Abstract
Autophagy is a complex cellular process having multiple roles, depending on tissue, physiological, or pathological conditions. Major post-translational regulators of autophagy are well known, however, they have not yet been collected comprehensively. The precise and context-dependent regulation of autophagy necessitates additional regulators, including transcriptional and post-transcriptional components that are listed in various datasets. Prompted by the lack of systems-level autophagy-related information, we manually collected the literature and integrated external resources to gain a high coverage autophagy database. We developed an online resource, Autophagy Regulatory Network (ARN; http://autophagy-regulation.org), to provide an integrated and systems-level database for autophagy research. ARN contains manually curated, imported, and predicted interactions of autophagy components (1,485 proteins with 4,013 interactions) in humans. We listed 413 transcription factors and 386 miRNAs that could regulate autophagy components or their protein regulators. We also connected the above-mentioned autophagy components and regulators with signaling pathways from the SignaLink 2 resource. The user-friendly website of ARN allows researchers without computational background to search, browse, and download the database. The database can be downloaded in SQL, CSV, BioPAX, SBML, PSI-MI, and in a Cytoscape CYS file formats. ARN has the potential to facilitate the experimental validation of novel autophagy components and regulators. In addition, ARN helps the investigation of transcription factors, miRNAs and signaling pathways implicated in the control of the autophagic pathway. The list of such known and predicted regulators could be important in pharmacological attempts against cancer and neurodegenerative diseases.
Keywords: autophagy, estrogen receptors, miRNA, network, protein-protein interactions, regulation, resource, signaling pathway, transcription factors
Abbreviations: miRNA, microRNA; NHR, nuclear hormone receptor; PPI, protein-protein interaction; TFs, transcription factors
Introduction
Since the discovery of autophagy in the 1960s, and the discovery of autophagy-related genes in yeast in the 1990s, our knowledge of the regulation of autophagy expanded significantly. Major post-translational regulators of the autophagic machinery are well known, compared to the transcriptional and post-transcriptional regulators, where only limited information is available currently.1,2 Autophagy is essential in homeostasis and stress-response as well as in macromolecular turnover and development.3 Both its insufficient and overdriven functions can hinder cell survival.4 Thus, the regulation of autophagy is critical, with high medical importance. The autophagic machinery, consisting of a complex interplay between more than 30 initiator and executor proteins, must be under constraints of precise, context-dependent and systems-level regulatory mechanisms at post-translational, transcriptional, and post-transcriptional levels.
The proteins involved in the process of autophagy are organized into interacting complexes, having different functions in the autophagic process (e.g., initiation, membrane sequestration, and in targeting the materials to degrade in the forming phagophore). Most of the interactions within the core machinery of autophagy are well known, however, there are some unanswered questions that are needed to be resolved in order to better understand the mechanism.5 Interestingly, direct connections between the initiation and execution complexes were only found in the past year: it has been proved that ULK1/2, the major initiator could activate autophagy by phosphorylating another key autophagic protein, BECN1/Beclin 1.6 A similar important finding was obtained from yeast, where Atg1 (yeast ortholog of ULK1/2) phosphorylates Atg9 and Atg2, enhancing the membrane trafficking to the phagophore assembly site.7 These recent and key findings indicate that post-translational regulation of autophagy could still provide unexpected and undiscovered connection with high evolutionary or biomedical relevance. To facilitate such discoveries in silico, structure-based predictions could guide experimental researchers to validate and identify such connections.
There is no doubt that post-translational regulation of autophagy is only one part of the story. Autophagic activity also depends on the expression of autophagy-related genes and is regulated by certain transcription factors (TFs) and microRNAs (miRNAs).1,2 These regulatory influences can be realized on different time scales, be driven by external signals, and constitute feedback loops. Considering the transcriptional regulation of autophagy, some elements have already been highlighted in the literature, such as the transcription factors TFEB, FOXO, and SREBFs/SREBPs.8-10 By modulating autophagy, these TFs take part in the cellular response to starvation, stress, or lipid depletion, and are also involved in the pathomechanism of several diseases.1,11 TFEB is activated upon starvation, and facilitates the transcription of many autophagy and lysosome related genes and maintains the regeneration of lysosomes.1 FOXO1 and FOXO3 act as effectors of the insulin signaling pathway, to regulate autophagic activity.1,11 Analogously, SREBF2/SREBP2 activates autophagy in case of sterol depletion.10 Beyond the role of the few TFs extensively examined and highlighted in the literature, further transcriptional regulatory components are expected to regulate autophagy in certain context. Given the advances of novel high-throughput techniques in protein-DNA interaction discovery, such as ChIP-Seq, PBA, and SELEX,12–14 numerous candidate TFs have been discovered. In addition, with resources containing TF binding site information, like JASPAR,15 potential target genes for a given TF can be predicted on a genome-wide scale. One may think that the current limitation in the search for autophagy regulators is the available data and computational expertise to evaluate and analyze data sets.
Several miRNAs downregulate mRNAs of autophagy-related genes by specific binding. However, little is known about their systems-level role. A recent review listed more than 16 miRNAs regulating autophagy genes post-transcriptionally.2 These miRNAs are able to block specific steps of autophagy (e.g.,, MIR376B acts on ATG4 and BECN1, while MIR630 acts on ATG12 and UVRAG). Remarkably, most of these miRNAs affect the early stage of autophagic vacuole formation, possibly because this way miRNAs could prevent the accumulation of autophagosomes.2 The growing number of experimental data on miRNA-driven regulation necessitates repositories for the post-transcriptional regulation of autophagy. Such resources could facilitate our understanding on the context-dependent role of these regulators.
The importance of identifying such context-dependent regulators is also supported by the fact that autophagy is a promising therapeutic target in several pathologies, especially in cancer and neurodegenerative diseases.16 Because autophagy has an ambiguous role in cancer, described by the ‘double-edged sword’ metaphor, therapies targeting the process need to be specific and context-dependent.17 Considering the complexity of autophagy and its regulation, searching for therapeutic targets without a systems-level analysis is like looking for needle in a haystack. The first step on the way to investigate the regulation of autophagy as a system is to collect all the available knowledge, including all levels of regulation. Currently elements of this knowledge are scattered in huge number of articles and bioinformatics resources, like databases of protein-protein interactions, transcriptional regulation, or post-transcriptional regulation.18 An integrated and precisely compiled interaction network could allow mapping feedback loops at all levels of regulation; to investigate differences by tissue, physiological or pathological state, drug effect, or gender; to build models using different mathematical formalisms, and thus simulate different conditions, and verify the models experimentally.
Until now few systems-level resources about autophagy have been published. The Human Autophagy Database19 (HADb) is a collection of 234 autophagy-related genes, containing references to major genome and protein databases.19 It does not intend to provide an interaction network, so it completely lacks interaction data. Another database named Autophagy Database20 (ADB) contains orthologs from 40 species, and gives a comparative list of them, including a total of 206 proteins in human. For some proteins, it also collects a list of interactions—641 interactions in human—but the sources of those data and the scope of the collection is not clearly defined. A large-scale LC-MS (liquid chromatography and mass spectrometry) study provided a network of 751 interactions between 409 autophagy-related proteins.21 The advantage of this dataset is the uniform methodology and the relatively wide range of proteins involved in the study. However, this resource contains only the interactions detectable by the LC-MS method, and omits other interactions described in the literature. The 2 mentioned autophagy-focused resources lack data on transcriptional and post-transcriptional regulation.
Prompted by the lack of a proper bioinformatics database that extensively collects available data from the literature, from protein-protein interaction databases, and prediction methods, and contains data on several levels of regulation, we developed Autophagy Regulatory Network (ARN; http://autophagy-regulation.org), a novel resource to help both in silico and wet lab researchers in their investigation of the human autophagic process.
Results
The Autophagy Regulatory Network (ARN) database
The ARN database (http://autophagy-regulation.org) contains proteins involved in the mechanisms of autophagy, their regulators, and their TF and miRNA regulators as well as connections between all these components and signaling pathways (Fig. 1). Six main layers build up the structure of ARN: (1) autophagy proteins, (2) their direct regulators from autophagy specific resources, (3) post-translational regulators that directly regulate proteins in the first 2 layers, (4) transcriptional regulators of the first 3 layers, (5) post-transcriptional regulators of the first 4 layers, (6) signaling pathways and protein-protein interactions connecting pathways to autophagy regulators. ARN contains interactions from manual curation, 19 external databases, and 4 prediction methods (listed in Table 1). For basic statistics, please see Figure 2. Users are able to filter interactions by sources, and use resources in a comparative way, according to their requirements. Interactions may have confidence scores, users can filter by the data set, setting preferable level of confidence, using the customizable download module.
Table 2.
Basic statistics of the Autophagy Regulatory Network
| Data sources and layers | Identical nodes | Identical edges |
|---|---|---|
| Core autophagy proteins | 38 | 238 |
| ADB20 | 31 | 114 |
| ARN manual curation | 20 | 26 |
| Behrends et al.21 | 26 | 38 |
| BioGRID44 | 30 | 76 |
| ELM-based prediction38 | 16 | 64 |
| HPRD47 | 11 | 11 |
| InnateDB45 | 28 | 40 |
| IntAct46 | 34 | 136 |
| Post-translational regulators | 13,803 | 197,167 |
| ADB | 206 | 381 |
| ARN manual curation | 47 | 46 |
| Behrends et al. | 398 | 441 |
| BioGRID | 12,051 | 71,496 |
| Domain-domain based prediction | 166 | 1,138 |
| ELM-based prediction | 937 | 78,824 |
| HPRD | 7,290 | 29,617 |
| InnateDB | 2,816 | 6,125 |
| IntAct | 9,862 | 45,745 |
| Transcriptional regulators | 13,340 | 170,245 |
| ABS40 | 23 | 14 |
| ARN manual curation | 31 | 35 |
| ENCODE41 | 2,209 | 9,217 |
| HTRI29 | 12,209 | 39,477 |
| JASPAR15 | 12,813 | 119,873 |
| ORegAnno42 | 908 | 932 |
| PAZAR43 | 1,940 | 3,018 |
| Post-transcriptional regulators | 7,633 | 20,186 |
| miR2Disease48 | 171 | 124 |
| miRDeathDB49 | 126 | 108 |
| miRecords50 | 664 | 760 |
| miRTarBase51 | 7,203 | 19,177 |
| TarBase52 | 1,798 | 2,584 |
| Transcriptional regulation of miRNAs | 646 | 6,911 |
| ENCODE | 195 | 590 |
| PuTmiR 1.153 | 413 | 3,034 |
| PuTmiR 2.0 | 288 | 3,095 |
| TransmiR54 | 291 | 542 |
| Signaling pathways and interactions | 1,199 | 3,287 |
| SignaLink 2 manual curation55 | 1,199 | 3,287 |
| BioGRID | 535 | 697 |
| HPRD | 481 | 611 |
| InnateDB | 9 | 51 |
| IntAct | 12 | 69 |
Data sources of each layer are listed with the corresponding number of nodes (i.e., proteins or miRNAs) and edges (i.e., protein-protein interactions, TF-gene, miRNA-mRNA, or TF-miRNA regulatory connections). The number of identical nodes shows both connecting component pairs (i.e., TFs and target genes as well). For each major layer we highlighted the total number of nodes and edges in ARN that is generally less than the sum of the components due to the overlap among the resources. Note that the highlighted numbers in each layer are higher than those in any of the sources.
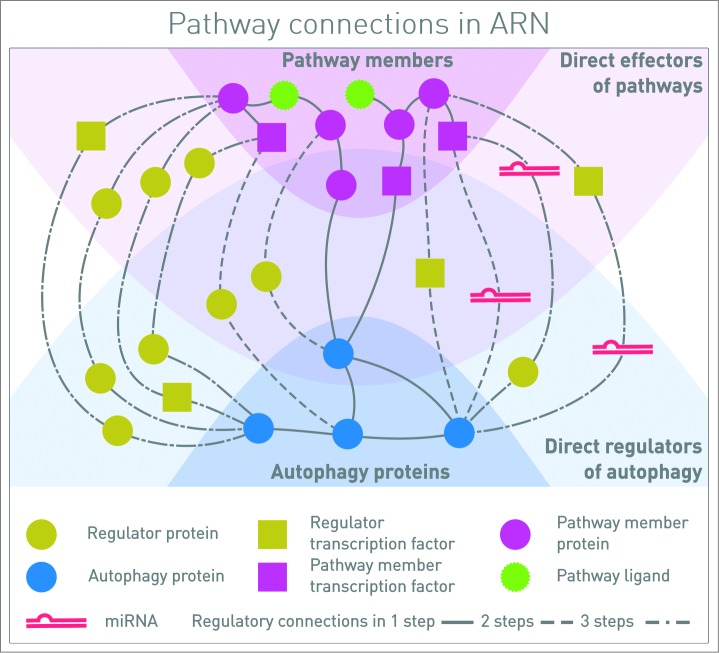
Figure 1.
Connections between autophagy components and signaling proteins in one, 2 or 3 steps. One-step connections are direct protein-protein interactions (PPIs), or a pathway member TF regulates the transcription of an autophagy protein. Two-step connections also can include PPIs and TF-gene interactions, but TF-miRNA-mRNA interactions as well. Three-step interactions are combinations of all these types of interactions, involving 4 molecular species. In this representation, signal is coming from the signaling pathway receptors binding ligands, toward the proteins executing autophagy. By analyzing the whole network, feedback circuits and network motifs can be identified along the paths.
Table 1.
The data sources of the Autophagy Regulatory Network
| Type of interactions | Data sources |
|---|---|
| Protein-protein interactions | ADB20 |
| ARN manual curation | |
| Behrends et al.21 | |
| BioGRID44 | |
| HPRD47 | |
| InnateDB45 | |
| IntAct46 | |
| ELM based prediction38 | |
| Prediction based on domain-domain interaction | |
| Signalink 2.0 manual curation55 | |
| Transcriptional regulation | ABS40 |
| ARN manual curation | |
| ENCODE distal41 | |
| ENCODE proximal filtered41 | |
| HTRI29 | |
| JASPAR15 | |
| ORegAnno42 | |
| PAZAR43 | |
| miRNA-mRNA interactions | miR2Disease48 |
| miRDeathDB49 | |
| miRecords50 | |
| miRTarBase51 | |
| TarBase52 | |
| Transcriptional regulation of miRNAs | ENCODE41 |
| PuTmiR 1.153 | |
| PuTmiR 2.053 | |
| TransmiR 1.254 |
ARN contains data from manual curation and from 23 external resources. From SignaLink we included 3,287 manually curated interactions; we used 4 prediction methods in ARN (domain-domain based prediction (using data from Pfam,39 DOMINE60 and Negatome,61) domain-motif based prediction using the structure filter for ELM,38 TF-promoter binding prediction using the JASPAR15 algorithm, and TF-miRNA gene regulation from PuTmiR53); the remaining 18 databases, including all the miRNA-mRNA data sets, contains data mainly from high-throughput experiments.
Figure 2.
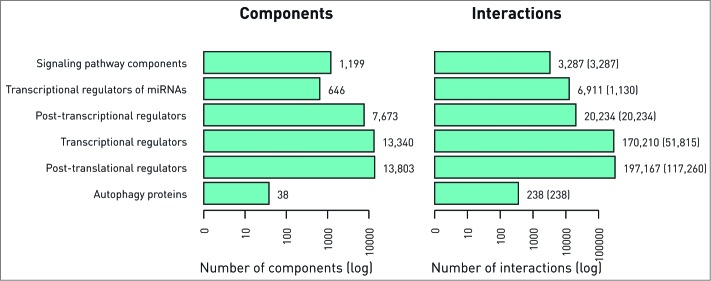
Basic statistics of ARN. Number of components (A) and interactions (B) in different layers of ARN is shown. The numbers of experimentally verified interactions are indicated in parenthesis next to the total number of interactions, which also includes predicted ones.
The ARN website
ARN's website is available at http://autophagy-regulation.org. The website is designed to give a comfortable way to browse interactions, providing hyperlinks to original sources and PubMed references of each interaction. The download section of the website gives an opportunity to customize the data to download: select between layers, and filter interactions by source, or by confidence score.
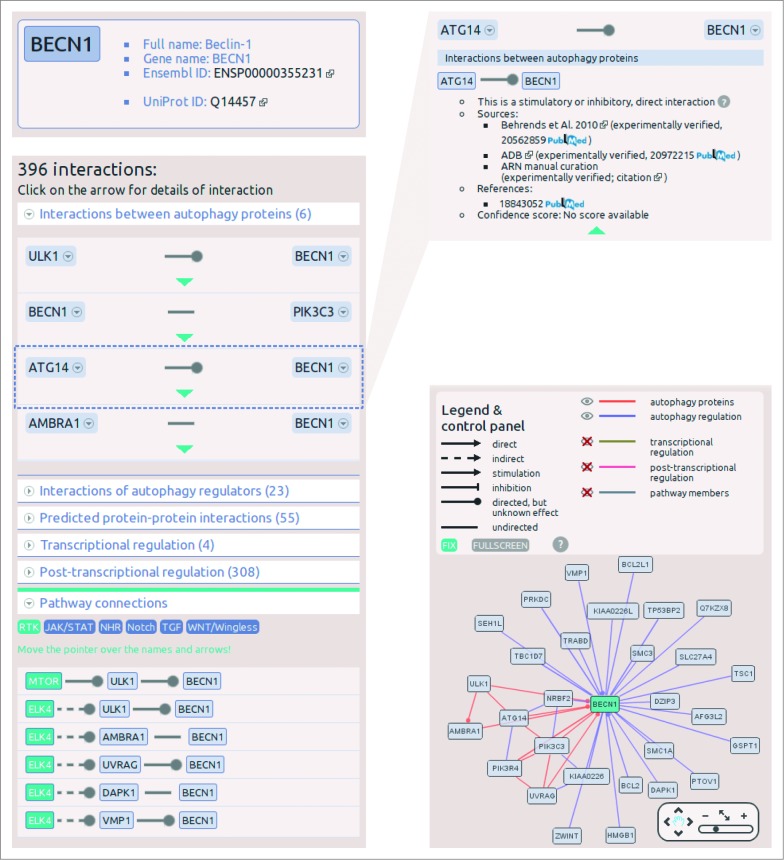
The search field on the main page autocompletes the search term, and understands several different database IDs and accession numbers. If the search is successful, the page navigates to the datasheet of the selected protein. The protein datasheet shown in Figure 3 illustrating the interactions of a key autophagy protein, BECN1, contains 4 main sections. At the top of the page, in a box the full name, gene name, UniProt ID and Ensembl ID of the protein are available. Below the names, a list of related diseases and cancer types can be found. On the left side, a list of interactions enumerates all the first neighbors of the protein, grouped by layers. The lists of the layers are expandable, and within these lists, detailed information (e.g., sources, references, confidence scores) can be obtained about an individual interaction. Below the list of the interactions, the connections between signaling pathways and the autophagy system are listed. We defined this pathway connection either in one or 2 steps, where one of the proteins is a member of a given pathway, and the other one is a present in the ARN database. On the right side of the protein datasheet, an interactive view of the first neighbors’ network is presented. In this view, interactors can be filtered by layers of ARN, and users are able to get more information on proteins and interactions by clicking on them (Fig. 3).
Figure 3.
Screenshots from the protein datasheet of BECN1 from the ARN webpage. (A) At the top of the datasheet the name, gene name, UniProt ID, and Ensembl protein ID of the selected protein is shown, with hyperlinks to the UniProt and Ensembl webpages. Below this box, the potential signaling properties and disease related information with a special highlight on cancer types is listed. (B) The interactions of the selected protein are listed, grouped by layers. In addition, at the bottom of the list, the pathway connections can be browsed by pathways. (C) Information on sources, references and confidence scores of each interaction can be obtained by clicking on the green triangles. (D) On the right side of the datasheet, an interactive network image of the first neighbors of the selected proteins is available. Note that unlike ULK1, ULK2 is not present in the BECN1 network as ARN contains only those interactions that were specifically identified between exact proteins, and no publications were curated that experimentally verified the likely connection between ULK2 and BECN1.
Comparison with other resources
Compared with general protein-protein interaction (PPI) databases, BioGRID44 contains 76 interactions between 30 autophagy proteins, while in IntAct46 136 interactions between 34 autophagy proteins can be found. ADB20 contains 114 interactions between 31 autophagy proteins. ARN as an integrated resource contains 238 interactions between 38 autophagy proteins. Note that nearly all of the PPIs in ARN are present in other sources but it is ARN that contains them together in a single resource. Thanks to our manual curation we could increase the number of well-referenced interactions with 18, which are not present in the other sources.
Similar comparison with transcriptional and post-transcriptional resources is shown in Table 2. Note that many of these connections might be false positives or highly context specific. However, similarly to PPI predictions, these potential connections could also serve as a pool of possible autophagy-related regulatory mechanisms that should be examined and confirmed experimentally. The ARN resource contains 98 known and predicted TFs for 37 autophagy genes with 557 TF-gene connections; 35 of them are manually curated, and cannot be found in other resources. Of note, we found only a few TFs present in multiple bioinformatics resources, indicating the importance of different approaches to discover TFs capable to regulate autophagy, and the usefulness of ARN as an integrated single resource.
We extracted the interactions relevant in the regulation of autophagy from all constituting databases, while we have integrated different types of molecular interactions (protein-protein, TF-gene, miRNA-mRNA) into a uniform data scheme. Overall, ARN contains more regulatory interactions for the autophagy proteins than any of the constituting databases. Interactions from 23 sources have been integrated into one comprehensive database, giving the opportunity for comparison and selection between the data sources. Note that the total numbers in each ARN layer at Table 2 are higher than any of the sources. This indicates the increased amount of data in ARN, compared to other resources.
Application
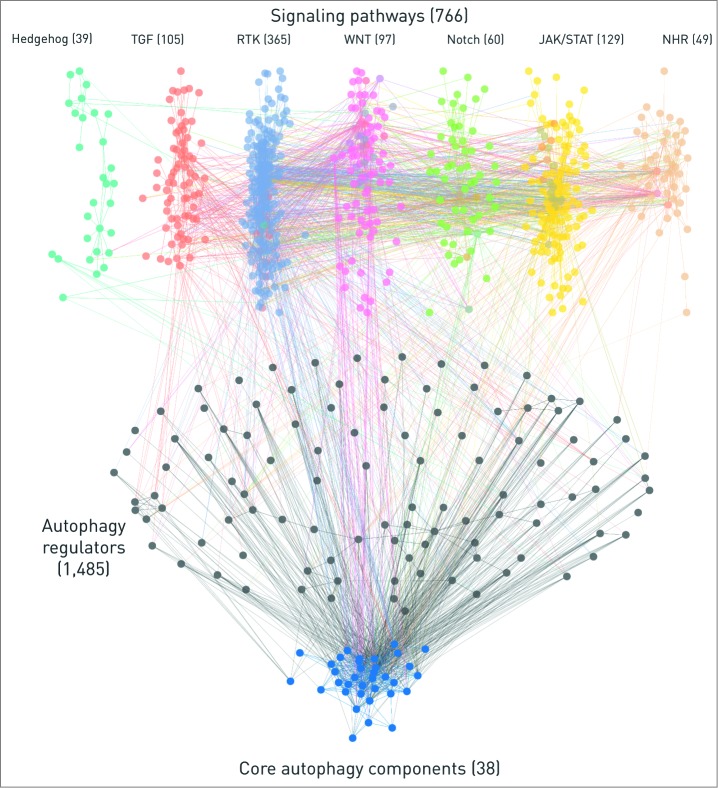
ARN can be used to examine the autophagy system in humans for both a global analysis or for gene-specific studies. For both cases, different levels of the regulation can be examined, validated or experiments can be evaluated. Here, we highlight another key feature of ARN that is its immersive connection with signaling pathways: ARN connects autophagy proteins directly and indirectly with 7 major signaling pathways taken from SignaLink 2. We included all connections up to 3 steps (4 elements) length, considering PPIs, TF-gene, and miRNA-mRNA interactions as well. There are 357 direct connections between pathway member proteins and autophagy proteins, indicating the robust and context specific regulation of autophagy by signaling pathways. On the Figure 4 one- and 2-step long connections between pathways and autophagy components are shown. This is a global map that could be specifically analyzed or zoomed in by users who download ARN.
Figure 4.
The network of 7 signaling pathways with direct autophagy regulators and core autophagy proteins. The numbers represent the total number of components in each section but for clarity, only components with the highest confidence, one- or 2- step long connections are shown on this figure. We also omitted the connections through transcription factors or miRNAs. Edges between autophagy proteins are blue. Intermediate components (i.e., direct autophagy regulators) in the 2-step connections and their edges are colored with black. Pathways are color-coded, multipathway proteins and edges between different pathways have the colors of the involved pathways mixed. Edges directly connecting pathways and autophagy proteins have the color of the source pathway.
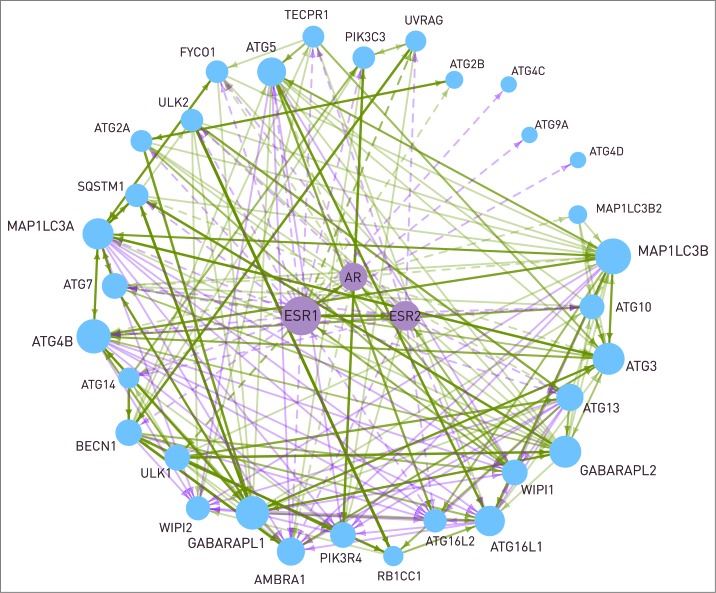
In the following, we illustrate the power of multilayered connection between autophagy and signaling pathways with the example of the nuclear hormone receptor (NHR) pathway. Most of the transcription factors regulating autophagy proteins belong to the NHR pathway. Using ARN data, we found potential androgen or estrogen receptor binding sites in the promoters of 2-thirds of the autophagy proteins (32). Though gender differences at the level of autophagy are observed in many diseases, little is known about the mechanisms underlying this phenomenon.22 For example, in cardiomyocytes and neurons, following ischemia and reperfusion, autophagy mediates in part the cytoprotective effect of estrogen,23-25 resulting in a higher level of apoptosis in males.26,27 Also in neurodegenerative diseases, gender differences in autophagy have been described.28 In addition, almost all neurodegenerative diseases have higher incidence in females.22 At certain prostate cancer cell types, androgen signaling plays a cardinal role in the choice between autophagy and apoptosis, former helping survival and metastasis formation, while latter delaying tumor growth.4 ARN could help to find the connection between sex steroid signaling and autophagy. In Figure 5, the transcriptional regulation of autophagy proteins by the androgen and estrogen receptors is presented. Regulation of ULK1/2 and UVRAG by ESR1 is experimentally verified,29 according to the HTRI database. All the other connections were predicted in ARN using the JASPAR algorithm.15 Another autophagy protein, WIPI1 can also be transcriptionally regulated by sex steroid receptors. In addition, WIPI1 contains an LXXLL motif, which enables it to bind to ESR1, ESR2, and AR in a hormone independent way.30 This connection is important, because localization of WIPI1 depends on autophagic activity, and at the same time it regulates sex steroid signaling, affecting the transcription of several autophagy proteins, including WIPI1 itself. As it is shown in Figure 5, 84% of the core autophagy proteins are transcriptionally regulated by sex steroid receptors. ESR1, ESR2, and AR regulate different but overlapping sets of autophagy proteins (23, 12, and 12 proteins, respectively). AR is also able to heterodimerize and activate ESR1, as well as ESR1 and ESR2 each other. Further research studies might reveal the role of these mechanisms in a gender-specific regulation of the autophagic activity.
Figure 5.
Interactions between the 2 estrogen receptors (ESR1 and ESR2), the androgen receptor (AR), and 32 autophagy proteins. Dashed line represents transcriptional regulation, while continuous line is for post-translational regulation. The width of the lines shows the number of data sources where the interaction can be found. The size of a node is proportional with the number of its connections. WIPI1 is able to bind to the estrogen receptors. AR and ESR2 are able to heterodimerize with ESR1. The interactions between the autophagy proteins are shown with a continuous line.
Discussion
Here we present a novel resource on the regulation of autophagy in human. Autophagy Regulatory Network (ARN; http://autophagy-regulation.org) is a comprehensive interaction database featuring a manually curated core dataset, integrated and predicted data from numerous sources, and direct connection to literature curated interactions of 7 major signaling pathways. Directions, signs, confidence scores, and references are available for each interaction. ARN is accessible through a user-friendly webpage, and the data can be downloaded in all major bioinformatics standard formats, including simple text/table files and visualized Cytoscape networks.
To achieve a better understanding of context-dependent regulation of autophagic activity, a systems-level analysis of regulatory mechanisms is necessary. External stimuli processed by the signaling network can modulate autophagy at post-translational, transcriptional, and post-transcriptional level. Applying this approach in research studies can lead to the identification of key regulatory circuits, which are responsible for specificities in the regulation of autophagy, in different tissues, and under pathologic or therapeutic conditions. We created ARN with the aim to support the systems-level analysis of context-dependent regulation of autophagy, and also to facilitate the large-scale examination of a single autophagy-related protein.
Primary data on post-translational, transcriptional, and post-transcriptional regulation of autophagy proteins can be obtained from various resources. However, to use data from multiple resources in one analysis can be tedious because of the different data formats and molecular database IDs. Furthermore, many resources often contain erroneous interactions between proteins, derived from high-throughput methods or predictions. To address this problem, ARN involves manually curated interactions between autophagy proteins, their post-translational regulators, and between signaling components. Most of the interactions in ARN have confidence values allowing the user to set an own cut-off value (or use the default value calculated by ROC analysis, using manually curated interactions as gold standard set). For all protein-protein interactions we offer the Gene Ontology semantic similarity score.31 This score is based on the assumption, that proteins involved in similar biological processes are more likely to interact in vivo. This way we can decrease the ratio of erroneous interactions from high-throughput screenings or predictions.
Before ARN, 2 autophagy-focused resources have been published. The Human Autophagy Database19 (HADb) contains only sequence data of genes from an autophagy-dedicated microarray. Autophagy Database20 (ADB) provides orthology data from 41 species, and for some proteins also a list of interactions. However, the source of these interactions and the scope of the curation are not clearly defined. Indeed, the main aim of ADB is to serve a comprehensive collection of orthologs of autophagy-related genes. The interaction data are not available for download in a single file, but can be browsed only on the webpage. Compared to HADb and ADB, in ARN the data sources are well defined, and the size of the network is determined by the principles of its design. ARN provides data not only on post-translational regulators, but also on transcriptional and post-transcriptional regulators. In addition, beside the direct regulators of the proteins involved in autophagy initiation and execution, ARN makes a connection between the cellular signaling network and the regulation of autophagy. The directions, signs, and confidence scores of the interactions are supplied in format ready for computational processing.
ARN serves as a good basis for various kinds of bioinformatics approaches, while it also effectively supports wet lab research work. Using the ARN website, researchers are able to search for potential interactors or regulators affecting their subject of interest. ARN database contains many potential regulators of the entire autophagic process and even for a single component that allows researchers to combine expression or mutation data sets and analyze autophagy in context-specific states. For example, ARN data can be used to point out important alterations in autophagy regulation upon a disease.17 Therefore, ARN can support experiment design and evaluation for both basic and translational research works.
Furthermore, network data of ARN can be analyzed with graph topological methods, modularization methods, perturbation simulations, and models can be built using different mathematical formalisms. Having an appropriate, good quality a priori knowledge as a starting point is a crucial requirement of successful modeling.32 ARN aims to support modeling approaches by serving as a good basis for a variety of methods, such as Boolean and rule-based modeling.33,34 Combining with gene expression or mutation data, comparative analyses can be carried out to investigate differences in autophagy regulation by tissue, physiological or pathological conditions, gender, and many other aspects. With the inclusion of drug compound and target interaction data, ARN is suitable to support network-based pharmacological attempts, such as multi-target and allo-network drug design.35
Knowing that the list of components and interactions in each layer is not complete, we will include further experimentally validated data every year. We also intend to include tissue-specific localization information to future versions of ARN. In addition, we will work on the extension of ARN for other species, for example, yeast, Drosophila, and zebrafish. In the form of the feedback option of the ARN website, we are looking for comments and suggestions from autophagy researchers on how we can improve ARN.
In conclusion, the Autophagy Regulatory Network reported here is a novel, extensive bioinformatics resource focusing on the regulation of autophagy. It opens up new opportunities in autophagy research, both for experimental and in silico research work, as well as for small-scale and systems-level studies. On the ARN website (http://autophagy-regulation.org), possible post-translational, transcriptional, and post-transcriptional regulators of autophagy related proteins can be examined easily. Key disease and cancer-related information are also listed to highlight the medical relevance of the proteins. ARN database can be downloaded in a user specific content and format allowing a customizable and efficient way to assist the community. ARN is a gap-filling integrative resource, and we hope that it will enable the autophagy research community to analyze more easily the already available data, guide future research projects, and facilitate autophagy-related conceptualizations of biomedical processes.
Methods
Compilation of the Autophagy Regulatory Network
We developed an onion-like, multilayered database structure to integrate and utilize the different regulatory layers of the Autophagy Regulatory Network. The core of the network contains autophagy executor proteins based on reviews. Within the core module, interactions between the proteins are from manual curation of the literature. First, we systematically checked every autophagy related protein-protein interactions mentioned in the review articles. Next, we searched for the original research articles experimentally verifying the interactions. We also used iHop36 and Chilibot37 web services to supplement the review-based information and cite experimental evidence. For each manually curated interaction, we listed the following information on the interaction: 1) PubMed ID of the primary first-time verifying article; 2) direction; 3) effect type (stimulatory/inhibitory); 4) molecular mechanism (if available). We searched for interactions among autophagy core proteins and between autophagy core proteins and their regulating proteins. We collected exclusively and very strictly interactions between 2 human proteins; interspecies, or even uncertain human-protein interactions, were omitted.
We considered interactions as direct if chemical reaction or physical binding occur between the 2 molecules (e.g., a protein phosphorylates another). Interactions presumably without such chemical or physical mechanism are denoted as indirect (e.g., interaction between a transcription factor and the protein, whose gene is targeted by the transcription factor, or in case of 2 members of a complex without direct binding to each other). Similarly, all miRNA interactions are indirect, because the miRNA does not regulate directly the protein's concentration or activity, but only its translation process. ARN is a network database, where nodes represent primarily proteins, not genes or mRNAs. That is why in the ARN database interactions taking effect with interposition of more molecules, are indirect.
In the first layer, the direct protein regulators of the core autophagy machinery are collected. The first layer is from 3 sources: (a) from manual curation of the literature, (b) data acquired from the Autophagy Database20 (ADB), and (c) from a proteomic analysis of the autophagy network.21 In the second layer, potential protein regulators are listed that have not yet been found to regulate the core autophagy proteins or their known regulators but in silico methods predicted their enzymatic reaction or protein binding to them. For this purpose, we used the ELM server38 and searched for enzymes (i.e., phosphatases, ubiquitin-ligases, peptidases, etc.) that can directly or indirectly modify autophagy components. We also used protein domain information from PFAM39 to predict a protein-protein interaction (PPI) based on domain-domain interactions.
The next 3 layers contain information on the transcriptional and post-transcriptional regulators of the above described inner-layers (i.e., autophagy components, their known and predicted protein regulators). The transcriptional regulatory layer contains transcription factors that are known or predicted to transcriptionally regulate the inner layers. These regulatory connections were integrated from databases such as ABS,40 ENCODE,41 HTRIdb,29 ORegAnno42 and PAZAR,43 or predicted with JASPAR.15 We also performed manual curation to collect TFs directly regulating autophagy proteins. In addition, to add the known complexity of transcriptional regulation, this layer also contains PPIs between the TFs from BioGRID,44 InnateDB,45 IntAct46 and HPRD47 databases. In the next layer, we integrated miRNAs as post-transcriptional regulators of the inner-layers (autophagy components and their direct regulators, including enzymes and TFs) from experimentally verified miRNA-mRNA interaction databases: miR2Disease,48 miRDeathDB,49 miRecords,50 miRTarBase,51 and Tarbase.52 The third regulatory layer contains the transcriptional regulators of these miRNAs (i.e., TFs known to regulate the expression of the miRNAs known to downregulate autophagy component or regulators). We used ENCODE,41 PuTmiR53 1.1 and 2.0 versions and TransmiR v1.254 to integrate this information. Data from the integrated resources were downloaded in the spring of 2013.
In the last step of the compilation, we connected signaling pathways from SignaLink 2 (http://signalink.org),55 a resource we recently developed, containing manually curated data of signaling pathways. SignaLink 2 contains 7 major signaling pathways: RTK (receptor tyrosine kinase), TGFB/TGF-β (transforming growth factor β), WNT, Hedgehog, JAK-STAT, NOTCH and NHR. Connections between signaling pathways and autophagy were derived in 3 different ways: (a) predicted or experimentally verified direct PPIs between a signaling protein and an autophagy protein; (b) via the transcriptional regulation of a signaling pathway related TFs and its autophagy-related target; and (c) through post-transcriptional regulation, where a signaling pathway affects a TF of a miRNA, which regulates a protein involved in autophagy or its regulation. Note that we also added further protein-protein interactions from BioGRID,44 InnateDB,45 IntAct,46 HPRD,47 and predictions between all the already included protein components.
For every integrated data source containing interactions collected with different methods, quality control is highly important. From each source databases we included the available confidence scores, maintaining the possibility for the users to exclude low confidence interactions from their analysis. However, these scores are only available for the subset of interactions derived from the specific source. To obtain a general confidence score for all protein-protein interactions, we calculated semantic similarity score31 between the Gene Ontology Biological Process56 properties of the interacting pairs of proteins. In case of PPIs inferred from domain-domain based prediction, we performed a ROC analysis to minimize the false positive rate. With the domain-motif based prediction, we used the cut-off value suggested by the authors of the ELM Structure Filter38 algorithm.
For each protein in ARN we included disease and cancer type annotations. We obtained diseases from GAD (The Genetic Associations Database),57 and OMIM (Online Mendelian Inheritance in Man),58 and cancer-type mutation patterns from COSMIC (Catalog of Somatic Mutations in Cancer).59
Database implementation and structure
Data storage is based on MySQL, which serves data to the webpage by a PHP interface. The webpage uses jQuery on the client side to offer a high interactivity. Information can be loaded asynchronously by small http requests, giving an efficient and comfortable browsing experience through hundreds of interactions. We wrote a separate data export module in Python language that offers various choices to download data in CSV, BioPAX, PSI-MI TAB, PSI-MI XML, SBML, and Cytoscape's CYS format. Several options are available to customize the network to download: users are able to filter by interaction types (e.g., PPIs, transcriptional regulation), as well as by sources. There is also an option to separate experimentally verified and predicted interactions. The customized network files are generated according to the selected options by the export module running in the background. This process can take few minutes. Then, for each download, we generate a URL, where users can access the data for 14 days Optionally, users can provide their email addresses to which files smaller than 10 MB will be emailed. The whole dataset is also available as a standard SQL dump, so any complex query or modification can be applied using SQL statements.
The core of the ARN database is the interaction table. In the interaction table source and target fields are integers pointing to the primary keys of protein or "mirna" tables. The layer field denotes the type of the interaction, and its value determines if the source or the target refers to a protein or miRNA. The meanings of the values in the layer field are the followings: 0: interactions between autophagy executor proteins; 1: PPIs between autophagy proteins and their direct regulators from our manual curation, ADB20 and the ChIP-Seq study of Behrends et al.;21 2: direct and indirect regulators of autophagy proteins from general PPI resources44-47 and from predictions based on domain-domain39 and domain-motif38 interactions; 3: value not used due to technical reasons; 4: TF-target connections; 5: miRNA-mRNA connections, 6: PPIs in the signaling pathways, imported from SignaLink 2;55 7: TF-miRNA connections; 8: PPIs between TFs, signaling pathways and autophagy regulators, from the same sources as layer 2. Each interaction has 3 main attributes: is_directed (0: undirected; 1: directed; 2: direction is predicted), is_direct (0: indirect; 1: direct) and is_stimulation (0: unknown; 1: stimulation, -1: inhibition). In addition, interactions have one or more sources. Sources are listed in the source table, and the interaction_source table contains their assignment to the interaction table. Manually curated interactions have literature references, contained by the interaction_reference table. In the interaction_reference table, articles are identified by their Pubmed IDs. Most of the interactions have confidence scores. These are stored as float values in the interaction_weight table, the different types of scores are listed in weight table. Components of ARN are listed in the protein and "mirna" tables. The protein table contains the uniprot_name field, which is unique, and it contains the UniProt accession number of proteins. All records imported from other databases, as well protein names from articles are mapped to their primary UniProtKB ID. Proteins may have signaling topological properties and pathway assignments, available in protein_topology and protein_pathway tables. In the "mirna" table we used miRBase AC and miRNA name to identify miRNAs.
Acknowledgments
We thank members of the Vellai lab, the NetBiol group, and the LINK-Group for helpful discussions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the European Union and the European Social Fund (TAMOP-4.2.2/B-10/1–2010–0013) and the Hungarian Scientific Research Fund (OTKA K83314, K109349′, NK78012). TK is a grantee of the János Bolyai Scholarship of the Hungarian Academy of Sciences, and is supported by a fellowship in computational biology at The Genome Analysis Center, in partnership with the Institute of Food Research, and strategically supported by BBSRC.
References
- 1. Hamacher-Brady A. Autophagy regulation and integration with cell signaling. Antioxid Redox Signal 2012; 17:756-65; PMID:22149388; http://dx.doi.org/ 10.1089/ars.2011.4410 [DOI] [PubMed] [Google Scholar]
- 2. Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis 2012; 33:2018-25; PMID:22902544; http://dx.doi.org/ 10.1093/carcin/bgs266 [DOI] [PubMed] [Google Scholar]
- 3. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen S, Niu Y, Lee SO, Chang C. Androgen receptor (AR) positive vs negative roles in prostate cancer cell deaths including apoptosis, anoikis, entosis, necrosis and autophagic cell death. Cancer Treat Rev 2014; 40:31-40; PMID:23993415; http://dx.doi.org/ 10.1016/j.ctrv.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reggiori F. Autophagy: New questions from recent answers. ISRN Mol Biol 2012; 2012:1-12; http://dx.doi.org/ 10.5402/2012/738718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russell RC, Tian Y, Yuan H, Park HW, Chang Y-Y, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741-50; PMID:23685627; http://dx.doi.org/ 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 2014; 53:471-83; PMID:24440502; http://dx.doi.org/ 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salih DAM, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008; 20:126-36; PMID:18394876; http://dx.doi.org/ 10.1016/j.ceb.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seo Y-K, Jeon T-I, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab 2011; 13:367-75; PMID:21459322; http://dx.doi.org/ 10.1016/j.cmet.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lavallard VJ, Meijer AJ, Codogno P, Gual P. Autophagy, signaling and obesity. Pharmacol Res 2012; 66:513-25; PMID:22982482; http://dx.doi.org/ 10.1016/j.phrs.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 12. Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106-17; PMID:18555785; http://dx.doi.org/ 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- 13. Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science 2009; 324:1720-3; PMID:19443739; http://dx.doi.org/ 10.1126/science.1162327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell 2013; 152:327-39; PMID:23332764; http://dx.doi.org/ 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 15. Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 2010; 38:D105-110; PMID:19906716; http://dx.doi.org/ 10.1093/nar/gkp950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther 2011; 11:157-68; PMID:21228626; http://dx.doi.org/ 10.4161/cbt.11.2.14622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kubisch J, Türei D, Földvári-Nagy L, Dunai ZA, Zsákai L, Varga M, Vellai T, Csermely P, Korcsmáros T. Complex regulation of autophagy in cancer - integrated approaches to discover the networks that hold a double-edged sword. Semin Cancer Biol 2013; 23:252-61; PMID:23810837; http://dx.doi.org/ 10.1016/j.semcancer.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 18. Santra T, Kolch W, Kholodenko BN. Navigating the multilayered organization of eukaryotic signaling: a new trend in data integration. PLoS Comput Biol 2014; 10:e1003385; PMID:24550716; http://dx.doi.org/ 10.1371/journal.pcbi.1003385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, et al. The acquisition of resistance to TNFα in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy 2011; 7:760-70; PMID:21490427; http://dx.doi.org/ 10.4161/auto.7.7.15454 [DOI] [PubMed] [Google Scholar]
- 20. Homma K, Suzuki K, Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res 2011; 39:D986-990; PMID:20972215; http://dx.doi.org/ 10.1093/nar/gkq995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lista P, Straface E, Brunelleschi S, Franconi F, Malorni W. On the role of autophagy in human diseases: a gender perspective. J Cell Mol Med 2011; 15:1443-57; PMID:21362130; http://dx.doi.org/ 10.1111/j.1582-4934.2011.01293.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhupathy P, Haines CD, Leinwand LA. Influence of sex hormones and phytoestrogens on heart disease in men and women. Womens Health 2010; 6:77-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bouma W, Noma M, Kanemoto S, Matsubara M, Leshnower BG, Hinmon R, Gorman JH, Gorman RC. Sex-related resistance to myocardial ischemia-reperfusion injury is associated with high constitutive ARC expression. AJP Heart Circ Physiol 2010; 298:H1510-H1517; http://dx.doi.org/ 10.1152/ajpheart.01021.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen C, Hu L-X, Dong T, Wang G-Q, Wang L-H, Zhou X-P, Jiang Y, Murao K, Lu S-Q, Chen J-W, et al. Apoptosis and autophagy contribute to gender difference in cardiac ischemia-reperfusion induced injury in rats. Life Sci 2013; 93:265-70; PMID:23827240; http://dx.doi.org/ 10.1016/j.lfs.2013.06.019 [DOI] [PubMed] [Google Scholar]
- 26. Weis SN, Toniazzo AP, Ander BP, Zhan X, Careaga M, Ashwood P, Wyse ATS, Netto CA, Sharp FR. Autophagy in the brain of neonates following hypoxia-ischemia shows sex- and region-specific effects. Neuroscience 2014; 256:201-9; PMID:24184979; http://dx.doi.org/ 10.1016/j.neuroscience.2013.10.046 [DOI] [PubMed] [Google Scholar]
- 27. Zhu C, Xu F, Wang X, Shibata M, Uchiyama Y, Blomgren K, Hagberg H. Different apoptotic mechanisms are activated in male and female brains after neonatal hypoxia-ischaemia. J Neurochem 2006; 96:1016-27; PMID:16412092; http://dx.doi.org/ 10.1111/j.1471-4159.2005.03639.x [DOI] [PubMed] [Google Scholar]
- 28. Barbati C, Pierdominici M, Gambardella L, Malchiodi Albedi F, Karas RH, Rosano G, Malorni W, Ortona E. Cell surface estrogen receptor alpha is upregulated during subchronic metabolic stress and inhibits neuronal cell degeneration. PLoS ONE 2012; 7:e42339; PMID:22860116; http://dx.doi.org/ 10.1371/journal.pone.0042339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bovolenta LA, Acencio ML, Lemke N. HTRIdb: an open-access database for experimentally verified human transcriptional regulation interactions. BMC Genomics 2012; 13:405; PMID:22900683; http://dx.doi.org/ 10.1186/1471-2164-13-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004; 23:9314-25; PMID:15602573; http://dx.doi.org/ 10.1038/sj.onc.1208331 [DOI] [PubMed] [Google Scholar]
- 31. Alvarez MA, Yan C. A graph-based semantic similarity measure for the gene ontology. J Bioinform Comput Biol 2011; 9:681-95; PMID:22084008; http://dx.doi.org/ 10.1142/S0219720011005641 [DOI] [PubMed] [Google Scholar]
- 32. Kremling A, Saez-Rodriguez J. Systems biology - An engineering perspective. J Biotechnol 2007; 129:329-51; PMID:17400319; http://dx.doi.org/ 10.1016/j.jbiotec.2007.02.009 [DOI] [PubMed] [Google Scholar]
- 33. Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry (Mosc) 2010; 49:3216-24; http://dx.doi.org/ 10.1021/bi902202q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maus C, Rybacki S, Uhrmacher AM. Rule-based multi-level modeling of cell biological systems. BMC Syst Biol 2011; 5:166; PMID:22005019; http://dx.doi.org/ 10.1186/1752-0509-5-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Csermely P, Korcsmáros T, Kiss HJM, London G, Nussinov R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery. Pharmacol Ther 2013; 138:333-408; PMID:23384594; http://dx.doi.org/ 10.1016/j.pharmthera.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics 2004; 5:147; PMID:15473905; http://dx.doi.org/ 10.1186/1471-2105-5-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet 2004; 36:664; PMID:15226743; http://dx.doi.org/ 10.1038/ng0704-664 [DOI] [PubMed] [Google Scholar]
- 38. Via A, Gould CM, Gemünd C, Gibson TJ, Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics 2009; 10:351; PMID:19852836; http://dx.doi.org/ 10.1186/1471-2105-10-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res 2011; 40:D290-D301; PMID:22127870; http://dx.doi.org/ 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blanco E, Farré D, Albà MM, Messeguer X, Guigó R. ABS: a database of Annotated regulatory Binding Sites from orthologous promoters. Nucleic Acids Res 2006; 34:D63-67; PMID:16381947; http://dx.doi.org/ 10.1093/nar/gkj116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012; 489:91-100; PMID:22955619; http://dx.doi.org/ 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Griffith OL, Montgomery SB, Bernier B, Chu B, Kasaian K, Aerts S, Mahony S, Sleumer MC, Bilenky M, Haeussler M, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res 2008; 36:D107-113; PMID:18006570; http://dx.doi.org/ 10.1093/nar/gkm967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Portales-Casamar E, Arenillas D, Lim J, Swanson MI, Jiang S, McCallum A, Kirov S, Wasserman WW. The PAZAR database of gene regulatory information coupled to the ORCA toolkit for the study of regulatory sequences. Nucleic Acids Res 2009; 37:D54-60; PMID:18971253; http://dx.doi.org/ 10.1093/nar/gkn783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chatr-aryamontri A, Breitkreutz B-J, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 2012; 41:D816-D823; PMID:23203989; http://dx.doi.org/ 10.1093/nar/gks1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ. InnateDB: systems biology of innate immunity and beyond-recent updates and continuing curation. Nucleic Acids Res 2012; 41:D1228-D1233; PMID:23180781; http://dx.doi.org/ 10.1093/nar/gks1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 2013; 42:D358-D363; PMID:24234451; http://dx.doi.org/ 10.1093/nar/gkt1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database-2009 update. Nucleic Acids Res 2009; 37:D767-772; PMID:18988627; http://dx.doi.org/ 10.1093/nar/gkn892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 2009; 37:D98-104; PMID:18927107; http://dx.doi.org/ 10.1093/nar/gkn714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu J, Li Y-H. miRDeathDB: a database bridging microRNAs and the programmed cell death. Cell Death Differ 2012; 19:1571; PMID:22743998; http://dx.doi.org/ 10.1038/cdd.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009; 37:D105-110; PMID:18996891; http://dx.doi.org/ 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsu S-D, Tseng Y-T, Shrestha S, Lin Y-L, Khaleel A, Chou C-H, Chu C-F, Huang H-Y, Lin C-M, Ho S-Y, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42:D78-85; PMID:24304892; http://dx.doi.org/ 10.1093/nar/gkt1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res 2012; 40:D222-229; PMID:22135297; http://dx.doi.org/ 10.1093/nar/gkr1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bandyopadhyay S, Bhattacharyya M. PuTmiR: a database for extracting neighboring transcription factors of human microRNAs. BMC Bioinformatics 2010; 11:190; PMID:20398296; http://dx.doi.org/ 10.1186/1471-2105-11-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res 2010; 38:D119-122; PMID:19786497; http://dx.doi.org/ 10.1093/nar/gkp803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fazekas D, Koltai M, Türei D, Módos D, Pálfy M, Dúl Z, Zsákai L, Szalay-Bekő M, Lenti K, Farkas IJ, et al. SignaLink 2 - a signaling pathway resource with multi-layered regulatory networks. BMC Syst Biol 2013; 7:7; PMID:23331499; http://dx.doi.org/ 10.1186/1752-0509-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/ 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet 2004; 36:431-2; PMID:15118671; http://dx.doi.org/ 10.1038/ng0504-431 [DOI] [PubMed] [Google Scholar]
- 58. McKusick-Nathans Institute of Genetic Medicine , Johns Hopkins University; (Baltimore, MD: ). Online Mendelian Inheritance in Man, OMIM. ; Internet. 2014 ; cited 2014 Aug 1; Available from: http://www.ncbi.nlm.nih.gov/omim [Google Scholar]
- 59. Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res 2010; 39:D945-D950; PMID:20952405; http://dx.doi.org/ 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raghavachari B, Tasneem A, Przytycka TM, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Res 2007; 36:D656-D661; PMID:17913741; http://dx.doi.org/ 10.1093/nar/gkm761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smialowski P, Pagel P, Wong P, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Rattei T, Frishman D, et al. The Negatome database: a reference set of non-interacting protein pairs. Nucleic Acids Res 2009; 38:D540-D544; PMID:19920129; http://dx.doi.org/ 10.1093/nar/gkp1026 [DOI] [PMC free article] [PubMed] [Google Scholar]