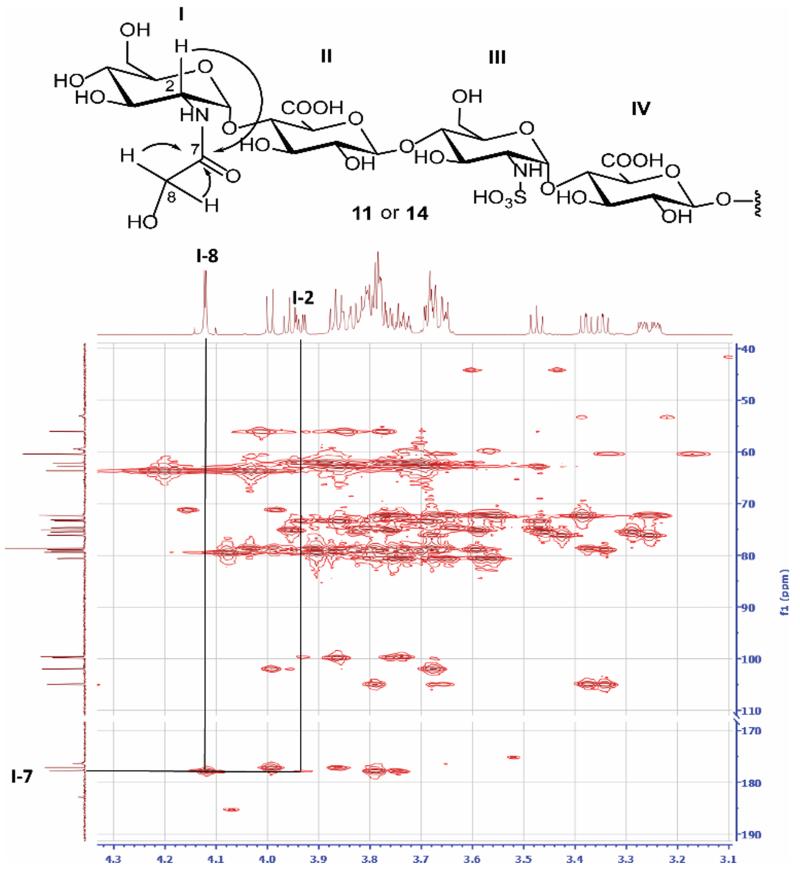
To establish the product structure, two disaccharide standards (12 and 13) were synthesized (Supplementary Figs S17 -S22). 12 has the N-carboxymethyl glucosamine residue as in 10, while 13 bears the N-glycolyl glucosamine residue, mimicking the structure of 11. From 1H-13C-HMBC (homonuclear multiple bond correlation) spectra, key correlations were observed from these disaccharides (Supplementary Fig S20). The signals from cross peak I-2 (3.97 ppm)/I-7(177.9 ppm) from 13 were similar to that observed from product of 1 (I-2(3.93 ppm)/I-7(177.8 ppm)) (Fig 2), hinting that 11 rather than 10 was the structure of main product from 1. To finally confirm the structure, hexasaccharide 14 was synthesized from 9 (Supplementary Fig S23 toS26) and glycolic acid. 1H-NMR spectrum of 14 matched well with that from acid treated 1 (Fig 2). Taken together, these results suggest that the major pathway for acid catalyzed conversion of 1 is through pathway 2 (Scheme 2b) rather than Wolff rearrangement.
Fig 2.
1H-13C HMBC key correlations of 11. The signals of featured groups are indicated. The cross peaks are connected with arrowed lines identifying correlated carbon and proton. The corresponding chemical structure is shown on top of each spectrum. The anticipated correlations are indicated in the structures by curved arrows.
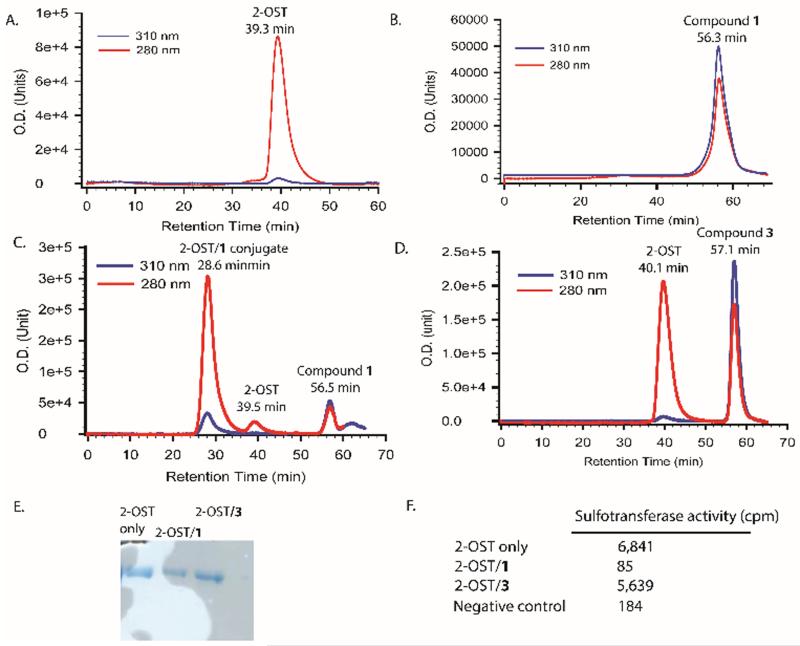
The abilities of compounds 1 and 2 to function as AHSBPs were investigated next using two HS biosynthesis enzymes, i.e., 2-OST and 3-OST.11 2-OST is an enzyme that transfers sulfo group to the 2-OH position of a GlcA residue that is flanked by two N-sulfated glucosamine residues as shown in hexasaccharide 1 (Fig 1). 1 was incubated with 2-OST at pH 5.5. The reaction was monitored with gel-permeation chromatography HPLC (GPC-HPLC) at two wavelength, 280 nm and 310 nm, characteristic for the protein and p-nitrophenyl chromophore, respectively. 2-OST (MW ~70 KDa) was eluted at 39.3 min (Fig 3A), and 1 (MW = 1379.4 Da) was eluted at 56.3 min (Fig 3B). The reaction mixture yielded a major new peak at 28.6 min, earlier than 2-OST alone (Fig 3C). Compared to 2-OST, the new peak has significantly higher absorbance in 310 nm, suggesting that the coupling of 2-OST and 1 occurred. In a control experiment, incubation of 2-OST with compound 3 bearing no GlcNDaz did not change the elution time for 2-OST (Fig 3D).
Fig 3.
Analysis of the conjugate product of 2-OST and compound 1. Panel A shows the GPC-HPLC chromatogram of 2-OST protein only in the absence of compound 1. Panel B shows the GPC-HPLC chromatogram of compound 1 only. Panel C shows the GPC-HPLC chromatogram after incubating 2-OST with compound 1. Panel D shows the chromatogram of 2-OST and compound 3 after coupling reaction. Panel E shows the image of PAGE analysis of three proteins samples from GPC-HPLC analysis from Panel A-C. Panel F shows the results of the sulfotransferase activity measurement of three protein samples. The activity was determined based on the amount 35S-labeled sulfo group transferred to a polysaccharide substrate.
Additional evidence suggesting the cross-linking between 2-OST and 1 was demonstrated by the effect on the enzymatic activity. The protein peaks resolved from GPC-HPLC were collected for activity measurement. The presence of the desired protein in collected fractions from GPC-HPLC was confirmed by SDS-PAGE (Fig 3E). Both the protein alone and the protein incubated with 3 displayed excellent sulfotransferase activities, while the sample incubated with 1 lost the enzymatic activity completely (Fig 3F). In contrast, incubation of 2-OST with 3 did not change the sulfotransferase activity. The reason for the lost activity of 2-OST/1 conjugate is presumably because the hexasaccharide was covalently attached to or near the active site of the enzyme, preventing the substrate binding.
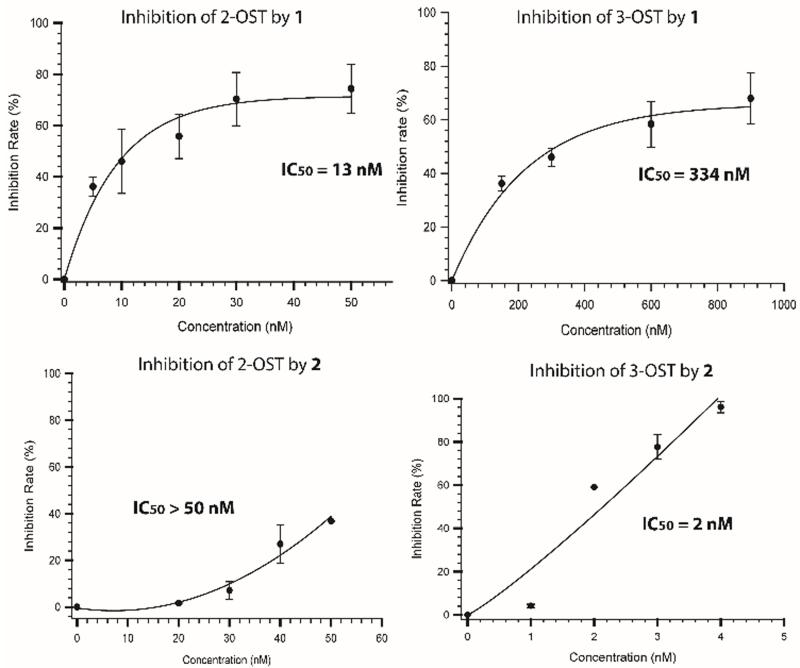
We next demonstrated that enzyme inhibition is based on the saccharide structures. 3-OST transfers a sulfo group to the 3-OH position of a glucosamine residue that is linked to a GlcA at the nonreducing end. 3-OST recognizes very different saccharide sequence from 1, preferring substrates with high level of sulfations.12 Based on the substrate specificity studies and crystal structures, only 2-OST can bind to the hexasaccharide 1,13 but not to 2.12 In contrast, 3-OST should bind to 2, but not to 1.12 In this experiment, 1 or 2 was incubated with 2-OST and 3-OST separately under an acidic condition to allow the compound to react with the enzymes. The activity measurement for each enzyme was then performed to assess the impact of the AHSBP (Fig 4). 1 displayed much higher potency toward the inhibition of 2-OST (IC50 = 13 nM) than that for 3-OST (IC50 = 334 nM). In contrast, 2 potently inhibited the activity of 3-OST (IC50 = 2 nM), but not against 2-OST (IC50 > 50 nM). Our data suggest that 1 and 2 selectively inhibit their respective target enzymes. This is the first time that small molecule inhibitors display selective inhibition effects towards different HS biosynthetic enzymes.
Fig 4.
Inhibitory effects of compounds 1 and 2 on 2-OST and 3-OST. 2-OST, or 3-OST, was incubated with different concentrations of compound 1 or 2 in an acidic buffer at 4°C for 1h to allow the cross-linking between the compound and proteins. The reactants were then subjected to the activity measurement following the methods described under “Supplementary Information”.
Conclusions
HS is an essential glycan with a wide range of biological functions. The saccharide sequences of HS determine the selectivity/specificity of their functions. Traditionally, the efforts for the identification of the contribution of specific saccharide structure to HS biological function have been hindered due to the lack of an effective method. The AHSBP approach takes advantage of the understanding of structural selectivity of HS receptors. Although using diazo group that is conjugated with peptide substrates has been used to study the biochemistry of proteases7, utilization of this chemistry in HS oligosaccharides has not been previously reported. The covalent linkage formed between the probe and the target protein enables irreversible inhibition of the activities in a sequence selective manner. This approach can be potentially applied to any HS binding proteins or enzymes. Conversely, a specific AHSBP molecule can be utilized to identify previously unknown binding partners of HS. With further development, the AHSBP method can become a powerful method to aid in the study of HS biology.
Supplementary Material
Acknowledgments
This work is supported in part by National Institutes of Health Grants HL094463 (to J.L.), GM102137 (to J.L.) and GM072667 (to X.H.). W.Z. was partially supported by a fellowship from the China Scholarship Council. T.R.O. is supported by Ruth L. Kirschstein Individual National Research Service Award from National Institutes of Health (HL120598).
Footnotes
Electronic Supplementary Information (ESI) available: Experimental section and compound characterizations can be found in Supplementary Information.
Notes and references
- 1.Patel VN, Lombaert IMA, Cowherd SN, Shworak N, Xu Y, Liu J, Hoffman MP. Dev. Cell. 2014;29:662–673. doi: 10.1016/j.devcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop J, Schuksz M, Esko JD. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Linhardt RJ. Nat. Prod. Rep. 2014;31:1676–1685. doi: 10.1039/c4np00076e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Thorp SC. Med. Res. Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 5.Xu D, Esko J. Annu. Rev. Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreuger J, Spillmann D, Li J.-p., Lindahl U. J. Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 8.Ouihia A, Rene L, Guilhem J, Pascard C, Badet B. J. Org. Chem. 1992;58:1641–1642. [Google Scholar]
- 9.Wentrup C, Bibas H, Kuhn A, Mitschke U, McMills MC. J. Org. Chem. 2013;78:10705–10717. doi: 10.1021/jo401695x. [DOI] [PubMed] [Google Scholar]
- 10.Chaimovich H, Vaughan RJ, Westheimer FH. J. Am. Chem. Soc. 1968;90:4088–4093. [Google Scholar]
- 11.Liu J, Moon AF, Sheng J, Pedersen LC. Curr. Opin. Struct. Biol. 2012;22:550–557. doi: 10.1016/j.sbi.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon AF, Xu Y, Woody S, Krahn JM, Linhardt RJ, Liu J, Pedersen LC. Proc. Natl. Acad. Sci. USA. 2012;109:5256–5270. doi: 10.1073/pnas.1117923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Sheng J, Krahn JM, Perera L, Xu Y, Hsieh P, Duo W, Liu J, Pedersen LC. J. Biol. Chem. 2014;289:13407–13418. doi: 10.1074/jbc.M113.530535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.