Highlights
-
•
Fungal infection of plants with long latent periods.
-
•
Fungal manipulation of plant defences.
-
•
Models for strictly extracellular mode of pathogenesis.
-
•
Current status of the Zymoseptoria vs wheat interaction.
Keywords: Plant pathogen, Dothideomycete, Plant defence, Virulence
Abstract
Zymoseptoria tritici (previously Mycosphaerella graminicola, teleomorph, Septoria tritici, anamorph) causes Septoria tritici blotch, one of the most economically important diseases of wheat (Triticum aestivum). The host pathogenic interaction, as currently understood, is intriguing, and may distinguish Z. tritici from many of the current models for plant pathogenic fungi. Many important questions remain which require a deeper understanding including; the nature and biological significance of the characteristic long latent periods of symptomless plant infection; how/why the fungus then effectively transitions from this to cause disease and reproduce? Elements of this transition currently resemble a putative “hijack” on plant defence but how is Z. tritici able to do this without any form of plant cell penetration? This commentary provides a summary of the recent history of research into the host-pathogen interaction, whilst highlighting some of the challenges going forwards, which will be faced by improved technologies and a growing research community.
This commentary on the Zymoseptoria tritici vs. wheat host-pathogen interaction seeks to address progress that has been made in the last decade of research, dating from approximately 2004. The main molecular genetic resources available at that time to study the susceptible interaction included two variable size fungal EST collections (Keon et al., 2005a; Kema et al., 2008), an approximate predicted one quarter genome cDNA microarray (Keon et al., 2005b), and ∼300,000 wheat ESTs (mostly not from leaves), some of which were just being used to fabricate early wheat microarrays (http://www.plexdb.org/). However, some very useful experimental tools/procedures had already been developed, in particular the Agrobacterium-mediated fungal transformation procedure (Zwiers and De Waard, 2001), which facilitated the first direct identification of a virulence gene from Z. tritici, namely the ABC transporter, MgAtr4 (Stergiopoulos et al., 2003). For wheat, reverse genetics was not so “easy”, and whilst stable transformation with RNAi constructs was being developed it was, and remains, a relatively time consuming methodology, which had not really addressed many pathogenic interactions.
This article covers only the susceptible disease interaction between Z. tritici and wheat and will not address resistant cultivar interactions, which are described elsewhere in this issue. For the “compatible” (or susceptible) interaction alone poses many intriguing questions still pertinent today. Many of these concern the extensive “latent period” of symptomless fungal colonisation, described as the period of time from which the fungus arrives on the host plant (inoculation) to the time when disease symptoms are macroscopically visible and sporulation has commenced (Leonard and Mundt, 1984). Long latent periods are now recognised as quite a conserved and peculiar feature of plant infection by most Mycosphaerella fungi, and can in some cases extend to several months. What purpose this serves, its genetic and biochemical basis, and why and how it suddenly ends with the induction of plant cell death remain some of the most scientifically interesting questions for this pathosystem, and others involving related Mycosphaerella fungi. This is even more remarkable when you also consider that Z. tritici does not appear to penetrate plant cells at any point during infection (Kema et al., 1996), instead exclusively colonising the intercellular spaces following initial entry through plant stomata right through to its exit, via the same route (Fig. 1). There is no such thing as “typical” for this system, but the symptomless latent period for Z. tritici on susceptible wheat is usually in the region of 7–14 days prior to leaf cells dying and the onset of fungal asexual sporulation. This suggests exquisite and dynamic communication mechanisms must exist throughout the interaction, and that the onset of wheat cell death is tightly regulated both temporally and spatially (Dean et al., 2012).
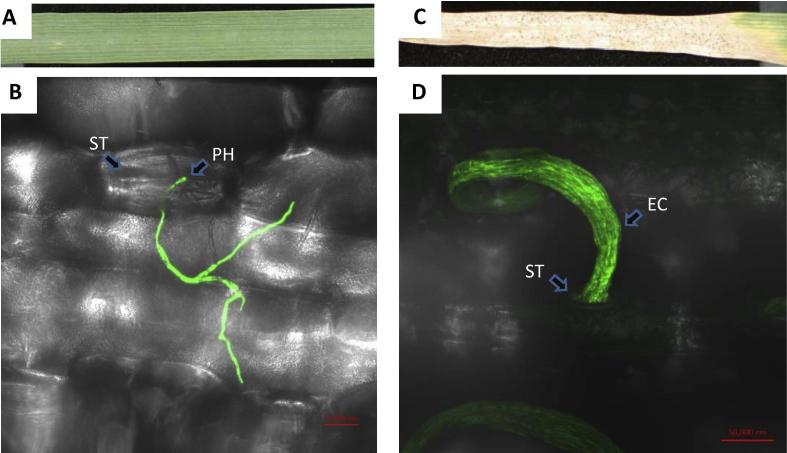
Fig. 1.
Z. tritici uses a strictly extracellular mode of plant pathogenesis with a long latent period for disease development. (A) Susceptible wheat leaf infected with a GFP expressing isolate of Z. tritici at 1 day post surface inoculation (1-dpi). (B) Stereomicroscope image of GFP tagged hyphal filaments developing on the leaf surface and penetrating the leaf through a stomatal aperture. Image taken at 1-dpi. ST = stomatal aperture; PH = penetrating hyphae. (C) Susceptible wheat leaf infected with a GFP expressing isolate of Z. tritici at 21 day post inoculation (21-dpi). (D) Stereomicroscope image of GFP tagged asexual spore masses exuding within a cirrus from below a leaf stomata. Image taken at 21-dpi. ST = stomatal aperture; EC = extracellular oozing cirrus containing new asexual pycnidiospores.
The overriding consensus a decade ago was that localised plant cell death in response to microbial pathogens functioned only as an exquisitely organised plant disease resistance response (Heath, 2000), which it probably is against biotrophs and some hemibiotrophs. In contrast cell death occurring during infection by necrotrophs and/or other hemibiotrophs was just some form of random disorganised collapse under the attack of arsenals of pathogen-derived hydrolytic enzymes and toxins. The aforementioned work from Kema and associates (1996) on the histology of infection had already shown that Z. tritici did not extensively penetrate plant cells during infection. But it still elicited plant cell death somehow from its extracellular location, and also took over a week to do it. This excellent study was performed using traditional scanning and transmission electron microscopy and first identified some form of host cell perception of fungal hyphae taking place around the onset of plant cell death. This was associated with specific sub-cellular alterations including the movement of particular organelles towards hyphae and irregular enlargement of chloroplast structures. This suggested that the plant cells had begun to “recognise” and respond to something associated with the encroaching extracellular fungal hyphae. Exactly what factors might be recognised is still unknown although a consensus is emerging in several labs that perhaps host selective protein toxins produced in a temporally regulated manner on the switch to necrotrophy might play a role. This would represent a slightly modified model to what has been shown for the wheat pathogens Stagonospora nodorum and Pyrenophora tritici-repentis in particular (Oliver and Solomon, 2010; Winterberg et al., 2014). This model is currently being tested for Z. tritici (see Ben M’Bareck et al., 2015 and Gohari et al., 2015), but it is clear for the plant side of the interaction that the transition to disease symptoms involves very specific changes in gene expression and the activation of signalling pathways which are more commonly associated with plant “defence” (Keon et al., 2007; Rudd et al., 2008, 2015; Yang et al., 2013). Wheat leaf cells ultimately appear to undergo a form of regulated programmed cell death (PCD) in the vicinity of Z. tritici hyphae (Keon et al., 2007), in response to these as yet unidentified pathogen cues. The fact that the pathogen effectively reproduces (asexual sporulation) in this environment suggests that the plants effort to “defend” itself has in some way been manipulated or “hijacked” by the pathogen to support its asexual reproduction. This is an attractive model but one which admittedly requires further testing.
Arguably the most significant recent progress has been made in the area of Z. tritici genomics. The first publically available genome resource for Z. tritici resulted from the actions of a research community led by Dutch and US scientists who lobbied for some time to get a reference genome of Z. tritici sequenced and assembled. The case was supported, eventually, by re-emphasizing the point that Z. tritici belonged to one of the largest groups of plant pathogenic fungi, the Dothideomycetes, and that numerous Mycosphaerella species within this group were responsible for causing many of the world’s most important crop diseases (Goodwin, 2004). Despite this there were few available sequenced genomes covering these organisms at that time (Goodwin, 2004). Moreover the sequencing and assembly, done in collaboration with the United States Department of Energy-Joint Genome Institute (US DOE-JGI) was such a great success that the project was extended to produce a “finished” genome of the reference isolate, IPO323 (Goodwin et al., 2011). The high quality of this reference genome (http://genome.jgi-psf.org/Mycgr3/Mycgr3.home.html) has since facilitated comparative studies with sequences from closely related species (Stukenbrock et al., 2011), along with members of the larger Dothideomycete class in general (Ohm et al., 2012) with the variable aims of understanding host adaptation, evolution and virulence mechanisms. Of the many notable observations arising from these genome sequences was that Z. tritici, and Mycosphaerella species in general, have relatively low numbers of predicted secreted plant cell wall attacking enzymes (Goodwin et al., 2011; do Amaral et al., 2012). This is intriguing and may have evolved as either a cause or a consequence of their extracellular lifestyles on their hosts.
It is arguably the search for new virulence and pathogenicity genes which will potentially gain most from the new tools for Z. tritici which are described in the range of accompanying articles. Relatively speaking, and despite its agricultural importance, the number of genes which have been shown to contribute to pathogenicity and virulence of Z. tritici is small (Table 1 lists 17 to date). Moreover this list is largely made up of global regulators of metabolism and cell signalling including components of mitogen-activated protein kinase pathways and cyclic nucleotide signalling for example. Only one secreted protein effector currently makes this list, the chitin binding protein 3LysM (Marshall et al., 2011). Many, if not all, of these genes have already been ascribed virulence roles in other plant pathogenic fungi (Perez-Nadales et al., 2014), which highlights that the unique determinants that allow Z. tritici to colonise wheat leaves remain to be discovered (if they exist at all). Overall this relatively small number of pathogenicity and virulence genes compares quite unfavourably with the numbers reported in literature for the more established models including, for example, Magnaporthe oryzae, Fusarium graminearum and Ustilago maydis. When you consider that the diseases caused by these fungi still affect global agriculture, this only serves to emphasize the task in hand for Z. tritici.
Table 1.
Current list of Z. tritici and T. aestivum genes which influence the outcome of the susceptible host-pathogen interaction.
| Z. tritici gene name | Gene function | Reference | T. aestivum gene name | Gene function | Reference |
|---|---|---|---|---|---|
| Atr4 | ABC Transporter | Stergiopoulos et al. (2003) | CERK1 | Putative chitin activated receptor kinase- competes genetically with Z. tritici3LysM | Lee et al. (2014) |
| Fus3 | Mitogen-activated protein kinase (MAPK) | Cousin et al. (2006) | CEBiP | Putative chitin binding protein- competes genetically with Z. tritici3LysM | Lee et al. (2014) |
| Slt2 | Mitogen-activated protein kinase (MAPK) | Mehrabi et al. (2006a) | PDS | Phytoene desaturase-Carotenoid biosynthesis | Lee et al. (2015a,b) |
| Hog1 | Mitogen-activated protein kinase (MAPK) | Mehrabi et al. (2006b) | ChlH | Magnesium chelatase sub-unit H-Chlorophyll biosynthesis | Lee et al. (2015a,b) |
| STE11 | MAPK kinase kinase | Kramer et al. (2009) | TaR1 | Homeodomain protein | Lee et al. (2015a,b) |
| STE50 | Scaffold protein for MAPK signalling | Kramer et al. (2009) | |||
| STE12 | Transcription factor target of MAPK signalling | Kramer et al. (2009) | |||
| STE7 | MAPK kinase | Kramer et al. (2009) | |||
| Alg2 | Protein N-glycosylation | Motteram et al. (2011) | |||
| Gpa1 | G-protein alpha sub-unit | Mehrabi et al. (2009) | |||
| Gpa3 | G-protein alpha sub-unit | Mehrabi et al. (2009) | |||
| Gpb1 | G-protein beta sub-unit | Mehrabi et al. (2009) | |||
| Tpk2 | Protein kinase A catalytic sub-unit | Mehrabi and Kema (2006c) | |||
| Bcy1 | Protein kinase A regulatory sub-unit | Mehrabi and Kema (2006c) | |||
| 3LysM | Chitin binding effector protein | Marshall et al. (2011) | |||
| MCC1 | c-type cyclin | Choi and Goodwin (2011) | |||
| Wor1 | Transcription factor | Mirzadi Gohari et al. (2014) |
What has been the reason behind such comparatively slow progress in building a catalogue of virulence / pathogenicity genes from Z. tritici and corresponding wheat genes which function during the interaction? I would argue that the relatively small size of the research community has played the greatest role, so it is now exciting to see a significant increase in the number of groups now undertaking research on this system from both fungal and plant perspectives. Whilst well established methods to generate Z. tritici mutant strains have been around and used for some time (Zwiers and De Waard, 2001; Motteram et al., 2009), including the availability of a Ku70 modified strain of IPO323 (Bowler et al., 2010), there was room for further improvement, particularly in generating gene disruption/deletion constructs and performing parallel fungal transformations in higher throughput. Similarly, the recent development of a Barley Stripe Mosaic Virus (BSMV) – viral induced gene silencing system (VIGs) system for wheat which can be combined with Z. tritici inoculations promises to rapidly speed up gene discovery in the host plant, as exemplified by five published examples in the last 2 years alone (Table 1).
However, possibly the single most important technical element which has to date been lacking from studies on the host-pathogen interaction, is the use of advanced cell biological techniques. From the many welcome technical advances described in this issue, I believe that improved cell biology may have the greatest impact. To substantiate this I will offer one example where we understand reasonably well the genetics of the interaction, but where their currently exist gaps in the cell biology. Together with collaborators we had previously demonstrated the importance for Z. tritici of evading host recognition through its cell wall chitin, by way of producing a specific effector protein, during the early phases of leaf infection (Marshall et al., 2011). Recent data suggest the genetics of this interaction involve as few as three genes in total which can dictate whether a leaf becomes diseased or resists infection. These include the secreted (or more accurately predicted secreted) chitin binding Z. tritici effector 3LysM, which is a homologue of the secreted LysM effectors ECP6 from Cladosporium fulvum (Bolton et al., 2008; de Jong et al., 2010; Sánchez-Vallet et al., 2013) and Slp1 from Magnaporthe oryzae (Mentlak et al., 2012), along with two predicted wheat plasma membrane receptors TaCEBiP and TaCERK1 (Marshall et al., 2011; Lee et al., 2014). The available genetic and biochemical data suggests that 3LysM sequesters elicitor active chitin fragments in the wheat leaf apoplast and prevents activation of the two putative wheat chitin receptors. This supports the symptomless phase of infection by preventing premature activation of selective plant defences. A similar observation was also made for the interaction of Slp1 from M. oryzae with CEBiP from rice (Mentlak et al., 2012) highlighting conservation of an important virulence mechanism.
The fact that as few as three genes can dictate the outcome of the Z. tritici on wheat interaction is quite remarkable and clearly manipulation of chitin signalling represents a possible future target for Z. tritici control. Nevertheless there are still many gaps in this story which could be addressed with advanced cell biology including, somewhat obviously, demonstrating that all three proteins localise as predicted. But perhaps a more interestingly question would be to determine the precise spatial and temporal dynamics of localisation of the three proteins. For example, exactly where during colonisation of leaf tissue do the Z. tritici hyphae need to effectively hide their chitin? Performing such an experiment represents a significant technical challenge which I hope some of the accompanying articles might address. A key consideration will be to demonstrate how deep within infected wheat leaves we can go and still visualise in real-time the dynamics of host and pathogen protein production during the various phases of the interaction? I hope this proves technically possible as it would then allow us to address some rather fundamental questions relating to the effector biology of Z. tritici and putative host resistance towards it. For example do exclusively non-cell penetrating pathogenic fungi like Z. tritici produce effectors which are internalised into plant cells or not? The published data on disease resistance interactions involving C. fulvum (a Mycosphaerella fungus with a non-cell penetrating mode of pathogenesis) on tomato may suggest not, as all published resistance genes recognising C. fulvum avirulence effectors encode plasma membrane localised (predicted) proteins (Stergiopoulos and de Witt, 2009). This special issue describes many new cell biology and other tools which will soon be available to an expanded research community and will help test this and many other hypotheses, further establishing the Z. tritici – wheat interaction as an emerging model system and leading to future applications relevant to disease control.
Acknowledgments
J.J.R. is supported by the Biotechnology and Biological Sciences Research Council of the UK (BBSRC) through the Institute Strategic Program Grant “20:20 Wheat®” (BB/J/00426X/1) awarded to Rothamsted Research and through the Rothamsted Institute Fellowship Program. J.J.R. would like to thank Kirstie Halsey in Rothamsted Bioimaging and Fabianne Brito (University of Brasilia) for generating images of GFP-tagged Z. tritici.
References
- Ben M’Barek S., Cordewener J.H.G., Tabib Ghaffary S.M., van der Lee T.A.J., Liu Z., Mirzadi Gohari A., Mehrabi R., America A.H.P., Robert O., Friesen T.L., Hamza S., Stergiopoulos I., de Wit P.J.G.M., Kema G.H.J. FPLC and liquid-chromatography mass spectrometry identify candidate necrosis-inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici. Fungal Genet. Biol. 2015;79:54–62. doi: 10.1016/j.fgb.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Bolton M.D., van Esse H.P., Vossen J.H. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol. 2008;69:119–136. doi: 10.1111/j.1365-2958.2008.06270.x. [DOI] [PubMed] [Google Scholar]
- Bowler J., Scott E., Tailor R. New capabilities for Mycosphaerella graminicola research. Mol Plant Pathol. 2010;11:691–704. doi: 10.1111/j.1364-3703.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.E., Goodwin S.B. Gene encoding a c-type cyclin in Mycosphaerella graminicola is involved in aerial mycelium formation, filamentous growth, hyphal swelling, melanin biosynthesis, stress response, and pathogenicity. Mol. Plant-Microbe Interact. 2011;24:469–477. doi: 10.1094/MPMI-04-10-0090. [DOI] [PubMed] [Google Scholar]
- Cousin A., Mehrabi R., Guilleroux M., Dufresne M. The MAP kinase-encoding gene MgFus3 of the non-appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 2006;7:269–278. doi: 10.1111/j.1364-3703.2006.00337.x. [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H.P., Kombrink A. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- Dean Ralph, Kan Van., Jan A.L., Pretorius Zacharias A. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral Alexandre Morais, Antoniw John, Rudd Jason J. Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049904. Article Number: e49904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohari, A.M.1, Ware, S.B., Wittenberg, A.H., et al., 2015. Effector discovery in the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. http://dx.doi.org/10.1111/mpp.12251 (Epub ahead of print). [DOI] [PMC free article] [PubMed]
- Goodwin S.B. Minimum phylogenetic coverage: an additional criterion to guide the selection of microbial pathogens for initial genomic sequencing efforts. Phytopathology. 2004;94:800–804. doi: 10.1094/PHYTO.2004.94.8.800. [DOI] [PubMed] [Google Scholar]
- Goodwin Stephen B., Ben M’Barek Sarrah, Dhillon Braham. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002070. Article Number: e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Kema G.H.J., Yu D.Z., Rijkenberg F.H.J. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology. 1996;86:777–786. [Google Scholar]
- Kema Gert H.J., van der Lee Theo A.J., Mendes Odette. Large-scale gene discovery in the septoria tritici blotch fungus Mycosphaerella graminicola with a focus on in planta expression. Mol. Plant-Microbe Interact. 2008;21:1249–1260. doi: 10.1094/MPMI-21-9-1249. [DOI] [PubMed] [Google Scholar]
- Keon J., Antoniw J., Rudd J. Analysis of expressed sequence tags from the wheat leaf blotch pathogen Mycosphaerella graminicola (anamorph Septoria tritici) Fungal Genet Biol. 2005;42:376–389. doi: 10.1016/j.fgb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Keon J., Rudd J.J., Antoniw J. Metabolic and stress adaptation by Mycosphaerella graminicola during sporulation in its host revealed through microarray transcription profiling. Mol. Plant Pathol. 2005;6:527–540. doi: 10.1111/j.1364-3703.2005.00304.x. [DOI] [PubMed] [Google Scholar]
- Keon John, Antoniw John, Carzaniga Raffaella. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant-Microbe Interact. 2007;20:178–193. doi: 10.1094/MPMI-20-2-0178. [DOI] [PubMed] [Google Scholar]
- Kramer B., Thines E., Foster A.J. MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet Biol. 2009;46:667–681. doi: 10.1016/j.fgb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Lee Wing-Sham, Rudd Jason J., Hammond-Kosack Kim E. Mycosphaerella graminicola LysM Effector-Mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP Homologues in Wheat. Mol. Plant-Microbe Interact. 2014;27:236–243. doi: 10.1094/MPMI-07-13-0201-R. [DOI] [PubMed] [Google Scholar]
- Lee, W.S., Devonshire, J.B., Hammond-Kosack, K.E., et al., 2015. Deregulation of plant cell death through disruption of chloroplast functionality affects asexual sporulation of Zymoseptoria tritici on wheat. Mol. Plant-Microbe Interact., Epub ahead of print. http://dx.doi.org/10.1094/MPMI (11.12.14). [DOI] [PubMed]
- Lee J., Orosa B., Millyard L. Functional analysis of a Wheat Homeodomain protein, TaR1, reveals that host chromatin remodelling influences the dynamics of the switch to necrotrophic growth in the phytopathogenic fungus Zymoseptoria tritici. New Phytol. 2015;206:598–605. doi: 10.1111/nph.13323. [DOI] [PubMed] [Google Scholar]
- Leonard K.J., Mundt C.C. Methods for estimating epidemiological effects of quantitative resistance to plant diseases. Theor. Appl. Genet. 1984;67:219–230. doi: 10.1007/BF00317041. [DOI] [PubMed] [Google Scholar]
- Marshall Rosalind, Kombrink Anja, Motteram Juliet. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011;156:756–769. doi: 10.1104/pp.111.176347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi R., Kema G.H. Protein kinase A subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Mol. Plant Pathol. 2006;7:565–577. doi: 10.1111/j.1364-3703.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Van der Lee T., Waalwijk C. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol Plant-Microbe Interact. 2006;19:389–398. doi: 10.1094/MPMI-19-0389. [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Zwiers L.H., de Waard M.A. MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant-Microbe Interact. 2006;19:1262–1269. doi: 10.1094/MPMI-19-1262. [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Ben M’Barek S., van der Lee T.A. G(alpha) and Gbeta proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola. Eukaryot. Cell. 2009;8:1001–1013. doi: 10.1128/EC.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlak T.A., Kombrink A., Shinya T. Effector-mediated suppression of chitin-triggered immunity by magnaporthe oryzae is necessary for rice blast disease. Plant Cell. 2012;24:322–335. doi: 10.1105/tpc.111.092957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadi Gohari A., Mehrabi R., Robert O. Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 2014;15:394–405. doi: 10.1111/mpp.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motteram J., Küfner I., Deller S. Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant-Microbe Interact. 2009;22:790–799. doi: 10.1094/MPMI-22-7-0790. [DOI] [PubMed] [Google Scholar]
- Motteram J., Lovegrove A., Pirie E. Aberrant protein N-glycosylation impacts upon infection-related growth transitions of the haploid plant-pathogenic fungus Mycosphaerella graminicola. Mol. Microbiol. 2011;81:415–433. doi: 10.1111/j.1365-2958.2011.07701.x. [DOI] [PubMed] [Google Scholar]
- Ohm R.A., Feau N., Henrissat B. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003037. article e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R.P., Solomon P.S. New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 2010;13:415–419. [PubMed] [Google Scholar]
- Perez-Nadales E., Nogueira M.F., Baldin C. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd Jason J., Keon John, Hammond-Kosack Kim E. The wheat mitogen-activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 2008;147:802–815. doi: 10.1104/pp.108.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd Jason J., Kanyuka K., Hassani-Pak K. Transcriptome and metabolite profiling the infection cycle of Zymoseptoria tritici on wheat (Triticum aestivum) reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions, and a variation on the hemi-biotrophic lifestyle definition. Plant Physiol. 2015;167:1158–1185. doi: 10.1104/pp.114.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A., Saleem-Batcha R., Kombrink A. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLIFE. 2013:e00790. doi: 10.7554/eLife.00790. 10.7554/eLife. 00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I., de Wit P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., Zwiers L.H., De Waard M.A. The ABC transporter MgAtr4 is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol. Plant-Microbe Interact. 2003;16:689–698. doi: 10.1094/MPMI.2003.16.8.689. [DOI] [PubMed] [Google Scholar]
- Stukenbrock E.H., Bataillon T., Dutheil J.Y. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2011;21:2157–2166. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterberg B., Du Fall L.A., Song X. The necrotrophic effector protein SnTox3 re-programs metabolism and elicits a strong defence response in susceptible wheat leaves. BMC Plant Biol. 2014;14 doi: 10.1186/s12870-014-0215-5. article 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Li W., Jørgensen H.J.L. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen septoria tritici during two phases of the compatible interaction. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0081606. article e81606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers L.H., De Waard M.A. Efficient Agrobacterium tumefaciens-mediated gene disruption in the phytopathogen Mycosphaerella graminicola. Curr. Genet. 2001;39:388–393. doi: 10.1007/s002940100216. [DOI] [PubMed] [Google Scholar]