SUMMARY
One of the major determinants of aging in organisms ranging from worms to man are FOXO family transcription factors, which are downstream effectors of Insulin/IGF-1 signaling (IIS). The molecular mechanisms that actively promote DAF16/FOXO stability and function are unknown. Here we identify the deubiquitylating enzyme MATH-33 as an essential DAF-16 regulator in IIS, which stabilizes active DAF-16 protein levels and, as a consequence, influences DAF-16 functions, such as metabolism, stress response and longevity in C. elegans. MATH-33 associates with DAF-16 in cellulo and in vitro. MATH-33 functions as a deubiquitylase by actively removing ubiquitin moieties from DAF-16, thus counteracting the action of the RLE-1 E3-ubiquitin ligase. Our findings support a model in which MATH-33 promotes DAF-16 stability in response to decreased IIS by directly modulating its ubiquitylation state, suggesting that regulated oscillations in the stability of DAF-16 protein play an integral role in controlling processes such as metabolism and longevity.
INTRODUCTION
FOXO proteins are part of an evolutionarily conserved family of transcription factors associated with the regulation of a wide-range of biological processes, including the rate, onset, and severity of aging and age-related diseases. In model organisms, activation of FOXO transcription factors results in as much as a three-fold increase in median lifespan (Kenyon, 2010). More recently, genome-wide association studies (GWAS) have identified single nucleotide polymorphisms in the FOXO3a gene in humans who lived to an extreme age (greater than 95) (Wheeler and Kim, 2011). All together this would suggest an evolutionary conservation of FOXO proteins in regulating the aging process.
FOXO activity is modulated by various post-translational modifications (PTMs) including phosphorylation, acetylation and ubiquitylation. PTMs on FOXO proteins occur in response to diverse upstream signaling networks, which in turn regulate FOXO subcellular localization, activity and protein turnover (Calnan and Brunet, 2008). Previous studies have extensively characterized the mechanisms associated with phosphorylation-dependent regulation of FOXO/DAF-16 in the context of Insulin/IGF-1 signaling (IIS) (Huang and Tindall, 2007). Insulin/IGF-1 receptors initiate a cascade of conserved protein kinases resulting in phosphorylation and cytoplasmic retention of FOXO/DAF-16 transcription factors, major effectors of IIS (Calnan and Brunet, 2008). In contrast, reduction of IIS signaling results in dephosphorylation of FOXOs, promoting their nuclear translocation and their ability to activate or repress transcriptional targets. While the mechanisms surrounding nuclear translocation of FOXO proteins are well established, processes required for stabilizing the nuclear FOXO pool are widely unknown.
In C. elegans, IIS activity can be reduced by mutations in core components of the pathway, such as the insulin/IGF-1-like receptor daf-2 or the PI3K catalytic subunit orthologue age-1. Reduction of IIS results in nuclear translocation and activation of the FOXO transcription factor DAF-16 and is critical for regulating certain developmental decisions, metabolism and longevity in nematodes (Wolff and Dillin, 2006).
FOXO transcription factors are also controlled at the level of protein stability and activity by both poly- and monoubiquitylation (Huang and Tindall, 2011). In C. elegans, the RING finger-containing E3 ubiquitin ligase RLE-1 (regulation of longevity by E3) catalyzes DAF-16 polyubiquitylation and promotes its proteasomal degradation (Li et al., 2007). A loss of RLE-1 is sufficient to increase DAF-16 stability, which affects development, stress resistance and lifespan in nematodes (Li et al., 2007). In mammals several ubiquitin E3 ligases have been found to play a role in the polyubiquitylation and degradation of FOXO proteins (Zhao et al., 2011), however, the counteracting deubiquitylating enzymes (DUBs) promoting protein stability by antagonizing polyubiquitylation have not been discovered.
The complexity of PTMs found on DAF-16/FOXO such as phosphorylation, acetylation, ubiquitylation and methylation suggests that a network of modifiers must coordinately work to ensure the dynamic regulation of DAF-16 activity (Calnan and Brunet, 2008). DAF-16 is directly phosphorylated by many protein kinases, including AKT-1, AKT-2, SGK-1 (Cahill et al., 2001; Hertweck et al., 2004; Lin et al., 2001), AMP-activated protein kinase (AMPK/AAK-1) (Greer et al., 2007), Ca2+/calmodulin-dependent kinase type II (CAMKII) (Tao et al., 2013), Jun-N-terminal kinase (JNK/JNK-1) (Oh et al., 2005) and the Ste20-like protein kinase (MST1/CST-1) (Lehtinen et al., 2006). In addition, DAF-16 stability and functions are regulated by ubiquitylation (Li et al, 2006), acetylation (Chiang et al., 2012) and methylation (Takahashi et al., 2011). We took an unbiased approach towards the identification of novel proteins responsible for modulating PTMs on DAF-16 with the goal of identifying proteins that play a role in stabilizing DAF-16. By purifying DAF-16 protein complexes from C. elegans and multi-dimensional protein identification MS technology (MudPIT), we have identified the DUB MATH-33 as a novel interacting factor for DAF-16. MATH-33 shares 31% sequence identity with human USP7/HAUSP. USP7/HAUSP has been previously shown to deubiquitylate monoubiquitylated FOXO proteins under acute oxidative stress conditions and serum starvation, which negatively regulates FOXO1 and FOXO4 activity (Hall et al., 2014; van der Horst et al., 2006). In contrast, we find MATH-33 functions as a positive regulator for DAF-16 stability in the context of reduced IIS and is essential for various DAF-16-mediated phenotypic readouts such as metabolism, stress response and lifespan determination. We demonstrate that MATH-33 acts as a deubiquitylase and antagonizes RLE-1-mediated polyubiquitylation of DAF-16, providing a novel mechanism for how FOXO levels are stabilized when IIS is reduced. The divergence of activities of MATH-33 from USP7 in the regulation of FOXO proteins highlights the complexity required to regulate this crucial transcription factor family.
RESULTS
Identification of DAF-16 regulators by MudPIT and reporter based screening
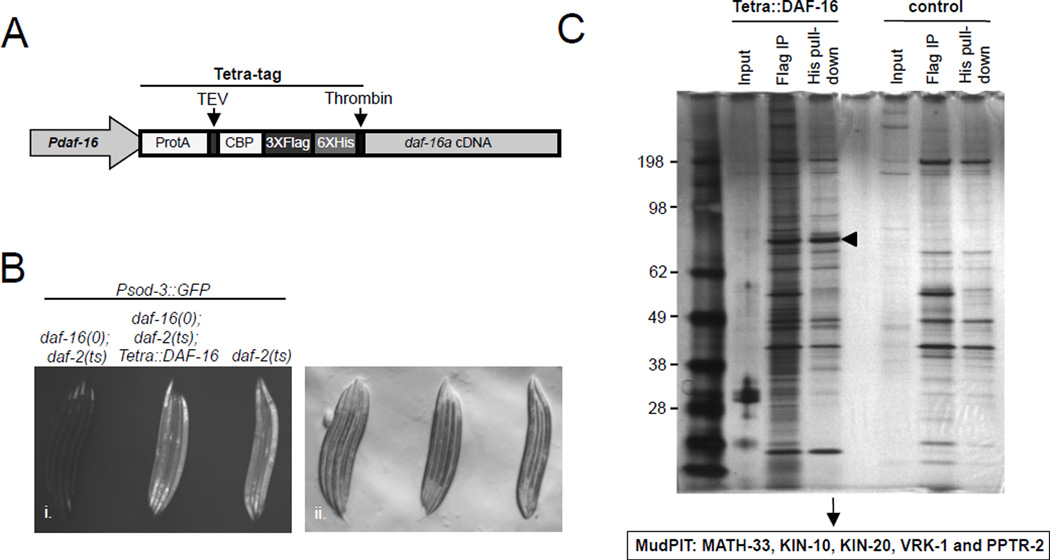
Alterations in IIS modulate PTMs on FOXO proteins, including phosphorylation, acetylation and ubiquitylation (Calnan and Brunet, 2008). Therefore, we hypothesized that an analysis of DAF-16 binding proteins under conditions of reduced IIS might reveal novel regulators of DAF-16 post-translational modifications. To test this, we established a tandem affinity purification method for C. elegans DAF-16 protein complexes combined with multidimensional protein identification MS technology (MudPIT) (Link et al., 1999; Washburn et al., 2001). The DAF-16a isoform was fused to four sequential epitope tags (Figure 1A) (Yang et al., 2006) and stably integrated into nematodes containing both the temperature sensitive daf-2(e1370) allele of the insulin/IGF-1 like receptor and a null allele of daf-16 (daf-16(mu86)). We verified that the tagged DAF-16 version was expressed and functional, as it could activate transcription of a DAF-16 responsive reporter (Figure 1B). DAF-16 complexes were purified from nematodes grown under conditions where IIS was downregulated using the temperature sensitive daf-2(e1370) allele (Figure 1C).
Figure 1. Isolation of post-translational modifiers for DAF-16 by Tandem Affinity Purification and MudPIT.
(A) Tandem Affinity Purification tagging strategy for DAF-16 using a Tetra tag. (B) Tetra::DAF-16a activates the Psod-3::GFP reporter in daf-16(mu86); daf-2(e1370) animals shifted to 25°C. Fluorescent (i.) and DIC (ii.) micrographs are presented. (C) SDS-PAGE/silver staining analysis of Tetra::DAF-16 and potential binding partners after FLAG and His epitope-based Tandem Affinity Purification. Control, isogenic strain without Tetra::DAF-16; arrowhead denotes Tetra::DAF-16. MudPIT results of post-translational modifiers for DAF-16 are presented.
Because we were primarily interested in identifying proteins capable of affecting PTMs of DAF-16, of the proteins that were selectively identified in association with DAF-16, we prioritized the characterization of binding partners traditionally associated with PTMs and identified five candidate proteins. These included three protein kinases (KIN-10, KIN-20 and VRK-1), one protein phosphatase (PPTR-2) and one deubiquitylating enzyme (MATH-33). We then subjected each of these candidates to a secondary reporter-based screen and found that only math-33 was required for DAF-16 activity (Figure S1A and data not shown). Our findings suggest that math-33 is a potential regulator for DAF-16 activity and function.
MATH-33 physically interacts with and regulates DAF-16 dependent on IIS
In our secondary reporter screen, based on a Psod-3::GFP transcriptional reporter responsive to DAF-16 (Libina et al., 2003), we found that RNAi-mediated inactivation of math-33 significantly diminished the transcriptional activity of DAF-16 when IIS was downregulated in a daf-2(e1370) temperature-sensitive mutant (Figure S1A). Reduction of math-33 function did not diminish the basal Psod-3::GFP reporter activity in a wild type (N2) background, indicating that math-33 is essential for DAF-16 activity primarily when IIS is reduced. To test the specificity of math-33, we asked if math-33 was required for the induction of general stress responsive networks. We found that RNAi-mediated knockdown of math-33 did not affect the induction of the hsp-4 (ER stress), hsp-16.2 (heat stress) or hsp-6 (mitochondrial stress) promoters (Figures S1B–S1D), suggesting that math-33 specifically acts as a critical regulator for IIS-mediated transcriptional readouts rather than affecting general stress response pathways.
The effect of math-33 inactivation on the Psod-3::GFP reporter activity was further analyzed using the math-33(tm3561) allele, which has been previously characterized as a loss-of-function allele in the context of early embryonic polarity establishment (McCloskey and Kemphues, 2012). In our experiments, math-33 inactivation by the loss-of-function math-33(tm3561) allele in daf-2(e1370) nematodes reduced Psod-3::GFP reporter activity to a level similar to that observed in daf-16(mu86); daf-2(e1370) control animals (Figure 2A). These results suggest that math-33 is required for daf-16-mediated Psod-3::GFP reporter activation when IIS is compromised.
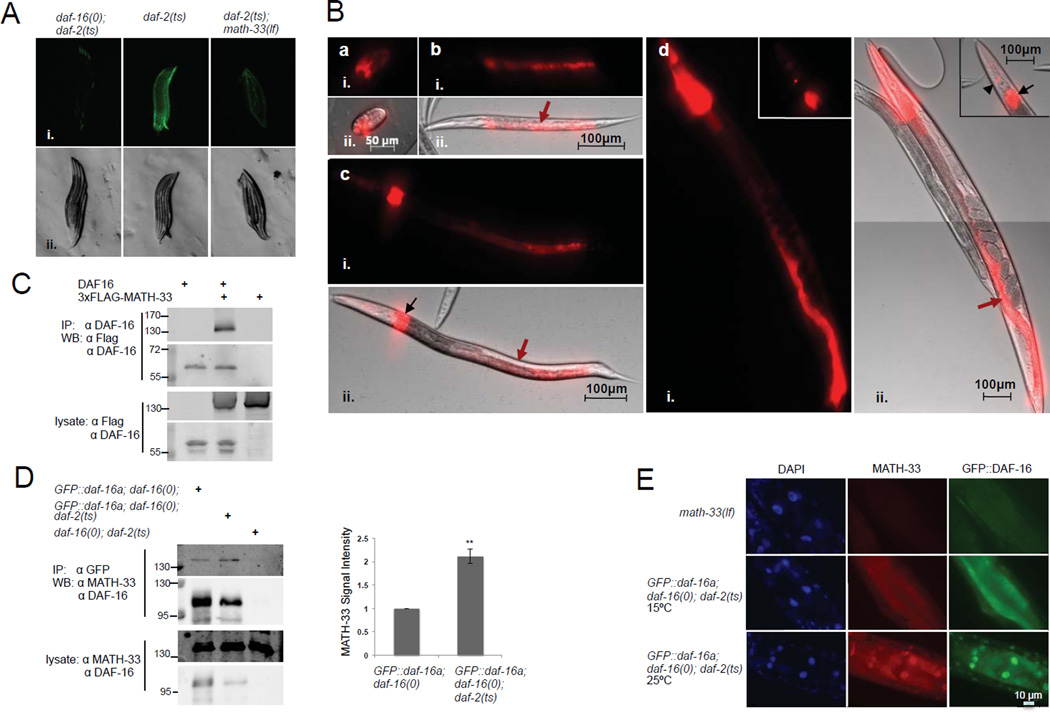
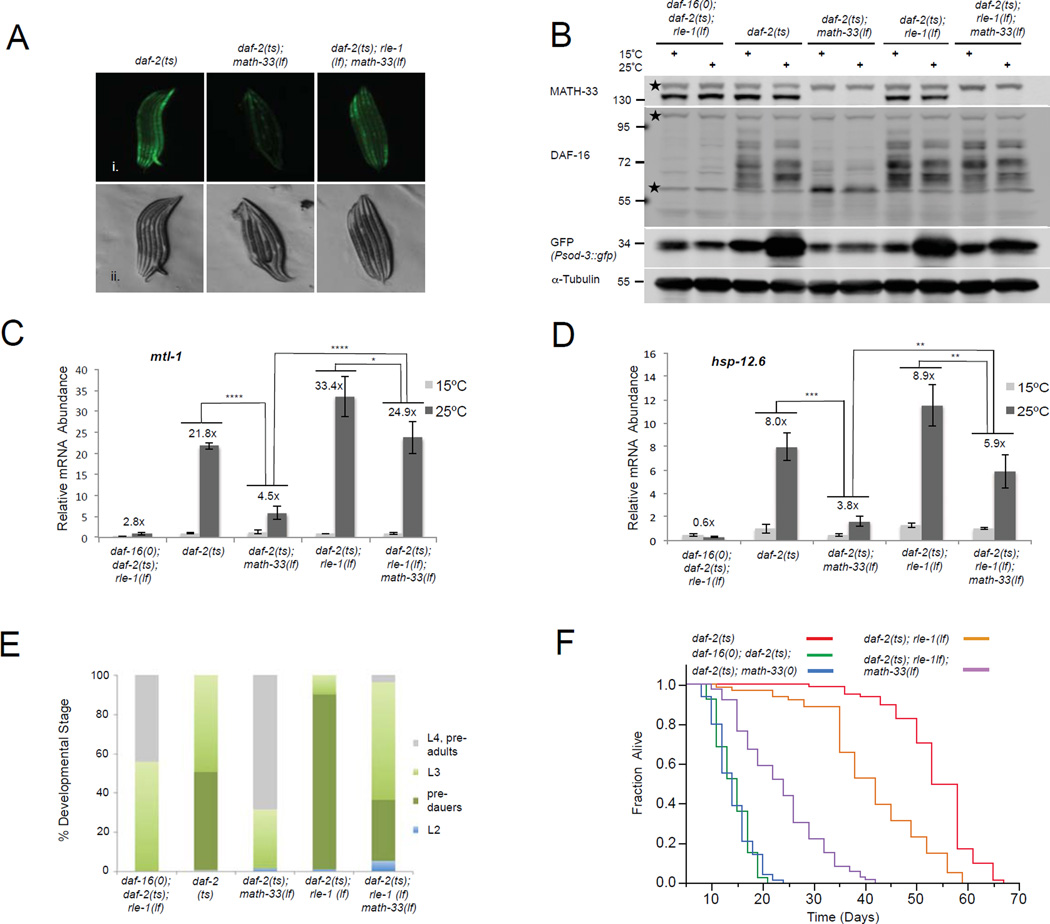
Figure 2. MATH-33 is required for DAF-16 activity, physically interacts and co-localizes with DAF-16 dependent on IIS.
(A) Reporter assays of C. elegans strains containing a Psod-3::GFP reporter responsive to DAF-16. Animals were raised at 25°C and analyzed 24 hr after hatching. (B) Expression analysis of math-33 based on a Pmath-33::tdTomato transcriptional reporter. Reporter activity was detected in daf-2(e1370) mutant embryos (a), L2 and L3 larval stages (b and c) and adults (d). Reporter fluorescence was apparent in the intestinal tissue (red arrows), a group of undefined cells located at the dorsal side of the anterior intestine (black arrows) and in head neurons (black arrowhead). Fluorescent (i.), DIC (A ii.) and composite fluorescent/DIC images (B ii.) are presented. (C, D) Co-immunoprecipitation analysis of MATH-33 and DAF-16 in HEK293T cells and C. elegans. DAF-16 and 3xFLAG-MATH-33 were expressed in HEK293T cells (C). Lysates were immunoprecipitated with a DAF-16 specific antibody and probed with antibodies against FLAG and DAF-16. Lysates from nematode strains expressing GFP::DAF-16 (D) were immunoprecipitated using a GFP-Trap resin and probed with antibodies against MATH-33 and DAF-16. Signal intensities of co-immunoprecipitated MATH-33 were quantified relative to GFP::DAF-16. p-Values were calculated using two-tailed Student's t-test. **: p<0.05; (E) Immunostaining of MATH-33 and GFP::DAF-16 in intestinal cells using affinity-purified anti-MATH-33 and anti-GFP antibodies. DAPI staining indicates localization of nuclei. (A–E). The daf-2(e1370) temperature sensitive allele, the daf-16(mu86) null allele and the math-33(tm3561) loss-of-function allele were used.
In previous studies, the intestine has been described as one of the major tissues where DAF-16 is localized and mediates its lifespan extending function (Libina et al., 2003). To analyze the expression pattern of math-33 in C. elegans andpotential changes when IIS is compromised, we generated a daf-2(e1370) transgenic strain in which the 1.7 kbp upstream promoter region of math-33 was fused to a tdTomato reporter gene. We did not observe any obvious changes in the activity of the tdTomato reporter when IIS was reduced (data not shown). However, the transcriptional reporter detected expression of math-33 in intestinal cells and a group of cells located anterior of the intestine during larval and adult stages in daf-2 mutant animals (Figure 2B). Our data indicate that math-33 is predominantly expressed in the intestinal tissue, in which DAF-16 has been previously found to extend lifespan.
The closest mammalian orthologue of MATH-33, USP7, might target substrates by physical association rather than recognizing specific ubiquitin chains (Faesen et al., 2011). To analyze the physical interaction between MATH-33 and DAF-16 indicated by our MuDPIT analysis, we performed co-immunoprecipitation studies of MATH-33 and DAF-16 transiently expressed in human HEK293T cells and detected a physical association of MATH-33 with DAF-16 (Figure 2C). Furthermore, we tested whether endogenous MATH-33 expressed in C. elegans physically binds to DAF-16 in vivo. Interestingly, endogenous MATH-33 co-immunoprecipitated with transgene-expressed GFP-DAF-16 in nematodes when IIS activity was reduced (Figure 2D). When the nuclear translocation of GFP::DAF-16 was induced at 25°C in temperature sensitive daf-2(e1370) mutants, association of endogenous MATH-33 with GFP::DAF-16 was enhanced compared to a control strain harboring the wild type daf-2 allele in which GFP::DAF-16 is predominantly cytoplasmic. Our data suggest that IIS regulates the physical association of DAF-16 with MATH-33. To analyze the subcellular distribution of MATH-33 and DAF-16 in C. elegans, we performed immunostainings and found that endogenous MATH-33 and transgene expressed GFP::DAF-16 co-localized in the nucleus of intestinal cells when IIS was diminished and DAF-16 nuclear translocation was induced (Figure 2E). In addition, we observed a nuclear enrichment of MATH-33 when IIS was compromised (Figure 2E, Figure S2A and S2B). Importantly, these assays indicated that MATH-33 colocalizes with active, nuclear DAF-16, and that MATH-33 binding to DAF-16 is increased when IIS is downregulated.
MATH-33 regulates DAF-16 stability by antagonizing ubiquitylation of DAF-16
In mammalian cells, the closest orthologue of MATH-33, USP7, is capable of deubiquitylating monoubiquitylated FOXO transcription factors under conditions of acute cellular oxidative stress and serum starvation. Removal of monoubiquitin residues by USP7 causes a decrease in FOXO1 and FOXO4 activity (Hall et al., 2014; van der Horst et al., 2006). By analogy, loss of MATH-33 would be predicted to enhance, rather than suppress, transcription of DAF-16 targets. The downregulated levels of SOD-3 in math-33 loss-of-function mutants suggests, however, that deubiquitylation in the context of reduced IIS has a distinct and opposing effect on DAF-16 function, as compared to USP7 upon FOXO1/4.
We therefore hypothesized that MATH-33 might instead serve as a deubiquitylating enzyme to reverse polyubiquitylation of DAF-16, protecting DAF-16 from degradation by the ubiquitin-proteasome system and increasing its stability. Thus, we analyzed the effects of math-33 mutation on steady-state levels of DAF-16, using a DAF-16 antibody raised against the C-terminus of DAF-16 that is capable of detecting multiple DAF-16 isoforms (Kwon et al., 2010). Strikingly, reduction of IIS in daf-2(e1370) young adult animals resulted in a decrease of several DAF-16 isoforms in the absence of a functional math-33 (Figures 3A and 3B). The effect upon the DAF-16 isoforms was already observed at the semi-permissive temperature (15°C), but was not observed in a math-33 single mutant, a background in which IIS functions at normal levels. In addition, immunostaining detected a reduction of endogenous nuclear DAF-16 levels in a daf-2(e1370) strain at 25°C when math-33 was mutated (Figure 3C). Our data suggest that MATH-33 specifically stabilizes DAF-16 levels in the context of reduced IIS. To test if MATH-33 could regulate DAF-16 protein levels, we analyzed whether DAF-16 isoform transcript levels were altered in daf-2(e1370); math-33(tm3561) mutants. As predicted, no overall change in daf-16a and b transcript levels were observed when comparing the daf-2(e1370) single mutant with the daf-2(e1370); math-33(tm3561) double mutant (Figure S3A and S3B) indicating that MATH-33 might directly regulate DAF-16a and b protein levels. Surprisingly, however, we did detect a reduction of daf-16d/f transcript levels when math-33 was inactive (Figure S3C). This likely indicates a potential DAF-16-dependent transcriptional auto-regulation of the daf16d/f isoform (see Discussion).
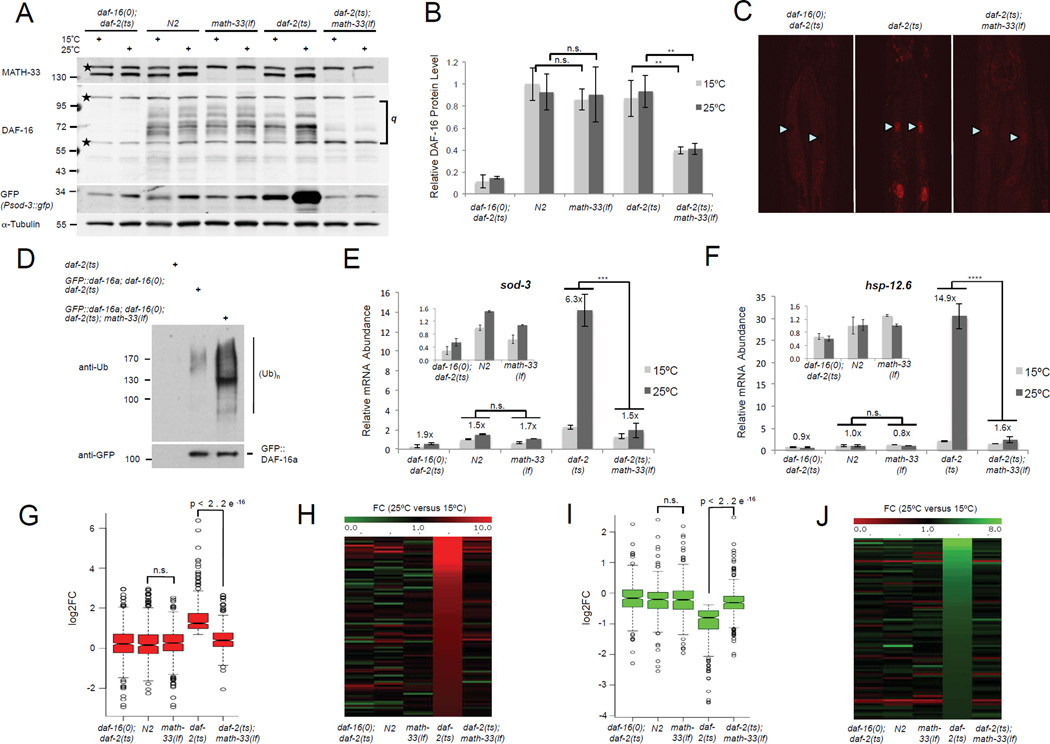
Figure 3. MATH-33 is required for controlling DAF-16 ubiquitylation and protein levels affecting DAF-16 target gene regulation in the context of IIS.
(A) Endogenous DAF-16 isoform levels and Psod-3::GFP reporter activation detected by immunoblotting. Membrane was probed with anti-DAF-16 and anti-MATH-33 antibodies followed by reprobing with GFP and alpha tubulin specific antibodies. Stars denote nonspecific bands. (B) Quantification of endogenous DAF-16 isoform levels relative to tubulin signals from immunoblots in (A). q, quantified area. Lower background band overlays with a DAF-16 specific band and was included. (C) Immunostaining of endogenous nuclear DAF-16 displayed for anterior intestinal cells when IIS is reduced. Arrowheads denote intestinal nuclei. Lipid droplet accumulation in daf-2(e1370) intestinal cells causes cytoplasmic background. (D) DAF-16 ubiquitylation in C. elegans detected by immunoblotting. GFP::DAF-16a was immunoprecipitated using a GFP-Trap resin and ubiquitin conjugation to DAF-16 was detected by immunoblotting with an ubiquitin-specific antibody (FK2). Equal amounts of GFP::DAF16 were loaded for comparison of DAF-16 ubiquitylation levels. (E, F). qRT-PCR of endogenous sod-3 and hsp-12.6 transcripts. Insets show expanded histograms for controls. Numbers above bars indicate relative induction on 25°C versus 15°C. The mean normalized levels and STDEV of three biological repeats are shown (B, E and F, see methods). (G–J) Analysis of math-33 inactivation for DAF-16 target gene regulation using RNA sequencing. (G, I) Box plots display fold changes (log2FC) in DAF-16 target gene activation (G, 441 targets) and repression (I, 362 targets) based on FPKM values on 25°C versus 15°C for indicated strains. (H, J) Heat maps illustrate MATH-33-mediated alterations in the top 90 IIS induced (H) or repressed (J) DAF-16 targets. p-Values were calculated using two-tailed Student's t-test (B, G, I) or Two-Way ANOVA (E, F). ****: p<0.0001; ***: p<0.001; **: p<0.01; n.s.: not significant.
To address whether MATH-33 is involved in the regulation of DAF-16 ubiquitylation in vivo, we analyzed the ubiquitylation state of DAF-16 when math-33 activity was absent and IIS downregulated. Endogenous DAF-16 isoform protein levels are severely reduced in a daf-2(e1370); math-33(tm3561) background (Figure 3A), which precludes a study of their ubiquitylation state. Overexpressed GFP::DAF-16a protein levels were significantly downregulated as well in daf-16(mu86); daf-2(e1370); math-33(tm3561) animals (Figure S3D). However, sufficient protein could be recovered by immunoprecipitation experiments to analyze ubiquitin conjugated on the GFP::DAF-16a fusion protein. Using the daf-16(mu86); daf-2(e1370); math-33(tm3561) triple mutant background with the GFP::DAF-16a transgene, we found increased ubiquitylation of DAF-16 in the absence of math-33 compared to a control strain where math-33 is endogenously expressed (Figure 3D). These data clearly indicate that MATH-33 counteracts DAF-16 polyubiquitylation in vivo.
MATH-33 is required for DAF-16 target gene expression
Since DAF-16 is a key transcription factor downstream of IIS, we tested whether a reduction in DAF-16 steady state levels with loss of math-33 modulates the transcriptional readout of DAF-16. We found that the decrease of various endogenous DAF-16 isoforms in daf-2(e1370) animals on 25°C in the absence of functional math-33 resulted in diminished Psod-3::GFP reporter activation, which was detected in worm lysates by immunoblotting with an anti-GFP antibody (Figure 3A). Next, we tested whether inactivation of math-33 could affect the expression of IIS responsive, endogenous DAF-16 targets such as sod-3, the small heat shock protein encoding gene hsp-12.6 and the metallothionein encoding gene mtl-1 (Li et al., 2008; Murphy et al., 2003). We detected an upregulation of target gene transcript levels when IIS was reduced in young adult daf-2(e1370) animals (Figures 3E and 3F). However, loss of math-33 activity in daf-2(e1370) mutant animals resulted in a significant reduction in DAF-16 target transcript levels when IIS was downregulated. In addition, we found that math-33 was not required to maintain DAF-16 steady state levels and activity under oxidative stress conditions when IIS signals at normal physiological levels (Figure S3E and S3F). Our data suggest a distinct role of math-33 in DAF-16 regulation under oxidative stress conditions.
To address the effect of math-33 on DAF-16 target gene activation in the context of IIS on a global level, we conducted whole nematode RNA sequencing experiments, and identified 2,483 induced and 1,619 repressed genes in daf-2(e1370) mutant animals shifted to 25°C (edgeR FDR<0.05; RNA-seq expression data are available in GEO database). DAF-16 target genes were extensively analyzed in a previous study, which described 1,663 DAF-16 activated and 1,733 DAF-16 repressed target genes (Tepper et al., 2013). Our dataset displayed an overlap of 441 induced and 362 downregulated target genes with the list of published DAF-16 targets from the previous study. Using this set of characterized DAF-16 regulated genes, we found that the daf-2(e1370)-mediated induction and repression of DAF-16 target genes was decreased in daf-2(e1370); math-33(tm3561) mutant animals (Figure 3G, 3I, Table S4 and Table S5). We did not observe a statistical difference in DAF-16 target gene regulation between the wild type and the math-33 loss of function conditions (Figure 3G and 3I). In addition, when we visualized the top 90 DAF-16 induced and downregulated target genes on heat maps, we found that their daf-2 controlled induction or repression was clearly reduced in daf-2(e1370); math-33(tm3561) double mutants (Figure 3H and 3J). Our data suggest that the downregulation of endogenous DAF-16 isoforms in the absence of a functional MATH-33 severely affects the global expression of DAF-16 targets when IIS activity is reduced. Therefore, MATH-33 is essential for DAF-16-mediated target gene activation and repression in the context of IIS.
math-33 affects daf-16-dependent phenotypes for metabolism, stress resistance and aging
In C. elegans IIS is essential for the regulation of early development, stress resistance and metabolic processes, such as fat storage, and governs lifespan (Antebi, 2007; Kenyon, 2005; Wolff and Dillin, 2006). Since math-33 controls DAF-16 protein levels specifically in the context of reduced IIS, we next determined whether this deubiquitylase could affect daf-16 regulated phenotypes in C. elegans.
DAF-16 is an important regulator downstream of IIS for the formation of the dauer stage, a stage of diapause which C. elegans undergoes when growth conditions are unfavorable (Riddle, 1997). daf-2(e1370) mutants undergo daf-16-dependent dauer arrest at 25°C (Gottlieb and Ruvkun, 1994). Here we found that genetic inactivation of math-33 in a daf-2(e1370) background reduced the formation of dauer larvae and promoted development towards reproductive adulthood (Table 1, Table S3). We also determined whether math-33 affects the rate of early development. daf-2(e1370) animals grew slowly at 20°C, the semi-permissive temperature, which is dependent on daf-16 (Ruaud et al., 2011). Inactivation of math-33 in the daf-2(e1370) background reversed the growth delay- and pre-dauer formation phenotype (Figure 4A). These data suggest that math-33 is required for DAF-16-mediated functions during early development such as dauer formation and developmental speed in the context of reduced IIS.
Table 1.
math-33 is required for dauer formation induced by low IIS
| Genotype | % L2 arrest |
% pre- dauers |
% dauers | % L3- adults |
No. animals scored |
|---|---|---|---|---|---|
| daf-16(null); daf-2(ts) | 0 | 0 | 0 | 100 | 209 |
| daf-2(ts) | 0 | 2.3 | 97.7 | 0 | 397 |
| daf-2(ts); math-33(lf) | 60.4 | 11.9 | 0 | 27.7 | 235 |
Dauer and larval arrest phenotypes were scored at 24.5°C (see methods). Animals that resembled a pre-dauer L2 stage (L2D) appear darker due to accumulation of intestinal fat granules. The daf-2(e1370) temperature sensitive allele, the daf-16(mu86) null allele and the math-33(tm3561) loss-of-function allele were used for dauer assays.
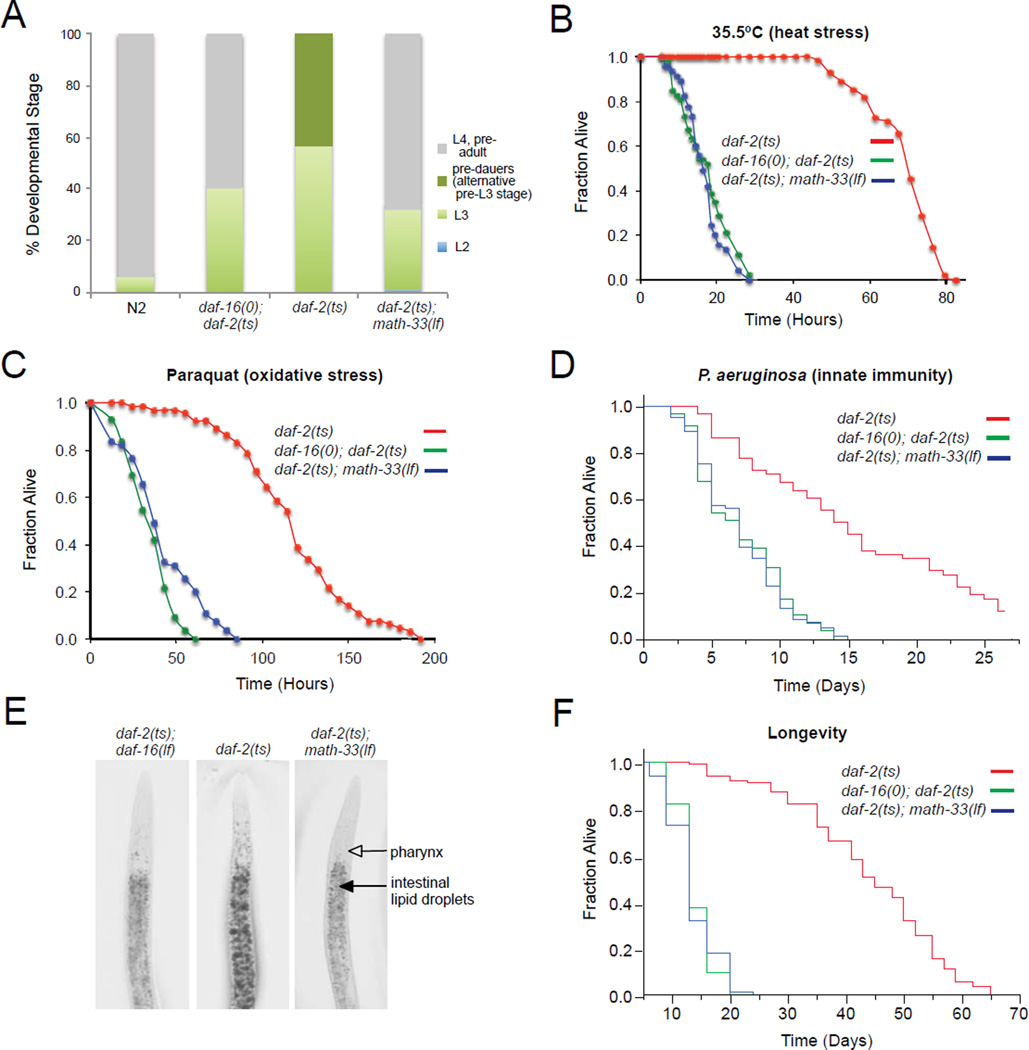
Figure 4. math-33 is required for multiple DAF-16-mediated phenotypes.
(A) Determination of developmental rate of indicated strains grown at 20°C and analyzed 52 hr after egg laying. (n > 450). (B–D) Analysis of math-33 requirement for stress and pathogen resistance of daf-2(e1370) mutants. Indicated strains were exposed to 35.5°C (thermal stress, n > 50) (B), paraquat (oxidative damage, n > 55) (C), and pathogenic challenge to Pseudomonas aeruginosa (innate immunity, n > 60) (D). (E) Detection of fat storage using Oil Red O staining. (F) Lifespan analysis of indicated mutant animals. Lifespan values are given in Supplementary information (Table S1). The daf-2(e1370) temperature sensitive allele, the daf-16(mu86) null allele and the math-33(tm3561) loss-of-function allele were used. Representative data from one of at least two independent experiments are presented.
daf-2 mutant animals are extremely resistant to various stresses including heat and oxidative stress, and pathogen challenge (Evans et al., 2008; Honda and Honda, 1999; Lithgow et al., 1995). Thus, we analyzed the effect of math-33 inactivation on the thermotolerance and oxidative stress resistance of daf-2(e1370) animals. The loss-of-function mutation of math-33 in daf-2(e1370) animals reduced stress resistance similar to that observed for daf-16(mu86); daf-2(e1370) double mutants when animals were exposed to heat stress or paraquat, an oxygen free-radical-producing drug (Figure 4B and 4C). In addition, we found that math-33 inactivation by the math-33(tm3561) loss-of-function allele decreased the lifespan of daf-2(e1370) animals exposed to the pathogenic bacterium P. aeruginosa (Figure 4D). Therefore, physiological evidence supports a requirement for math-33 in daf-16 conferred resistance to thermal stress, oxidative stress and bacterial infection in the context of compromised IIS.
In addition to elevated stress resistance and immune response, daf-2 mutants display increased fat storage (Ashrafi et al., 2003; Kimura et al., 1997). We next investigated whether math-33 could affect daf-16 regulated fat storage in daf-2(e1370) animals by using Oil Red O staining. The math-33(tm3561) loss-of-function allele significantly reduced the increased fat storage in daf-2(e1370) mutants similar to fat levels detected for a daf-16(mu86); daf-2(e1370) double mutant (Figure 4E). Together, our data suggest that math-33 regulates multiple daf-16 controlled phenotypes, which is consistent with our finding that MATH-33 is required for DAF-16 isoform stabilization in the IIS pathway.
Since MATH-33 regulates DAF-16 protein stability under conditions of reduced IIS where lifespan is extended, we tested whether math-33, like daf-16, was required for the lifespan extension observed in daf-2(e1370) mutants. Inactivation of math-33 by the math-33(tm3561) loss-of-function allele completely suppressed the long lifespan seen with daf-2(e1370) mutant animals to that observed for daf-16(mu86); daf-2(e1370) double mutants (Figure 4F and Table S1). The lifespan of math-33(tm3561) single mutant animals was similar to that of daf-16(mu86) null mutants (Table S1). Consistent with these observations, RNAi-mediated knockdown of math-33 also decreased the long daf-2 lifespan (Figures S4A, S4B and Table S2). RNAi knockdown of math-33 in wild type animals from hatching moderately shortened the lifespan, whereas reducing expression of math-33 in daf-16(null) animals did not further shorten lifespan (Table S2). In wild type animals subjected to math-33 RNAi and in math-33 loss-of-function mutants, which have a decreased lifespan, neither IIS nor DAF-16 isoform levels are reduced, indicating that the MATH-33 deubiquitylase might target additional substrates involved in lifespan regulation other than DAF-16.
However, our functional data for math-33 as a regulator of DAF-16 isoform levels combined with our lifespan studies suggest that math-33 is an important regulator for DAF-16-mediated lifespan extension.
The E3 ligase, rle-1, is epistatic to the DUB, math-33
In mammals five different ubiquitin E3 ligases have been identified to act on FOXO transcription factors (Zhao et al., 2011) suggesting that FOXO levels and activity are subject to a complex regulation. In C. elegans the ubiquitin E3 ligase RLE-1 plays an important role in regulating DAF-16 stability (Li et al., 2007). RLE-1 is able to polyubiquitylate DAF-16, targeting it for proteasomal degradation. However, the function of RLE-1 has not been linked to reduced IIS to date. We hypothesized that MATH-33 could counteract the ubiquitylation of DAF-16 by the RLE-1 ubiquitin E3 ligase when IIS is diminished. If so, rle-1 mutations in a daf-2 mutant background would be expected to suppress the math-33 phenotypes, such as reduced DAF-16 target gene expression, diminished dauer development, reduced lipid accumulation and longevity. To explore the capacity of rle-1 mutations to suppress math-33 mutant phenotypes under conditions when IIS was downregulated, we generated a daf-2(e1370); rle-1(cxTi510); math-33(tm3561) triple mutant containing the Psod-3::GFP reporter. In this background, DAF-16 cannot be polyubiquitylated by RLE-1 and simultaneous inactivation of the DUB activity should not affect DAF-16 stability. Indeed, the Psod-3::GFP reporter activity was partially restored in daf-2(e1370); rle-1(cxTi510); math-33(tm3561) triple mutants (Figure 5A). Strikingly, immunoblotting and immunostaining for DAF-16 detected a reappearance of DAF-16 isoform levels and nuclear DAF-16 (Figures 5B, S5A S5B and S5C), correlating with a partial rescue of the Psod-3::GFP reporter activation in the triple mutant (Figure 5B). In addition, we detected a partial restoration of mtl-1, hsp-12.6 and sod-3 target gene transcript levels in the triple mutant compared to daf-2(e1370); math-33(tm3561) double mutant animals at the restrictive temperature (Figures 5C, 5D and S5D). Moreover, the restoration of DAF-16 function in the triple mutant resulted in a partial rescue of the developmental delay phenotype (Figure 5E), dauer arrest (Figure 5E and Table S3), lipid storage (Figure S5E) and lifespan (Figure 5F and Table S1). Compared to shortlived daf-2(e1370); math-33(tm3561) animals, daf-2(e1370); rle-1(cxTi510); math-33(tm3561) triple mutants revealed a 42 to 66% increase in lifespan (Figure 5F and Table S1). Inactivation of rle-1 in a daf-2(e1370) mutant shortened the long lifespan of daf-2 animals indicating that rle-1 might have substrates in addition to DAF-16 required for IIS-mediated lifespan extension. Taken together, our data indicate that the E3 ubiquitin ligase rle-1 exerts its role in DAF-16 regulation epistatically to math-33.
Figure 5. Simultaneous genetic inactivation of both math-33 and rle-1 restores DAF-16 protein levels and partially rescues DAF-16 functions.
(A) Psod-3::GFP reporter activity of nematode larvae raised at 25°C and analyzed at the L2 stage. Fluorescent (i.) and DIC (ii.) micrographs are presented. (B) DAF-16 isoform levels and Psod-3::GFP reporter activation detected by immunoblotting. Membrane was probed with anti-DAF-16 and anti-MATH-33 antibodies followed by reprobing with GFP and alpha tubulin specific antibodies. Stars denote nonspecific bands. (C, D) qRT-PCR of endogenous mtl-1 and hsp-12.6 transcripts. The mean normalized levels and STDEV of three biological repeats are shown. p-Values were calculated using Two-Way ANOVA. ****: p<0.0001, ***: p<0.001, **: p<0.01 *: p<0.05. (E) Determination of developmental rate and pre-dauer formation. Animals were raised at 20°C and analyzed 52 hours after egg laying. (n>160). (F) Lifespan analysis for math-33(tm3561) and rle-1(cxTi510) loss-of-function effects in daf-2(e1370) mutants. Lifespan values are given in Supplementary information, Table S1.
MATH-33 functions as a deubiquitylase and deubiquitylates DAF-16
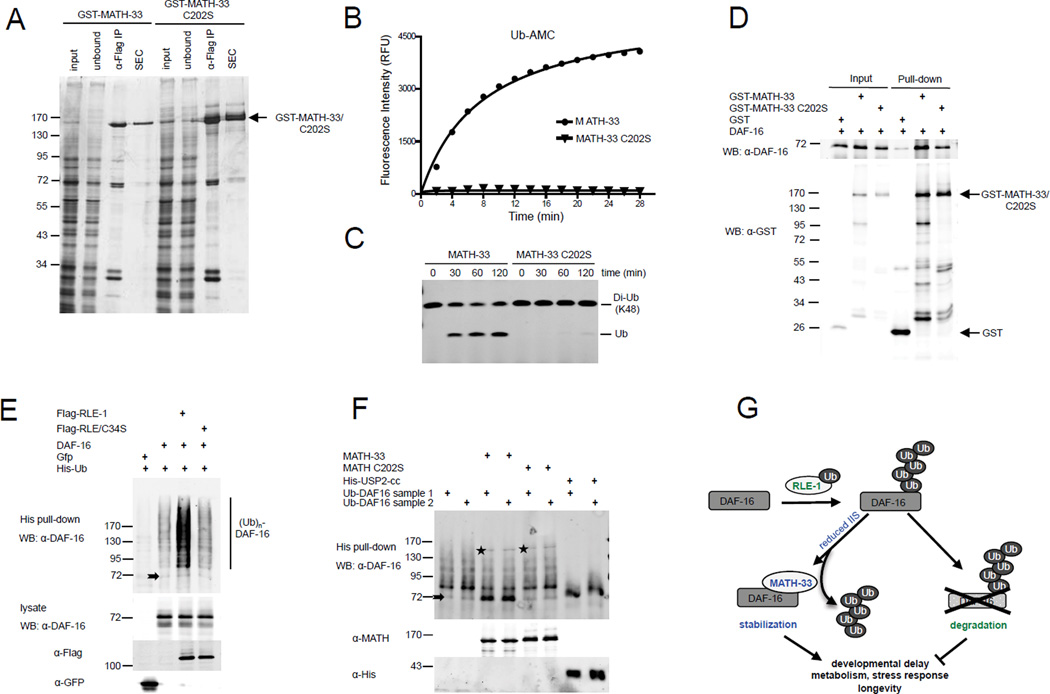
MATH-33 shares 31% overall sequence identity with human USP7. Although the catalytic domain is well conserved, the regulatory regions outside the catalytic domain reveal a sequence identity of only ~23 to 27% (Figure S6), indicating that MATH-33 might not necessarily act as a functional orthologue of USP7. Whether MATH-33 possesses deubiquitylase activity has not been studied yet. To test if MATH-33 has a DUB specific function, we purified insect cell derived, recombinant wild type MATH-33 and a MATH-33 mutant form in which the active site cysteine residue (C202) in the catalytic triad had been mutated to a serine (Figure 6A). We found that recombinant MATH-33 displayed DUB activity depending on its active site cysteine residue in a fluorometric assay, using ubiquitin-7-amido-4-methylcoumarin (Ub-AMC) as a substrate (Figure 6B). In addition, MATH-33 hydrolyzed K48-linked diubiquitin into monoubiquitin, whereas MATH-33(C202S) displayed no activity in this assay (Figure 6C). Thus MATH-33 functions as a deubiquitylase in vitro and its activity is dependent on the C202 catalytic active site.
Figure 6. MATH-33 functions as a deubiquitylase and actively removes ubiquitin moieties from DAF-16.
(A) SDS-PAGE/silver staining analysis of isolated, recombinant MATH-33 from insect cells following epitope-based purification and size-exclusion chromatography (SEC). (B, C) MATH-33 deubiquitylase activity detected in Ub-AMC and di-ubiquitin based assays. (D) GST-pull down in which GST-MATH-33 or GST alone bound to glutathione beads was incubated with recombinant DAF-16 isolated from bacteria. (E) In-vivo ubiquitylation of DAF-16 by RLE-1 in HEK293T cells. Denatured lysates were subjected to His pull-down assays and analyzed by immunoblot with an anti-DAF-16 antibody. (F) In-vitro deubiquitylation assay using insect cell-derived MATH-33 and bacterially expressed USP2-cc incubated with poly-ubiquitylated DAF-16 purified from HEK293T cells. Stars denote bands generated by cross reactivity of the DAF-16 antibody with MATH-33. Arrows denote non-ubiquitylated DAF-16 (E, F). (G) Model for MATH-33-mediated stabilization of DAF-16 regulating various DAF-16 controlled functions.
Our genetic analysis of daf-16, math-33 and rle-1 suggests that MATH-33 may target DAF-16 for deubiquitylation following RLE-1-mediated ubiquitylation. To establish a MATH-33 in vitro deubiquitylation assay for DAF-16 we first investigated whether MATH-33 was able to physically interact with DAF-16 in vitro. We performed pull-down studies using bacterially expressed DAF-16 and recombinant MATH-33 purified from insect cells. We found that GST-MATH-33 and its catalytically inactive derivative could pull down DAF-16 (Figure 6D). These results suggest that both MATH-33 and its catalytically inactive variant are able to form a complex with DAF-16.
To determine if the association of MATH-33 with DAF-16 results in deubiquitylation of DAF-16 we performed in vitro deubiquitylation assays. First, we generated a pool of ubiquitylated DAF-16 by transfecting HEK293T cells with His-tagged ubiquitin, DAF-16 and Flag-tagged RLE-1. Immunoblot analysis of His-tagged ubiquitylated DAF-16 isolated from denatured extracts revealed a RLE-1 dependent increase in a DAF-16-specific ladder of bands, which represents ubiquitylated DAF-16 (Figure 6E). The increase in DAF-16 polyubiquitylation was mediated by RLE-1, but not by its catalytically inactive mutant. To determine if MATH-33 directly deubiquitylates polyubiquitylated DAF-16, we performed in vitro deubiquitylation assays. Isolated and renatured ubiquitylated DAF-16 from HEK293T cells was incubated with purified MATH-33. MATH-33, but not its catalytically inactive derivative (C202S), caused an increase of free DAF-16 and a decrease of polyubiquitylated DAF-16 (Figure 6F). USP2cc, the highly active catalytic core of USP2, which cleaves all ubiquitin fusions, was used as a positive control. Taken together, our results indicate that MATH-33 directly deubiquitylates polyubiquitylated DAF-16 dependent on the catalytic activity of MATH-33.
DISCUSSION
Collectively, our data suggest a model in which MATH-33 is an essential regulator of DAF-16 by interacting physically with DAF-16 to regulate DAF-16 protein stability in C. elegans (Figure 6G). We propose that MATH-33 stabilizes DAF-16 by acting as a DUB to antagonize RLE-1-mediated polyubiquitylation and proteasomal degradation of DAF-16. Our genetic data indicate that math-33 is required for governing early developmental decisions, metabolism and longevity by controlling DAF-16 protein levels when IIS is diminished.
MATH-33 is the first described DUB regulating protein stability of a FOXO family member in any species. MATH-33 is related to the mammalian USP7/HAUSP, which has been previously found to regulate FOXO transcriptional activity (van der Horst et al., 2006), but not stability, by modulating monoubiquitylation. When cells are exposed to oxidative stress conditions FOXO4 is di-monoubiquitylated, resulting in increased nuclear localization and transcriptional activity of FOXO4. USP7 negatively regulates FOXO by antagonizing FOXO4 monoubiquitylation and activity without affecting protein stability. USP7 has also been described as a negative regulator of FOXO1 in hepatic gluconeogenesis in a recent study (Hall et al., 2014). Here we demonstrate that the C. elegans orthologue of USP7, MATH-33, functions as a positive, not negative, regulator to control DAF-16 protein levels when IIS is downregulated. Our genetic data indicate that math-33 is essential to maintain DAF-16 protein levels and genetically interacts with the E3 ubiquitin ligase RLE-1 in the context of IIS. We have found that MATH-33 association with DAF-16 is enhanced under diminished IIS conditions. Moreover, MATH-33 could antagonize DAF-16 polyubiquitylation in C. elegans and is able to remove ubiquitin moieties from polyubiquitylated DAF-16 in vitro. However, our genetic data indicate that MATH-33 might play a distinct role in the regulation of DAF-16 under oxidative stress, since inactivation of math-33 in wild type animals did not reduce DAF-16 isoform levels when nematodes were exposed to paraquat. We hypothesize that diverse upstream signaling networks, including stress and growth factor signaling, and differences in signaling intensities could impact how the DUB activity of MATH-33 affects DAF-16/FOXO. Such differences could influence the degree of FOXO/DAF-16 nuclear localization and activity as well as its interaction with the cellular ubiquitylation machinery. USP7/MATH-33 may act as a dual specificity DUB. In this case the DUB regulates FOXO/DAF-16 activity by reversing monoubiquitylation under one circumstance, but controls FOXO/DAF-16 stability by antagonizing its polyubiquitylation under other circumstances. Alternatively, C. elegans contains the single FOXO factor DAF-16, which is most homologous to FOXO3, while humans contain four FOXO members, FOXO1, 3, 4 and 6. Perhaps, USP7 specifically removes monoubiquitin from FOXO 1 and 4, leaving regulation of FOXO3 by polyubiquitylation. In addition, MATH-33 could have evolved a species-specific function for the regulation of DAF-16 polyubiquitylation and stability. The regulatory domains outside the catalytic domain are poorly conserved in C. elegans MATH-33 when compared to human USP7, indicating potential differences in the regulation of deubiquitylase function. More detailed studies are needed to determine potential species-specific differences of MATH-33 and USP7 deubiquitylases towards regulation of mono versus polyubiquitylation of DAF-16/FOXO transcription factors.
Our molecular data indicate that MATH-33 regulates DAF-16 on the protein level by antagonizing polyubiquitylation and degradation of DAF-16. By analogy, the transcript levels of two major DAF-16 isoforms, DAF-16a and DAF-16b, were not reduced in daf-2(e1370); math-33(tm3561) mutant animals compared to daf-2(e1370) single mutants. Interestingly, we could detect a reduction of DAF-16d/f isoforms in daf-2(e1370); math-33(tm3561) animals, and we hypothesize that this reduction is due to a DAF-16 transcriptional autoregulatory loop, since expression of DAF-16d/f isoforms is regulated by a different upstream promoter region than expression of DAF-16a and DAF-16b isoforms (Kwon et al., 2010). It has been previously reported in mammalian systems that FOXO transcription factors are able to regulate their own expression via a positive transcriptional autoregulatory feedback loop (Essaghir et al., 2009; Lutzner et al., 2012). ChIP experiments have identified a canonical DAF-16 Binding Element (DBE) located in close proximity to the DAF-16d/f transcriptional start site (modENCODE project) suggesting that a DAF-16-mediated transcriptional autoregulation occurs in C. elegans as well.
In C. elegans RLE-1 has been identified as an E3 ubiquitin ligase for DAF-16 catalyzing DAF-16 ubiquitylation and its degradation by the proteasome (Li et al., 2007). Genetic inactivation of RLE-1 in nematodes increased DAF-16 protein levels and extended the lifespan of animals. However, the role of rle-1 in DAF-16 regulation has not been studied under reduced IIS conditions. We uncovered an epistatic relationship of the ubiquitin E3 ligase RLE-1 and the deubiquitylase MATH-33 when IIS is downregulated. Strikingly, genetic inactivation of both rle-1 and math-33 under reduced IIS conditions was able to partially rescue DAF-16 functions, including its role in dauer formation, lipid storage and lifespan extension that were lost in daf-2(e1370); math-33(tm3561) double mutants. We postulate that additional ubiquitin E3 ligases regulate DAF-16, as simultaneous inactivation of rle-1 and math-33 only partially rescued DAF-16 functions. Further studies are needed to determine the role of additional E3 ubiquitin ligases in the regulation of DAF-16 by the ubiquitin proteasome system.
Overall our data suggest that MATH-33 is an essential deubiquitylase that promotes DAF-16 stability and function in regulating processes such as stress response, metabolism and longevity in the context of IIS. In the future, it will be imperative to elucidate whether USP7/HAUSP acts as a regulator for FOXO stability downstream of IIS in mammals. Since IIS is evolutionally conserved from C. elegans to humans, knowledge gained through these studies will have important implications for our understanding of the aging process in higher eukaryotes as well as for age-related diseases such as diabetes and cancer.
EXPERIMENTAL PROCEDURES
C. elegans strains and generation of transgenic lines
All strains were maintained at 15°C using standard C. elegans methods (Brenner, 1974) and fed on Escherichia coli OP50. Extrachromosomal array carrying transgenic strains were generated using standard microinjection methods (Mello et al., 1991).
Lifespan analysis
Lifespan analyses were performed as described (Dillin et al., 2002). Unless stated otherwise animals were grown at 15°C until the L4 stage, and then shifted to 20°C. Production of progeny and internal egg hatching of math-33(tm3561) animals was prevented by post-developmental exposure to 0.1 mg ml−1 FUdR.
Dauer formation and developmental delay assays
For dauer assays C. elegans strains were left to lay eggs at 20°C for 4 hr. Plates were transferred to 24.5°C for 3 days. Dauer formation was determined based upon morphology using a dissecting microscope and a Leica 6000B digital microscope. For developmental delay assays C. elegans strains were raised at 20°C for at least 2 generations and were left to lay eggs at 20°C for 2 hr. Developmental stages were determined based upon morphology at 52 hr grown at 20°C after egg laying.
RNA isolation and quantitative RT–PCR
Total RNA was isolated from semi-synchronized populations of approximately 3,000 worms. Worms were grown on 15°C and shifted to 25°C at the L4 stage for 24 hr. RNA isolation and quantitative RT–PCR was performed as described previously (Vilchez et al., 2012). SybrGreen real-time qPCR experiments were performed with a 1:20 dilution of cDNA using an ABI Prism 79000HT (Applied Biosystems) following the manufacturer’s instructions. Data were analyzed with the Standard Curve method using the geometric mean of cdc-42, pmp-3 and Y45F10D.4 as endogenous control (Hoogewijs et al., 2008).
RNA sequencing
RNA isolation, generation of stranded mRNA-seq libraries and sequencing using an Illumina HiSeq 2500 system was performed as described previously (Sun et al., 2014). Sequence reads were mapped to an annotated C. elegans genome (WS220/UCSC ce10) by using STAR 2.4.0j aligner (Dobin et al., 2013), after removing the low quality reads and the 3'-adaptor sequences. The gene expression levels, represented by fragments per kilo base per million reads (FPKM), were calculated by using HOMER v4.7 (homer.salk.edu/). The statistical significance of gene regulation was performed in edgeR, and the genes with FDR<0.05 were considered statistically significant. The heatmaps of FC (fold changes) were illustrated in MeV (Multi Experiment Viewer). A FPKM cut-off of 0.1 was applied to all the samples, and an additional FPKM of 1 was applied to samples originating from daf-2(ts1370) animals.
Tandem Affinity Purification for isolation of DAF-16 binding partners
Semi-synchronized nematode populations were shifted to 20°C at the late L4/ day 1 of adulthood stage for 2 to 6 hr and harvested. Nematodes were homogenized in a mortar with liquid nitrogen and resuspended in ice-cold lysis buffer [50 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Triton-X-100, 0.1 mM EDTA, 0.5 mM EGTA, 1mM PMSF supplemented with protease inhibitor tablets (Sigma) and phosphatase inhibitors (Calbiochem)]. FLAG and His based Tandem Affinity Purification was performed as described previously (Yang et al., 2006).
In-vivo ubiquitylation assay
HEK293T cells were transfected with expression plasmids for His6-Ub, Flag-tagged RLE-1 and DAF-16 using polyethylenimine (PEI) (Sigma) and collected 48 hr later. 20 µM MG-132 was added to cells 2.5 hr before lysis in 6 M GuHCl, 5 mM imidazole, 0.2% NP40, 50 mM HEPES (pH 7.6) and 10% glycerol. Ubiquitylated proteins were isolated using the TALON Metal Affinity Resin (Clontech).
In-vitro deubiquitylation assay
Ubiquitylated DAF-16 was isolated from HEK293T cells under denaturing conditions as described above and stepwise renatured by dialysis as described previously for p53 (van der Knaap et al., 2005). MATH-33 and its catalytic inactive mutant were co-expressed with Drosophila GMP Synthetase (GMPS) in Hi5 insect cells and purified as described in Supplemental Experimental Procedures. Drosophila GMPS has been previously found to stimulate the deubiquitylation activity of Drosophila USP7 (van der Knaap et al., 2005). 100 nM recombinant MATH-33 and bacterially expressed USP2-cc were used for deubiquitylation assays. Deubiquitylation reactions were assembled on ice in reaction buffer containing 50 mM HEPES (pH 7.6), 150 mM NaCl, 4 mM DTT, 20 mM EDTA and 5% glycerol, incubated 1 hr at 37°C and stopped by adding SDS-PAGE loading buffer.
Supplementary Material
Highlights.
DAF-16/FoxO polyubiquitylation and stability regulate metabolism and longevity
The DUB MATH-33 stabilizes DAF-16 protein levels in Insulin/IGF-1 signaling (IIS)
math-33 controls daf-16 mediated functions, such as metabolism and longevity in IIS
The E3-ubiquitin ligase rle-1 counteracts math-33 functions on DAF-16
Acknowledgements
T.H. is funded by a postdoctoral fellowship to the Salk Institute Glenn Center for Aging Research, the Austrian Science fund (FWF, grant J 2734) and by the National Institutes of Health (NIH R01DK070696, R01AG027463, R01ES021667, R01CA080100). A.D. is supported by HHMI. T. Hunter is supported by NIH R01CA080100, CA082683 and CA014195 from the National Cancer Institute. T. Hunter is a Frank and Else Schilling American Cancer Society Professor and holds the Renato Dulbecco Chair in Cancer Research. This work used the Functional Genomics Core at the Salk Institute, supported by the NCI Cancer Center Support Grant CA014195 and the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. We thank the Caenorhabditis Genetics Center, the National Bioresource Project for the Nematode and Malene Hansen for providing C. elegans strains. We are grateful to Heidi Tissenbaum and Sylvia Lee for providing anti-DAF-16 antibodies, Thomas Czerny for the pKC-3xFLAG expression vector and Sreekanth Chalasani for providing equipment. We also thank Suzanne Wolff, Malene Hansen and members of the A.D. and the T. Hunter laboratories for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions. T.H., T. Hunter and A.D. designed the experiments. T.H., Z.L., C.B., A.C.C., C.G.R., R.M., B.R.F., B.T. and C.E.R. performed the experiments. C.K., B.F.L., K.K., J.R.Y. and C.O. contributed reagents and analysis tools. The manuscript was written by T.H. and edited by A.C.C., A.D., and T. Hunter.
REFERENCES
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS genetics. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. The Journal of biological chemistry. 2009;284:10334–10342. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA, Chen WC, Tan MW. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging cell. 2008;7:879–893. doi: 10.1111/j.1474-9726.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, Sixma TK. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Tabata M, Rodgers JT, Puigserver P. USP7 attenuates hepatic gluconeogenesis through modulation of FoxO1 gene promoter occupancy. Mol Endocrinol. 2014;28:912–924. doi: 10.1210/me.2013-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim Biophys Acta. 2011;1813:1961–1964. doi: 10.1016/j.bbamcr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptorlike gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Developmental cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzner N, Kalbacher H, Krones-Herzig A, Rosl F. FOXO3 is a glucocorticoid receptor target and regulates LKB1 and its own expression based on cellular AMP levels via a positive autoregulatory loop. PloS one. 2012;7:e42166. doi: 10.1371/journal.pone.0042166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey RJ, Kemphues KJ. Deubiquitylation Machinery Is Required for Embryonic Polarity in Caenorhabditis elegans. PLoS genetics. 2012;8:e1003092. doi: 10.1371/journal.pgen.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Riddle D, Blumenthal T, Meyer B, Priess J. C. Elegans II. Cold Spring Harbor: Cold Spring Harbor Press; 1997. [Google Scholar]
- Ruaud AF, Katic I, Bessereau JL. Insulin/Insulin-like growth factor signaling controls non-Dauer developmental speed in the nematode Caenorhabditis elegans. Genetics. 2011;187:337–343. doi: 10.1534/genetics.110.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Liu Y, Hunter T. Multiple Arkadia/RNF111 structures coordinate its Polycomb body association and transcriptional control. Molecular and cellular biology. 2014;34:2981–2995. doi: 10.1128/MCB.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wheeler HE, Kim SK. Genetics and genomics of human ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:43–50. doi: 10.1098/rstb.2010.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Yang P, Sampson HM, Krause HM. A modified tandem affinity purification strategy identifies cofactors of the Drosophila nuclear receptor dHNF4. Proteomics. 2006;6:927–935. doi: 10.1002/pmic.200500230. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011;3:276–282. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.