Abstract
Current knowledge of molecules involved in immunology and allergic disease results from significant contributions of X-ray crystallography, a discipline that just celebrated its 100th anniversary. The histories of allergens and X-ray crystallography are intimately intertwined. The first enzyme structure to be determined was lysozyme, also known as the chicken food allergen Gal d 4. Crystallography determines the exact three-dimensional positions of atoms in molecules. Structures of molecular complexes in the disciplines of immunology and allergy have revealed the atoms involved in molecular interactions and in mechanisms of disease. These complexes include peptides presented by MHC class II molecules, cytokines bound to their receptors, allergen-antibody complexes, and innate immune receptors with their ligands. The information derived from crystallographic studies provides insights into the function of molecules. Allergen function is one of the determinants of environmental exposure, which is essential for IgE sensitization. Proteolytic activity of allergens or their capacity to bind lipopolysaccharides may also contribute to allergenicity. The atomic positions define the molecular surface that is accessible to antibodies. This surface in turn determines antibody specificity and cross-reactivity that are important factors for the selection of allergen panels used for molecular diagnosis and for the interpretation of clinical symptoms. This review celebrates the contributions of X-ray crystallography to clinical immunology and allergy, focusing on new molecular perspectives that influence the diagnosis and treatment of allergic diseases.
Keywords: Allergens, allergy, function, structure, cross-reactivity, X-ray crystallography
1. A little bit of history
In 1912, the German physicist Max von Laue published a demonstration of X-ray diffraction from a crystal of copper sulfate pentahydrate.1 This pioneering event led to a Nobel Prize two years later and marked the beginning of X-ray crystallography. The discipline celebrated its 100th anniversary in 2014. Crystallography started in hands of physicists with studies of non-biological molecules, and extended to the fields of chemistry, biology, and medicine. Crystal structures define the spatial location of atoms and the folding of macromolecules involved in biological processes. The first X-ray diffraction data from a small protein, pepsin, was collected by Bernal and Crowfoot in 1934 (Fig. E1A).2 Myoglobin and hemoglobin were the first protein structures obtained by John Kendrew in 1958, and by Max Perutz in 1960, respectively.3,4 In 1965, the first enzyme structure to be determined by David Phillips was hen egg-white lysozyme, which happens to be the chicken food allergen Gal d 4.5 With the advent of more powerful X-ray sources (such as synchrotrons), new detectors, new experimental protocols (such as reduction of radiation damage by cryo-cooling) and the development of modern software, the number of macromolecular structures has exponentially increased (Fig. E2).6,7 The bulk of macromolecular diffraction data collection has moved from in-house facilities to high flux sources, mainly at synchrotrons. Nowadays, the Cambridge Structural Database contains more than 750,000 structures of organic molecules, mostly determined by X-ray crystallography, and the Protein Data Bank (PDB) contains over 105,000 structures of biomacromolecules, almost 90% of which were determined by X-ray diffraction.8 This review is a tribute to the contributions of X-ray crystallography to clinical immunology and allergy. Notable achievements include the determination of the three-dimensional structure of allergens, antibodies, receptors, and other molecules involved in immunological processes and allergic disease. The review also highlights findings derived from structural studies, which revealed mechanisms that contribute to allergic sensitization.
2. The basics of three-dimensional structure determination
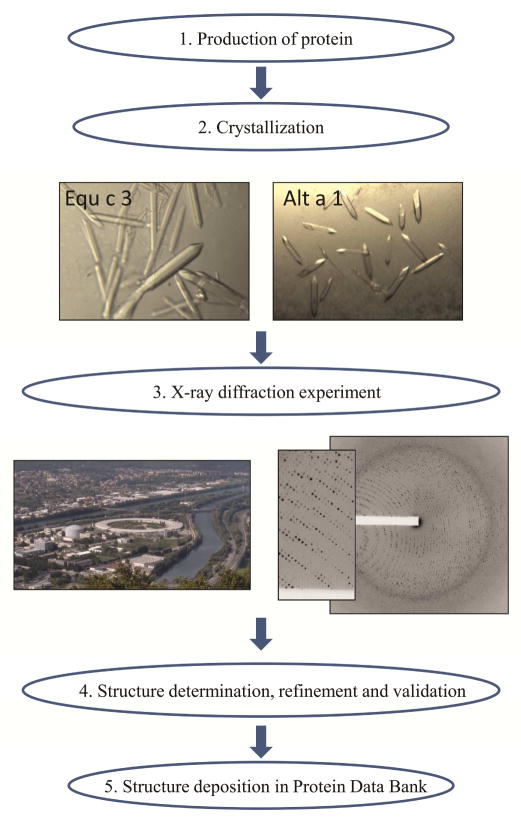
The determination of an X-ray crystal structure starts with the purification of a sufficient amount of highly concentrated and homogeneous protein (Fig. 1). Several factors that contribute to molecular variability need to be taken into consideration, including molecular variants, glycosylation, proteolysis, fragmentation, aggregation, precipitation, and/or molecular flexibility (see details in Online Repository). The expression of recombinant allergens, either alone or as fusion proteins, has often proven successful for obtaining homogeneous protein preparations.9–12 Molecules naturally embedded in membranes, such as the histamine receptor, may require strategies involving protein solubilization.13 Procuring soluble, properly folded, and pure protein is the most promising way to obtain diffraction quality crystals, and is the major bottleneck in the structure determination process. The difficulty in obtaining diffraction quality crystals promotes the development of complementary technologies such as cryo-electron microscopy.14 Screening of optimal conditions for crystal growth is performed either manually or by robotics.15 Hundreds of conditions are tested by mixing the highly concentrated protein with different precipitants. A commonly used technique is based on “hanging” or “sitting” drops in multi-well plates, where crystals form by slowly concentrating the protein and the precipitant through vapor diffusion. Once crystals are obtained, the X-rays to probe them are produced either by rotating anode generators found in most cyrstallography labs or at synchrotron stations. There are over 120 stations dedicated to macromolecular crystallography experiments worldwide.16 The diffraction pattern produced by exposing protein crystals to X-rays is analyzed to obtain an electron density map that is used for determination of a molecular model.17–19 The overall quality of structural models, as measured by various parameters used for structure validation, has significantly improved over time.20 Structure quality depends not only on the resolution of the collected diffraction data, but also on the processing of these data. Resolution is the smallest distance in angstroms (Å) between two atoms that can be shown to be separated (Fig. E3). The myoglobin structure determined in 1958 had a resolution of 6 Å. Nowadays, resolutions as high as 0.48 Å can be obtained.21 Crystallography has evolved from manual determination of molecular models to the use of hardware and software that greatly enhances the efficiency and quality of data collection, data reduction, model building and refinement. These transformative developments allow rapid data processing, model construction and refinement (optimization).22,23 Sophisticated software, such as HKL-3000, has enabled the determination of structures with “speed and finesse”.22 Indeed, a process that used to take years or months is now performed in days, or in hours in optimal cases. (Figure E2, Online Repository). The productivity of synchrotron stations, measured by structures determined, varies significantly and mostly depends on experimental protocols.24 There is no correlation between beamline productivity and any aspect of physical setup of data collection hardware, beamline flux or number of crystals used for diffraction. Frequent reports show that only 1–5 minutes of data collection time are needed to generate an entire data set sufficient for a good structure determination.25 The most productive synchrotron stations still need roughly 20 hours of data collection time to produce one structure. There is still room for improvement of experimental protocols and software in the future.
Figure 1. General outline of the process of structure determination using X-ray crystallography.
Crystals of horse Equ c 3 and Alt a 1 from Alternaria alternata grown using the vapor diffusion technique and used for X-ray diffraction experiments, are shown. The European Synchrotron Radiation Facility (ESRF) and Institute Laue-Langevin in Grenoble (France) (left; photographer: Christian Hendrich), is shown next to diffraction images obtained from a protein crystal used for structure determination (3N99) (right).
3. Three-dimensional structures of molecules involved in immunology and allergy
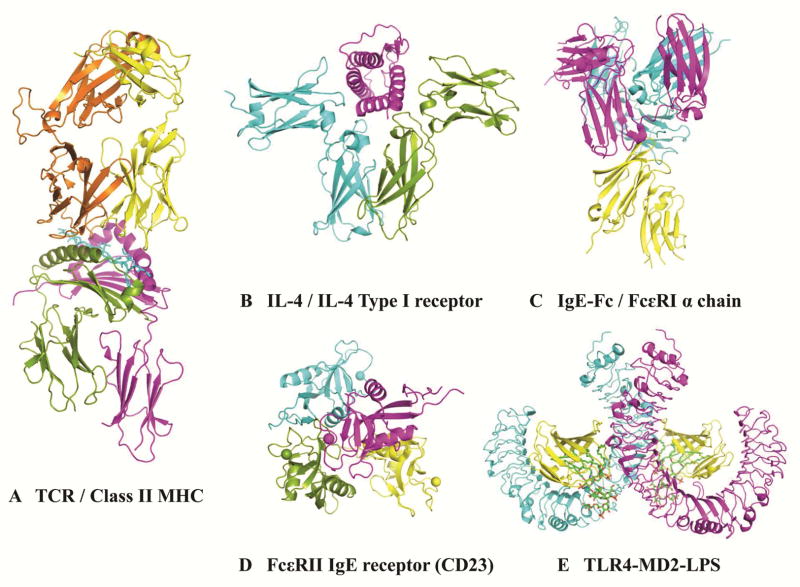
Immunological processes result from the interaction of specialized molecules, whose structure defines their function. Selected examples involved in adaptive and innate immunity are shown in Fig. 2. The structure of MHC class II molecules that are expressed in vivo on the surface of antigen presenting cells clearly revealed a groove that binds the peptides generated from antigen processing and presents them to T-cell receptors (Fig. 2A). This action, together with co-stimulatory signals, leads to activation of T cells to release cytokines that interact with cytokine receptors in B cells (Fig. 2B). Crystal structures of ovalbumin (Gal d 2) peptides bound to mouse MHC class II, and a peptide from Cry j 1, a major allergen from the Japanese red cedar (Cryptomeria japonica), bound to HLA-DP5 also aid in our understanding of molecular recognition of allergens.26,27 The structural basis of the interaction of cytokines with their receptors has been investigated to better understand what induces the production of antibodies by B cells. Cytokine receptors of the interleukin-4/13 system result from the assembly of different subunits. Interestingly, a single subunit can be shared by different cytokine receptors (i.e. IL4Rα shared by IL-4/IL-4Rα/γc and IL-13/IL-4Rα/IL-13Rα1) (Fig. 2B). These molecular interactions determine mechanisms of cytokine action, and influence disease phenotype and response to treatment.28,29
Figure 2. X-ray crystal structures of human molecules involved in immunology and allergic disease.
A) Complex of a human α/β-T cell receptor (orange and yellow), influenza HA antigen peptide (blue), and MHC Class II molecule, HLA-DR1 (green and fuchsia) (1FYT); B) cytokine receptor IL4 Type I (blue and green) bound to cytokine IL-4 (fuchsia) (3BPL); C) IgE-Fc (Cε2, Cε3 and Cε4 domains) (blue and fuchsia) bound to the extracellular domains of the high affinity IgE receptor (FcεRI) α chain (yellow) (2Y7Q); D) low affinity IgE receptors (CD23, FcεRII) (4G9A); and E) Toll-like receptor 4 (blue and fuchsia)-MD2 (yellow)-LPS (green) complex (3FXI).
IgE antibodies eventually bind to high affinity IgE receptors (FcεRI) on mast cells and basophils (Fig. 2C). X-ray crystal structures showed the flexibility of IgE-Fc (consisting of the Cε2, Cε3, and Cε4 domains). IgE-Fc adopts different conformations ranging from an acutely bent structure of the IgE-Fc when bound to the extracellular domains of the FcεRI α chain (Fig. 2C) to a diversity of conformations in solution, including a fully extended symmetrical one.30,31 The unique slow dissociation rate of IgE from FcεRI was attributed in part to these conformational changes. This observation provides a strategy for the design of asthma therapeutics using peptides that would disrupt the interaction between IgE and its high affinity IgE receptor.31
The low affinity IgE receptor (CD23, FcεRII) on B cells is a calcium-dependent molecule, with a C-type lectin domain, that contributes to the regulation of IgE levels. Calcium induces structural changes in CD23 that lead to additional interactions with IgE and a 30-fold increase in affinity for IgE (Fig. 2D),.32 The crystal structure of IgE bound to CD23 revealed a mechanism of reciprocal allosteric inhibition with the high affinity IgE receptor.33
Structures of molecules involved in innate immunity have also been determined, including the extracellular domain of Toll-like receptors (TLR). The structures of TLR differ from the “immunoglobulin-like” extracellular domains of B- and T-cell receptors mentioned above. For all these receptors, only the structures of the extracellular domains have been determined. Ten TLR are known in humans and they are formed by an N-terminal recognition domain, a single transmembrane helix, and a C-terminal signaling domain. The N-terminal domain recognizes pathogen-derived compounds or endogenous molecules released by the host in response to infection.34 This domain adopts a typical horseshoe-shape, made of tandem copies of a motif known as the leucine-rich repeat (LRR). Its concave surface has the ligand binding capacity in most LRRs. Two extracellular domains form a dimer upon ligand binding and activate signaling. The structure of the Toll-like receptor 4, for example, has revealed the importance of its interaction with MD-2 and endotoxin for signaling activation (Fig. 2E).
4. Allergen structure as a determinant of allergic disease
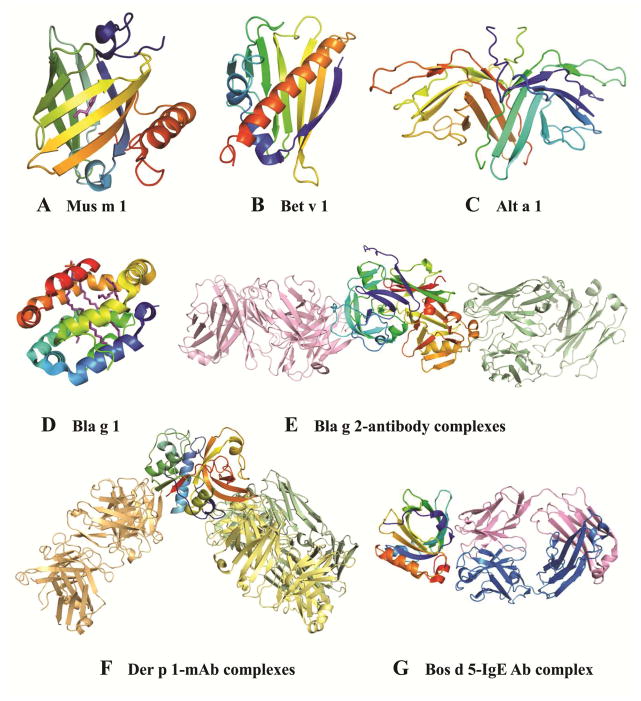
Immunological processes that lead to the development of allergic disease are strongly associated with structural features of the allergen (Fig. 3A–G). Allergen exposure, critical to IgE sensitization, is determined in part by the function of the allergen and the degree of molecular stability required for the protein to become an allergen, both of which are defined by the allergen structure. To access the immune system, allergens must be either released to the environment (e.g. pollens, spores, dander or fecal particles), or made accessible by other paths (ingestion of foods, injection of venoms, or contact through skin or infection). The function of the allergen can facilitate exposure in different ways. Some allergens are released to the environment because of their reproductive function in pollen or spores. Others have a digestive function and are excreted in fecal particles. Additional factors determine exposure, such as the aerodynamic properties of particles that carry inhalant allergens. One of the first functions gleaned from the X-ray crystal structure of allergens was obtained in 1992, for the major urinary proteins from mouse and rat. Both Mus m 1 and Rat n 1, which were not known as allergens at that time, are pheromone transporters, and belong to the lipocalin family of proteins. Lipocalins consist of a β-barrel with a hydrophobic ligand binding cavity (Fig. 3A) and are secreted in tears, urine, sweat, or saliva, which facilitates exposure.35 Lipocalins are common mammalian inhalant allergens, also found in cockroach (Bla g 4) and in cow’s milk (food allergen Bos d 5).
Figure 3. X-ray crystal structures of selected allergens.
A) Mus m 1 (1MUP); B) Bet v 1 (1BV1); C) Alt a 1 (3V0R); D) Bla g 1 (4JRB); E) Bla g 2 in complex with Fab of mAb 7C11 (2NR6; pink) superimposed with the mAb-Fab from the structure of Bla g 2 in complex with Fab of mAb 4C3 (3LIZ; green); F) Der p 1 in complex with 4C1 (pale yellow, 3RVW), superimposed with the mAb-Fab from the two structures of the allergen in complex with 10B9 (pale green; 4PP2) or 5H8 (pale orange, 4PP1); and G) Bos d 5 in complex with the Fab of a recombinant IgE antibody. Allergens are shown in rainbow colors from N (blue) to C-terminus (red), and ligands in magenta (A, D).
The first three-dimensional structure of a clinically important allergen was Bet v 1, a pathogenesis associated protein, from birch pollen (Fig. 3B).36 Bet v 1 is the most extensively studied allergen from pollen. The IgE prevalence in temperate climate areas of the northern hemisphere (i.e. Northern Europe), is high (>95%). Bet v 1 shows clinical cross-reactivity with homologous allergens from fruits and vegetables (i.e. apples, celery, carrot).37 A large number of variants (eighteen) have been identified in natural Bet v 1, sharing high amino acid sequence identity (~95%). Their nomenclature has recently been revised by the Allergen Nomenclature Sub-Committee from the World Health Organization and International Union of Immunological Societies (WHO/IUIS) (www.allergen.org).38,39 Bet v 1.0101 is the major component of natural Bet v 1 (>50%) and the main sensitizer, whereas other isoforms (Bet v 1.0401 and Bet v 1.1001) induce only minimal IgE antibody responses.40 Bet v 1.0101 and Bet v 1.0401 share the same fold, but differences in dimerization or aggregation could contribute to a decreased ability of the Bet v 1.0401 variant to elicit an allergic immune response. Interestingly, the fold of Bet v 1.0101 per se was demonstrated to be important for Th2 polarization and the induction of a strong IgE response, by comparison with an engineered folding variant.41
Nowadays, the Protein Data Bank (PDB) (www.rcsb.org) contains the three-dimensional structures of over 100 allergens, representing approximately 50 protein families, from approximately 800 allergens currently present in the WHO/IUIS official database of systematic Allergen Nomenclature (Tables E1 and E2, Online Repository). A database of allergen families (http://www.meduniwien.ac.at/allergens/allfam/) reports that allergens belong to a wide array of 186 protein families.42,43 The most frequent biochemical functions of allergens are hydrolysis of proteins, carbohydrate metabolism, binding of metal ions and lipids, storage and functions associated with the cytoskeleton42. Measurement of biological activity using a specific functional assay is the best way to confirm function, but this option is not always available. Sequence homology to a protein of known function may also be insufficient.44 Determination of the tertiary structure can delineate allergen function by defining the overall shape of the molecule and/or revealing specific functional residues.45 The major dust mite allergens, Der p 1 and Der f 1, are cysteine proteases. Their catalytic site has been identified in the three-dimensional structures of the native allergens (Fig 3F).46,47 In contrast, cockroach allergen Bla g 2 folds as a typical pepsin-like aspartic protease, but is devoid of aspartic protease activity due to specific substitutions in the catalytic site that were revealed by the crystal structure (Fig. 3E, E1A).48,49 In other cases, molecular flexibility confers a regulatory function, as seen in calcium-binding allergens commonly found in pollen and in troponin, which regulates muscle contraction (i.e. Bla g 6).50 The first three-dimensional structure of a representative of the 2 EF-hand allergen family was reported for Phl p 7 bound to calcium.51 Differences observed in IgE antibody binding depending on the allergen conformation suggest that conditions and conformation with optimal IgE reactivity should be selected for molecular diagnosis.
The storage peanut proteins Ara h 1 and Ara h 2 are food allergens with very different structural complexity. Ara h 1 is a trimer of 60 kDa bicupin-fold monomers, whereas Ara h 2 is a small α-helical protein, which is monomeric (17 kDa) (Fig. E4).11,52 It has been suggested that the quaternary structure of Ara h 1 may play a role in its allergenicity, by increasing molecular stability and preventing digestion of IgE antibody binding epitopes.53 However, there is no evidence that the differing tertiary structure of both allergens is responsible for differing allergenic potential. In fact, IgE antibody titers to Ara h 2, with a simpler structure, have been reported as the best predictor of peanut allergy.54–56 In general, three-dimensional structural complexity of allergens does not seem to be related to their allergenicity.
Finally, determination of the X-ray crystal structure of allergens has revealed the existence of new structural groups of proteins. Alt a 1, the major allergen from Alternaria alternata has a unique β-barrel structure that forms a “butterfly-like” dimer and is exclusively found in the Dothideomycetes and Sordariomycetes classes of fungi (Fig. 3C).57 Cockroach allergen Bla g 1 has an α-helical structure so far only found in insects (Fig. 3D).12,58 While the three-dimensional structures of these and other allergens have been determined, their functions still are not well understood.10,57,59 Cat allergen Fel d 1 has an uteroglobulin-like fold and consists of a dimer of heterodimers made exclusively of α-helices. Structures of recombinant Fel d 1 made by fusion of the monomers involved in each heterodimer have been determined.60,61 However, the native assembly of this major cat allergen remains unknown.
5. Influence of allergen function on allergenicity
An interesting example of how an X-ray crystal structure revealed a function associated with allergenicity is illustrated by Der p 2. This dust mite allergen resembles MD-2, the lipopolysaccharide (LPS)-binding component of the Toll-like receptor (TLR) 4 signaling complex, which is involved in activation of the innate immune system. Both proteins have an immunoglobulin-like fold, formed only of β-sheets (Fig. E1B, E4A).62,63 The surprisingly high structural similarity prompted an investigation as to whether Der p 2 would have a similar function. Der p 2 was not only able to mimic the function of MD-2 in mouse models of experimental asthma, but was also able to reconstitute LPS-driven TLR4 signaling in the absence of MD-2, suggesting a possible role of Der p 2 in activation of innate immunity.64 Results obtained with Der p 2 are supported by data demonstrating that low-level stimulation of toll-like receptors drives Th2 immune responses.64–68 This discovery marked an important step in our understanding of possible innate immune pathways that lead to allergic sensitization. Until then, activation of the adaptive immune system was the main recognized path for developing allergy. Proteolytic function had been considered to contribute to allergenicity, by cleaving molecules involved in the immune response.69,70 Der p 1 can contribute to allergenicity via non-canonical pathways. Der p 1 was reported to directly promote IgE synthesis through cleavage of the low affinity IgE receptor (CD23) on B cells, and, indirectly, through cleavage of the α-subunit of the interleukin-2 (IL-2) receptor (CD25) in T cells.71,72 Der p 1 can also contribute to inflammation by inducing cytokine production and disrupting gap junctions in the lung epithelium, which would increase membrane permeability and facilitate transepithelial allergen delivery and processing.69,70,73,74
Structural studies have shown that an increasing number of allergens, belonging to different structural families, bind hydrophobic ligands, and these are potent stimulators of the innate immune system.65 The crystal structure of Bla g 1 provided the first clues that this protein might be a lipid-binding protein (Fig. 3D). Lipids in cockroach frass were identified as fatty acids that are known to activate TLR2 and TLR4.75 Some allergens contain lipids in internal cavities that are formed by either β-sheets (Der p 2, lipocalins) or α-helices (Fel d 1, Bla g 1) (Figures E1B, 3A, D).12,61,62,76 Other allergens, lacking internal cavities, may also bind lipidic ligands in different ways. Der p 5 consists of three-helical bundles that tend to form multimers (Fig. E4B). A large hydrophobic cavity formed by each dimer has the potential to bind lipidic ligands.10 Der p 7 has a similar structure to a LPS-binding protein involved in TLR4 activation, and to the surfactant allergen Equ c 4.9,77 The fold consists of two 4-stranded antiparallel β-sheets that wrap around a long C-terminal helix (Fig. E4C).9 Although unable to bind LPS, Der p 7 bound the lipopeptide polymyxin B.9 The specific contribution of allergen-associated lipids to allergenicity has been recently reviewed.78 The interactions of these lipids with innate immunity are open to further investigation.
6. How crystallography contributes to clinical practice
The allergen structures determined by X-ray crystallography provide the basis for improved diagnosis and therapy. An undesired side-effect of immunotherapy is the induction of adverse reactions that may occur when administering increasing doses of the allergen during vaccination. To avoid these effects, hypoallergens with reduced IgE reactivity that preserve their capacity to induce T cell responses have been designed as candidates for vaccination. One strategy is the disruption of the overall allergen fold. This has proven efficacious with hypoallergenic chemically modified extracts (allergoids) that are successful for immunotherapy in Europe.79 Numerous variants of recombinant hypoallergens have also been designed, but only Bet v 1 variants have reached clinical trials where they have already shown promising results.80 Recently, a dose-ranging immunotherapy study of a new recombinant hypoallergenic folding variant of Bet v 1 showed efficacy in an environmental exposure chamber.81 Although knowledge of the three-dimensional structure of the allergen is not always necessary for the production of unfolded variants, the disruption of the overall allergen fold can be effectively designed based on the modification of specific structural features. For example, mite group 2 hypoallergens were produced by mutating cysteines involved in the formation of disulfide bonds that preserve the immunoglobulin-like structure.82,83 Another strategy to produce hypoallergens consists of modifying residues involved in IgE antibody binding, without affecting the fold of the allergen. In this case, knowledge of the three-dimensional structure of the allergen is required. The best way to precisely locate the epitope recognized by an antibody is the determination of the structure of the allergen in complex with the antibody. For example, the structure of Bet v 1 in complex with the Fab fragment of a monoclonal antibody (mAb) that interfered with IgE antibody binding identified a dominant epitope, also involved in IgE cross-reactivity with homologous allergens.84,85 The X-ray crystal structures of additional complexes revealed determinants of specificity and cross-reactivity.47,86–91 Bla g 2 and Der p 1 have been extensively analyzed through the structures of two and four allergen-antibody complexes, respectively (Fig. 3E, F).47,89–91 The structural basis of cross-reactivity was analyzed for mAb 4C1, which binds Der p 1 and Der f 1 in equivalent epitopes (Fig. 3F).47 It is not possible to obtain the mg amounts of pure native IgE antibodies required for X-ray crystallography, given their polyclonality and their low concentration in sera compared with IgG antibodies (0.05%). Therefore, recombinant IgE mAb, derived from combinatorial libraries from allergic patients, were used in complexes with bovine milk Bos d 5 (β-lactoglobulin) (Fig. 3G) and timothy grass pollen Phl p 2.87,88 This strategy, combined with site-directed mutagenesis, is a powerful tool to identify the main residues involved in IgE antibody recognition and to produce hypoallergens.47,85,92
Molecular allergy diagnosis has shown to improve patient diagnosis compared to exclusively using clinical history and skin prick test with allergen extracts.37,93–97 Structural analyses reveal relationships between homologous allergens and contribute to the design of panels of purified allergens for molecular diagnosis. Although a general rule suggests that cross-reactivity is likely among proteins that share a high degree of amino acid identity throughout the entire protein (>70%), and tends to be rare below 50% identity, exceptions do occur.98–102 Predictions of cross-reactivity based on the overall homology among allergens can only be used as guidelines, but lack precision. The three-dimensional structure of allergens allows identification of solvent exposed residues responsible for antibody recognition.103 Ideally, the selection of representative molecules from highly cross-reactive groups of allergens, and the inclusion of allergens with species-specific epitopes can simplify molecular diagnosis.104,105
Looking into the future
In the future, an increase in structure-functional studies is expected by combining methodologies from disciplines of medicine and structural biology. Cryo Electron Microscopy will enable study of larger complexes of antigens and antibodies without the need to crystallize them. Utilization of Free Electron Laser as an X-ray source will allow investigations using nanocrystals and lower the chances of structural changes due to radiation damage.106,107 NMR, currently only rarely used for structural studies of antigen-antibody complexes, is expected to have a higher impact on ligand screening and dynamic studies. As more structures become available, the ability to create informative homology models for hypothesis driven research will be enhanced. Tighter interactions between functional and structural studies are expected to impact more significantly the drug discovery process. The creation of a ‘big picture’ and better reproducibility will be possible by sophisticated database systems that will organize and analyze structural data in biomedical laboratories.108
X-ray crystallography has led to major contributions in clinical immunology and allergy. The three-dimensional structures of molecules involved in immunological processes and of allergens, alone or in complex with antibodies, have provided detailed information at the atomic level that reveals mechanisms of molecular interaction. Recombinant allergens are engineered to either preserve their native fold and amino acid composition, or to have specific residues and/or the overall fold modified for reduced IgE reactivity, which may diminish side-effects due to increasing allergen doses administered during therapy. Recombinant allergens have already shown promising results in immunotherapy clinical trials. The structural information is being used to increase the accuracy of diagnosis and to design new forms of immunotherapy that will complement and improve current approaches to treat allergic disease.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI077653 (A.P. contact PI); by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Research Project Number Z01-ES050147 (G.A.M) and ZIA ES102645 (L.C.P).
The authors thank Jill Glesner for her assistance in preparation of tables, and Dr. Susanna Keller for providing inspiring literature.
Abbreviations
- APC
Antigen presenting cells
- GST
Glutathione S-transferase
- GFP
Green fluorescent protein
- LRR
Leucine-rich repeat
- LPS
Lipopolysaccharide
- MHC
Major histocompatibility complex
- MBP
Maltose binding protein
- TLR
Toll-like receptor
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedrich W, Knipping P, Laue M. Interferenz-Erscheinungen bei Röntgenstrahlen. Sitz Bayer Akad Wiss. 1912:303–22. [Google Scholar]
- 2.Crowfoot D, Bernal JD. X-ray photographs of crystalline pepsin. Nature. 1934:794–5. [Google Scholar]
- 3.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662–6. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 4.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North AC. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185:416–22. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 5.Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206:757–61. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 6.Dauter Z, Jaskolski M, Wlodawer A. Impact of synchrotron radiation on macromolecular crystallography: a personal view. J Synchrotron Radiat. 2010;17:433–44. doi: 10.1107/S0909049510011611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coontz R, Fahrenkamp-Uppenbrink J, Lavine M, Vinson V. Crystallography at 100. Going from strength to strength. Introduction Science. 2014;343:1091. doi: 10.1126/science.343.6175.1091. [DOI] [PubMed] [Google Scholar]
- 9.Mueller GA, Edwards LL, Aloor JJ, Fessler MB, Glesner J, Pomés A, et al. The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. J Allergy Clin Immunol. 2010;125:909–17. doi: 10.1016/j.jaci.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller GA, Gosavi RA, Krahn JM, Edwards LL, Cuneo MJ, Glesner J, et al. Der p 5 crystal structure provides insight into the group 5 dust mite allergens. J Biol Chem. 2010;285:25394–401. doi: 10.1074/jbc.M110.128306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller GA, Gosavi RA, Pomés A, Wunschmann S, Moon AF, London RE, et al. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66:878–85. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller GA, Pedersen LC, Lih FB, Glesner J, Moon AF, Chapman MD, et al. The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. J Allergy Clin Immunol. 2013;132:1420–6. doi: 10.1016/j.jaci.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by C-corrected cryo-EM. Nature. 2015 doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 15.Cymborowski M, Klimecka M, Chruszcz M, Zimmerman MD, Shumilin IA, Borek D, et al. To automate or not to automate: this is the question. J Struct Funct Genomics. 2010;11:211–21. doi: 10.1007/s10969-010-9092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minor W, Tomchick D, Otwinowski Z. Strategies for macromolecular synchrotron crystallography. Structure. 2000;8:R105–R110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 17.Chruszcz M, Wlodawer A, Minor W. Determination of protein structures-a series of fortunate events. Biophys J. 2008;95:1–9. doi: 10.1529/biophysj.108.131789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275:1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chruszcz M, Borek D, Domagalski M, Otwinowski Z, Minor W. X-ray diffraction experiment-the last experiment in the structure elucidation process. Adv Protein Chem Struct Biol. 2009;77:23–40. doi: 10.1016/S1876-1623(09)77002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dauter Z, Wlodawer A, Minor W, Jaskolski M, Rupp B. Avoidable errors in deposited macromolecular structures: an impediment to efficient data mining. IUCrJ. 2014;1:179–93. doi: 10.1107/S2052252514005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt A, Teeter M, Weckert E, Lamzin VS. Crystal structure of small protein crambin at 0. 48 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:424–8. doi: 10.1107/S1744309110052607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 23.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H, Chordia MD, Cooper DR, Chruszcz M, Muller P, Sheldrick GM, et al. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9:156–70. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng H, Hou J, Zimmerman MD, Wlodawer A, Minor W. The future of crystallography in drug discovery. Expert Opin Drug Discov. 2014;9:125–37. doi: 10.1517/17460441.2014.872623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 1998;8:319–29. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 27.Kusano S, Kukimoto-Niino M, Satta Y, Ohsawa N, Uchikubo-Kamo T, Wakiyama M, et al. Structural basis for the specific recognition of the major antigenic peptide from the Japanese cedar pollen allergen Cry j 1 by HLA-DP5. J Mol Biol. 2014;426:3016–27. doi: 10.1016/j.jmb.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 28.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–72. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romeo MJ, Agrawal R, Pomés A, Woodfolk JA. A molecular perspective on TH2-promoting cytokine receptors in patients with allergic disease. J Allergy Clin Immunol. 2014;133:952–60. doi: 10.1016/j.jaci.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, et al. Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcvarepsilonRI. Nat Struct Mol Biol. 2011;18:571–6. doi: 10.1038/nsmb.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drinkwater N, Cossins BP, Keeble AH, Wright M, Cain K, Hailu H, et al. Human immunoglobulin E flexes between acutely bent and extended conformations. Nat Struct Mol Biol. 2014;21:397–404. doi: 10.1038/nsmb.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan D, Keeble AH, Hibbert RG, Fabiane S, Gould HJ, McDonnell JM, et al. Ca2+-dependent structural changes in the B-cell receptor CD23 increase its affinity for human immunoglobulin E. J Biol Chem. 2013;288:21667–77. doi: 10.1074/jbc.M113.480657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhaliwal B, Yuan D, Pang MO, Henry AJ, Cain K, Oxbrow A, et al. Crystal structure of IgE bound to its B-cell receptor CD23 reveals a mechanism of reciprocal allosteric inhibition with high affinity receptor FcepsilonRI. Proc Natl Acad Sci U S A. 2012;109:12686–91. doi: 10.1073/pnas.1207278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–59. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bocskei Z, Groom CR, Flower DR, Wright CE, Phillips SE, Cavaggioni A, et al. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature. 1992;360:186–8. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- 36.Gajhede M, Osmark P, Poulsen FM, Ipsen H, Larsen JN, Joost van Neerven RJ, et al. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat Struct Biol. 1996;3:1040–5. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- 37.Westman M, Lupinek C, Bousquet J, Andersson N, Pahr S, Baar A, et al. Early childhood IgE reactivity to pathogenesis-related class 10 proteins predicts allergic rhinitis in adolescence. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman MD, Pomés A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J Allergy Clin Immunol. 2007;119:414–20. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Radauer C, Nandy A, Ferreira F, Goodman RE, Larsen JN, Lidholm J, et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy. 2014;69:413–9. doi: 10.1111/all.12348. [DOI] [PubMed] [Google Scholar]
- 40.Wagner S, Radauer C, Bublin M, Hoffmann-Sommergruber K, Kopp T, Greisenegger EK, et al. Naturally occurring hypoallergenic Bet v 1 isoforms fail to induce IgE responses in individuals with birch pollen allergy. J Allergy Clin Immunol. 2008;121:246–52. doi: 10.1016/j.jaci.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Wallner M, Hauser M, Himly M, Zaborsky N, Mutschlechner S, Harrer A, et al. Reshaping the Bet v 1 fold modulates T(H) polarization. J Allergy Clin Immunol. 2011;127:1571–8. doi: 10.1016/j.jaci.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol. 2008;121:847–52. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 43.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llewellyn R, Eisenberg DS. Annotating proteins with generalized functional linkages. Proc Natl Acad Sci U S A. 2008;105:17700–5. doi: 10.1073/pnas.0809583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin AC, Orengo CA, Hutchinson EG, Jones S, Karmirantzou M, Laskowski RA, et al. Protein folds and functions. Structure. 1998;6:875–84. doi: 10.1016/s0969-2126(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 46.Chruszcz M, Chapman MD, Vailes LD, Stura EA, Saint-Remy JM, Minor W, et al. Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding. J Mol Biol. 2009;386:520–30. doi: 10.1016/j.jmb.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chruszcz M, Pomés A, Glesner J, Vailes LD, Osinski T, Porebski PJ, et al. Molecular determinants for antibody binding on group 1 house dust mite allergens. J Biol Chem. 2012;287:7388–98. doi: 10.1074/jbc.M111.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomés A, Chapman MD, Vailes LD, Blundell TL, Dhanaraj V. Cockroach allergen Bla g 2: structure, function, and implications for allergic sensitization. Am J Respir Crit Care Med. 2002;165:391–7. doi: 10.1164/ajrccm.165.3.2104027. [DOI] [PubMed] [Google Scholar]
- 49.Gustchina A, Li M, Wünschmann S, Chapman MD, Pomés A, Wlodawer A. Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition. J Mol Biol. 2005;348:433–44. doi: 10.1016/j.jmb.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 50.Hindley J, Wünschmann S, Satinover SM, Woodfolk JA, Chew FT, Chapman MD, et al. Bla g 6: a troponin C allergen from Blattella germanica with IgE binding calcium dependence. J Allergy Clin Immunol. 2006;117:1389–95. doi: 10.1016/j.jaci.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 51.Verdino P, Westritschnig K, Valenta R, Keller W. The cross-reactive calcium-binding pollen allergen, Phl p 7, reveals a novel dimer assembly. EMBO J. 2002;21:5007–16. doi: 10.1093/emboj/cdf526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chruszcz M, Maleki SJ, Majorek KA, Demas M, Bublin M, Solberg R, et al. Structural and immunologic characterization of Ara h 1, a major peanut allergen. J Biol Chem. 2011;286:39318–27. doi: 10.1074/jbc.M111.270132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maleki SJ, Kopper RA, Shin DS, Park CW, Compadre CM, Sampson H, et al. Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J Immunol. 2000;164:5844–9. doi: 10.4049/jimmunol.164.11.5844. [DOI] [PubMed] [Google Scholar]
- 54.Nicolaou N, Murray C, Belgrave D, Poorafshar M, Simpson A, Custovic A. Quantification of specific IgE to whole peanut extract and peanut components in prediction of peanut allergy. J Allergy Clin Immunol. 2011;127:684–5. doi: 10.1016/j.jaci.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Dang TD, Tang M, Choo S, Licciardi PV, Koplin JJ, Martin PE, et al. Increasing the accuracy of peanut allergy diagnosis by using Ara h 2. J Allergy Clin Immunol. 2012;129:1056–63. doi: 10.1016/j.jaci.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 56.Klemans RJ, van Os-Medendorp H, Blankestijn M, Bruijnzeel-Koomen CA, Knol EF, Knulst AC. Diagnostic accuracy of specific IgE to components in diagnosing peanut allergy: a systematic review. Clin Exp Allergy. 2015;45:720–30. doi: 10.1111/cea.12412. [DOI] [PubMed] [Google Scholar]
- 57.Chruszcz M, Chapman MD, Osinski T, Solberg R, Demas M, Porebski PJ, et al. Alternaria alternata allergen Alt a 1: A unique beta-barrel protein dimer found exclusively in fungi. J Allergy Clin Immunol. 2012;130:241–7. doi: 10.1016/j.jaci.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randall TA, Perera L, London RE, Mueller GA. Genomic, RNAseq, and molecular modeling evidence suggests that the major allergen domain in insects evolved from a homodimeric origin. Genome Biol Evol. 2013;5:2344–58. doi: 10.1093/gbe/evt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamiaux C, Maddumage R, Middleditch MJ, Prakash R, Brummell DA, Baker EN, et al. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J Struct Biol. 2014;187:276–81. doi: 10.1016/j.jsb.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 60.Kaiser L, Gronlund H, Sandalova T, Ljunggren HG, van Hage-Hamsten M, Achour A, et al. The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family. J Biol Chem. 2003;278:37730–5. doi: 10.1074/jbc.M304740200. [DOI] [PubMed] [Google Scholar]
- 61.Kaiser L, Velickovic TC, Badia-Martinez D, Adedoyin J, Thunberg S, Hallen D, et al. Structural Characterization of the Tetrameric form of the Major Cat Allergen Fel d 1. J Mol Biol. 2007;370:714–27. doi: 10.1016/j.jmb.2007.04.074. [DOI] [PubMed] [Google Scholar]
- 62.Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller GA, Rule GS, et al. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. J Mol Biol. 2002;318:189–97. doi: 10.1016/S0022-2836(02)00027-X. [DOI] [PubMed] [Google Scholar]
- 63.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 64.Trompette A, Divanovic S, Visintin A, Blanchard C, Hegde RS, Madan R, et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 2009;457:585–8. doi: 10.1038/nature07548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karp CL. Guilt by intimate association: what makes an allergen an allergen? J Allergy Clin Immunol. 2010;125:955–60. doi: 10.1016/j.jaci.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–51. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piggott DA, Eisenbarth SC, Xu L, Constant SL, Huleatt JW, Herrick CA, et al. MyD88-dependent induction of allergic Th2 responses to intranasal antigen. J Clin Invest. 2005;115:459–67. doi: 10.1172/JCI22462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redecke V, Hacker H, Datta SK, Fermin A, Pitha PM, Broide DH, et al. Cutting edge: activation of Toll-like receptor 2 induces a Th2 immune response and promotes experimental asthma. J Immunol. 2004;172:2739–43. doi: 10.4049/jimmunol.172.5.2739. [DOI] [PubMed] [Google Scholar]
- 69.Shakib F, Ghaemmaghami AM, Sewell HF. The molecular basis of allergenicity. Trends Immunol. 2008;29:633–42. doi: 10.1016/j.it.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 70.Chapman MD, Wunschmann S, Pomés A. Proteases as Th2 adjuvants. Curr Allergy Asthma Rep. 2007;7:363–7. doi: 10.1007/s11882-007-0055-6. [DOI] [PubMed] [Google Scholar]
- 71.Schulz O, Laing P, Sewell HF, Shakib F. Der p I, a major allergen of the house dust mite, proteolytically cleaves the low-affinity receptor for human IgE (CD23) Eur J Immunol. 1995;25:3191–4. doi: 10.1002/eji.1830251131. [DOI] [PubMed] [Google Scholar]
- 72.Hewitt CR, Brown AP, Hart BJ, Pritchard DI. A major house dust mite allergen disrupts the immunoglobulin E network by selectively cleaving CD23: innate protection by antiproteases. J Exp Med. 1995;182:1537–44. doi: 10.1084/jem.182.5.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.King C, Brennan S, Thompson PJ, Stewart GA. Dust mite proteolytic allergens induce cytokine release from cultured airway epithelium. J Immunol. 1998;161:3645–51. [PubMed] [Google Scholar]
- 74.Wan H, Winton HL, Soeller C, Tovey ER, Gruenert DC, Thompson PJ, et al. Der p 1 facilitates transepithelial allergen delivery by disruption of tight junctions. J Clin Invest. 1999;104:123–33. doi: 10.1172/JCI5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang S, Rutkowsky JM, Snodgrass RG, Ono-Moore KD, Schneider DA, Newman JW, et al. Saturated fatty acids activate TLR-mediated proinflammatory signaling pathways. J Lipid Res. 2012;53:2002–13. doi: 10.1194/jlr.D029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318 (Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vance SJ, McDonald RE, Cooper A, Smith BO, Kennedy MW. The structure of latherin, a surfactant allergen protein from horse sweat and saliva. J R Soc Interface. 2013;10:20130453. doi: 10.1098/rsif.2013.0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bublin M, Eiwegger T, Breiteneder H. Do lipids influence the allergic sensitization process? J Allergy Clin Immunol. 2014;134:521–9. doi: 10.1016/j.jaci.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Passalacqua G, Pasquali M, Ariano R, Lombardi C, Giardini A, Baiardini I, et al. Randomized double-blind controlled study with sublingual carbamylated allergoid immunotherapy in mild rhinitis due to mites. Allergy. 2006;61:849–54. doi: 10.1111/j.1398-9995.2006.01095.x. [DOI] [PubMed] [Google Scholar]
- 80.Cromwell O, Hafner D, Nandy A. Recombinant allergens for specific immunotherapy. J Allergy Clin Immunol. 2011;127:865–72. doi: 10.1016/j.jaci.2011.01.047. [DOI] [PubMed] [Google Scholar]
- 81.Meyer W, Narkus A, Salapatek AM, Hafner D. Double-blind, placebo-controlled, dose-ranging study of new recombinant hypoallergenic Bet v 1 in an environmental exposure chamber. Allergy. 2013;68:724–31. doi: 10.1111/all.12148. [DOI] [PubMed] [Google Scholar]
- 82.Smith AM, Chapman MD, Taketomi EA, Platts-Mills TA, Sung SS. Recombinant allergens for immunotherapy: a Der p 2 variant with reduced IgE reactivity retains T-cell epitopes. J Allergy Clin Immunol. 1998;101:423–5. doi: 10.1016/S0091-6749(98)70259-3. [DOI] [PubMed] [Google Scholar]
- 83.Takai T, Yokota T, Yasue M, Nishiyama C, Yuuki T, Mori A, et al. Engineering of the major house dust mite allergen Der f 2 for allergen-specific immunotherapy. Nat Biotechnol. 1997;15:754–8. doi: 10.1038/nbt0897-754. [DOI] [PubMed] [Google Scholar]
- 84.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, et al. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol. 2000;165:331–8. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 85.Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, Larsen JN. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J Immunol. 2003;171:3084–90. doi: 10.4049/jimmunol.171.6.3084. [DOI] [PubMed] [Google Scholar]
- 86.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I, et al. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol. 2007;368:742–52. doi: 10.1016/j.jmb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 87.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM, et al. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–21. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 88.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, et al. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–51. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 89.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, et al. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem. 2008;283:22806–14. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li M, Gustchina A, Glesner J, Wunschmann S, Vailes LD, Chapman MD, et al. Carbohydrates contribute to the interactions between cockroach allergen Bla g 2 and a monoclonal antibody. J Immunol. 2011;186:333–40. doi: 10.4049/jimmunol.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Osinski T, Pomés A, Majorek KA, Glesner J, Offermann LR, Vailes LD, et al. Structural analysis of Der p 1-antibody complexes and comparison with complexes of proteins or peptides with monoclonal antibodies. J Immuno. 2015 doi: 10.4049/jimmunol.1402199. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Glesner J, Wunschmann S, Li M, Gustchina A, Wlodawer A, Himly M, et al. Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding. PLoS ONE. 2011;6:e22223. doi: 10.1371/journal.pone.0022223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sastre J, Landivar ME, Ruiz-Garcia M, Andregnette-Rosigno MV, Mahillo I. How molecular diagnosis can change allergen-specific immunotherapy prescription in a complex pollen area. Allergy. 2012;67:709–11. doi: 10.1111/j.1398-9995.2012.02808.x. [DOI] [PubMed] [Google Scholar]
- 94.Stringari G, Tripodi S, Caffarelli C, Dondi A, Asero R, Di Rienzo BA, et al. The effect of component-resolved diagnosis on specific immunotherapy prescription in children with hay fever. J Allergy Clin Immunol. 2014;134:75–81. doi: 10.1016/j.jaci.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 95.Tripodi S, Frediani T, Lucarelli S, Macri F, Pingitore G, Di Rienzo BA, et al. Molecular profiles of IgE to Phleum pratense in children with grass pollen allergy: implications for specific immunotherapy. J Allergy Clin Immunol. 2012;129:834–9. doi: 10.1016/j.jaci.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 96.Le TM, Bublin M, Breiteneder H, Fernandez-Rivas M, Asero R, Ballmer-Weber B, et al. Kiwifruit allergy across Europe: clinical manifestation and IgE recognition patterns to kiwifruit allergens. J Allergy Clin Immunol. 2013;131:164–71. doi: 10.1016/j.jaci.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 97.Lieberman JA, Glaumann S, Batelson S, Borres MP, Sampson HA, Nilsson C. The utility of peanut components in the diagnosis of IgE-mediated peanut allergy among distinct populations. J Allergy Clin Immunol Pract. 2013;1:75–82. doi: 10.1016/j.jaip.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 98.Aalberse RC. Structural features of allergenic molecules. Chem Immunol Allergy. 2006;91:134–46. doi: 10.1159/000090277. [DOI] [PubMed] [Google Scholar]
- 99.Hazebrouck S, Ah-Leung S, Bidat E, Paty E, Drumare MF, Tilleul S, et al. Goat’s milk allergy without cow’s milk allergy: suppression of non-cross-reactive epitopes on caprine beta-casein. Clin Exp Allergy. 2014;44:602–10. doi: 10.1111/cea.12261. [DOI] [PubMed] [Google Scholar]
- 100.Saarelainen S, Rytkonen-Nissinen M, Rouvinen J, Taivainen A, Auriola S, Kauppinen A, et al. Animal-derived lipocalin allergens exhibit immunoglobulin E cross-reactivity. Clin Exp Allergy. 2008;38:374–81. doi: 10.1111/j.1365-2222.2007.02895.x. [DOI] [PubMed] [Google Scholar]
- 101.D’Avino R, Bernardi ML, Wallner M, Palazzo P, Camardella L, Tuppo L, et al. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy. 2011;66:870–7. doi: 10.1111/j.1398-9995.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- 102.Bublin M, Kostadinova M, Radauer C, Hafner C, Szepfalusi Z, Varga EM, et al. IgE cross-reactivity between the major peanut allergen Ara h 2 and the nonhomologous allergens Ara h 1 and Ara h 3. J Allergy Clin Immunol. 2013;132:118–24. doi: 10.1016/j.jaci.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 103.Power TD, Ivanciuc O, Schein CH, Braun W. Assessment of 3D models for allergen research. Proteins. 2013;81:545–54. doi: 10.1002/prot.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aalberse RC, Akkerdaas J, van Ree R. Cross-reactivity of IgE antibodies to allergens. Allergy. 2001;56:478–90. doi: 10.1034/j.1398-9995.2001.056006478.x. [DOI] [PubMed] [Google Scholar]
- 105.Mueller GA, Pedersen LC, Glesner J, Edwards LL, Zakzuk J, London RE, et al. Analysis of glutathione S-transferase allergen cross-reactivity in a North American population: relevance for molecular diagnosis. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.03.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kupitz C, Basu S, Grotjohann I, Fromme R, Zatsepin NA, Rendek KN, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–5. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McSweeney S, Fromme P. Crystallography: Sources of inspiration. Nature. 2014;505:620–1. doi: 10.1038/505620a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zimmerman MD, Grabowski M, Domagalski MJ, Maclean EM, Chruszcz M, Minor W. Data management in the modern structural biology and biomedical research environment. Methods Mol Biol. 2014;1140:1–25. doi: 10.1007/978-1-4939-0354-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.