Abstract
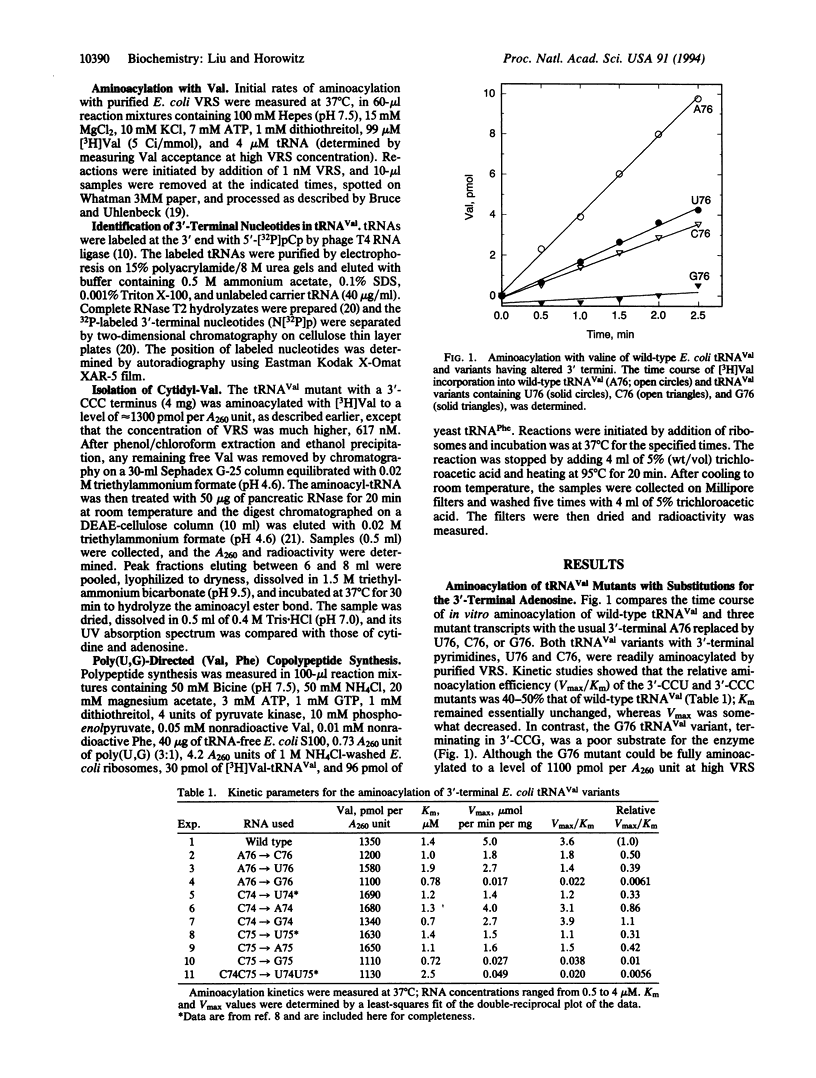
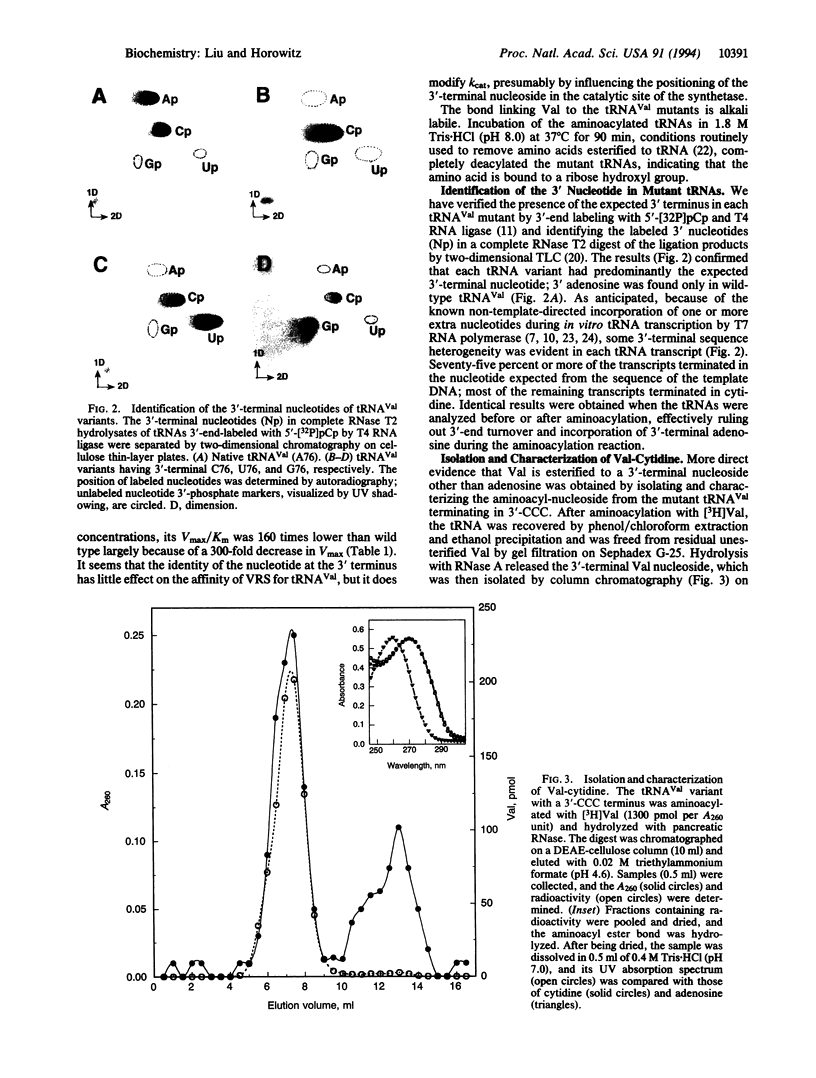
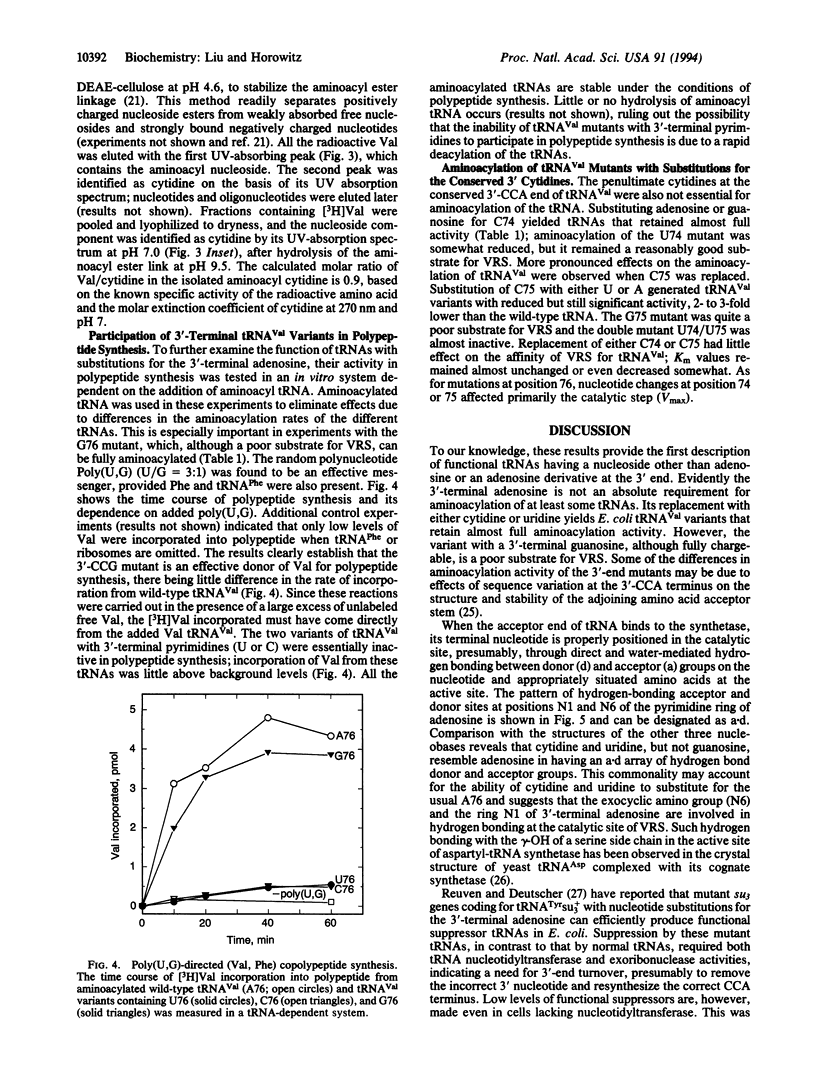
The trinucleotide CCA sequence is present at the 3' terminus of all mature tRNAs. Despite this high degree of conservation, we have been able to prepare in vitro transcripts of Escherichia coli tRNA(Val) with altered 3' termini that are readily aminoacylated and can function in polypeptide synthesis. Replacement of the 3'-terminal adenosine with either cytidine or uridine yields a tRNA(Val) variant that retains almost full aminoacylation activity, having specificity constants (Vmax/Km) 40-50% that of wild-type tRNA(Val). The tRNA(Val) variant with a 3'-terminal guanosine remains fully chargeable but is a poor substrate for valyl-tRNA synthetase, largely as the result of a decrease in the catalytic constant. End-group analysis revealed the absence of adenosine at the 3' end of the tRNA(Val) mutants and identified the nucleotide expected from the sequence of the DNA template as the predominant 3'-terminal residue; Val-cytidine was isolated from the aminoacylated C76 mutant. Val-tRNA(Val) with 3'-CCG is active in poly(U,G)-directed (Val, Phe) copolypeptide synthesis, whereas the tRNA(Val) mutants terminating in 3'-CCC and 3'-CCU, which are readily aminoacylated, are inactive. The differential effects of nucleotide substitution on aminoacylation and polypeptide synthesis suggest that the universally conserved 3'-CCA end of tRNAs is monitored at two or more steps in protein synthesis that have different nucleotide recognition specificities.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Eriani G., Rees B., Ruff M., Boeglin M., Mitschler A., Martin F., Gangloff J., Thierry J. C., Moras D. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994 Jan 15;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993 Jan;7(1):79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- Chu W. C., Horowitz J. 19F NMR of 5-fluorouracil-substituted transfer RNA transcribed in vitro: resonance assignment of fluorouracil-guanine base pairs. Nucleic Acids Res. 1989 Sep 25;17(18):7241–7252. doi: 10.1093/nar/17.18.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W. C., Horowitz J. Recognition of Escherichia coli valine transfer RNA by its cognate synthetase: a fluorine-19 NMR study. Biochemistry. 1991 Feb 12;30(6):1655–1663. doi: 10.1021/bi00220a031. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Erbe R. W., Nau M. M., Leder P. Translation and translocation of defined RNA messengers. J Mol Biol. 1969 Feb 14;39(3):441–460. doi: 10.1016/0022-2836(69)90137-5. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Gregory S. T., Lieberman K. R., Dahlberg A. E. Mutations in the peptidyl transferase region of E. coli 23S rRNA affecting translational accuracy. Nucleic Acids Res. 1994 Feb 11;22(3):279–284. doi: 10.1093/nar/22.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Lepier A., Schwägele F., Sprinzl M., Vogt H., Wintermeyer W. Specific recognition of the 3'-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol. 1988 Oct 5;203(3):699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson J. M., Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989 Dec 1;8(12):3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S., Hofmann H. P., Ott G., Sprinzl M. The 3'-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6199–6202. doi: 10.1073/pnas.90.13.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Horowitz J. In vitro transcription of transfer RNAs with 3'-end modifications. Biotechniques. 1993 Aug;15(2):264–266. [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Willis N. M., Bossi L., Gesteland R. F., Atkins J. F. Functional tRNAs with altered 3' ends. EMBO J. 1993 Jun;12(6):2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. 3. The formation of peptide bonds by ribosomes in the absence of supernatant enzymes. J Biol Chem. 1968 May 25;243(10):2810–2820. [PubMed] [Google Scholar]
- Reuven N. B., Deutscher M. P. Substitution of the 3' terminal adenosine residue of transfer RNA in vivo. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4350–4353. doi: 10.1073/pnas.90.10.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. A synthetic substrate for tRNA splicing. Anal Biochem. 1987 Oct;166(1):90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- SARIN P. S., ZAMECNIK P. C. ON THE STABILITY OF AMINOACYL-S-RNA TO NUCLEOPHILIC CATALYSIS. Biochim Biophys Acta. 1964 Dec 16;91:653–655. doi: 10.1016/0926-6550(64)90018-0. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Borén T., Johansen T. I., Lustig F. Properties of a transfer RNA lacking modified nucleosides. J Biol Chem. 1988 Sep 25;263(27):13692–13699. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Cramer F. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 1979;22:1–69. doi: 10.1016/s0079-6603(08)60798-9. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka M., Chládek S. Effect of nucleotide substitution on the peptidyltransferase activity of 2'(3')-O-(aminoacyl) oligonucleotides. Biochemistry. 1990 Jan 23;29(3):667–670. doi: 10.1021/bi00455a011. [DOI] [PubMed] [Google Scholar]
- WOLFENDEN R., RAMMLER D. H., LIPMANN F. ON THE SITE OF ESTERIFICATION OF AMINO ACIDS TO SOLUBLE RNA. Biochemistry. 1964 Mar;3:329–338. doi: 10.1021/bi00891a006. [DOI] [PubMed] [Google Scholar]