Abstract
Autophagy is a critical cellular homeostatic process that controls the turnover of damaged organelles and proteins. Impaired autophagic activity is involved in a number of diseases, including idiopathic pulmonary fibrosis suggesting that altered autophagy may contribute to fibrogenesis. However, the specific role of autophagy in lung fibrosis is still undefined. In this study, we show for the first time, how autophagy disruption contributes to bleomycin-induced lung fibrosis in vivo using an Atg4b-deficient mouse as a model. Atg4b-deficient mice displayed a significantly higher inflammatory response at 7 d after bleomycin treatment associated with increased neutrophilic infiltration and significant alterations in proinflammatory cytokines. Likewise, we found that Atg4b disruption resulted in augmented apoptosis affecting predominantly alveolar and bronchiolar epithelial cells. At 28 d post-bleomycin instillation Atg4b-deficient mice exhibited more extensive and severe fibrosis with increased collagen accumulation and deregulated extracellular matrix-related gene expression. Together, our findings indicate that the ATG4B protease and autophagy play a crucial role protecting epithelial cells against bleomycin-induced stress and apoptosis, and in the regulation of the inflammatory and fibrotic responses.
Keywords: ATG4B, autophagy, autophagin-1, epithelial cell, idiopathic pulmonary fibrosis, lung fibrosis
Abbreviations
- ACTA2
actin, α 2, smooth muscle, aorta
- ATG3
autophagy-related 3
- ATG4B
autophagy-related 4B
- cysteine
peptidase
- ATG5
autophagy-related 5
- ATG7
autophagy-related 7
- ATG9B
autophagy-related 9B
- BAX
BCL2-associated X protein
- CASP3
caspase 3, apoptosis-related cysteine peptidase
- CAV1
caveolin 1, caveolae protein, 22kDa
- CCL3
chemokine (C-C motif) ligand 3
- CXCL1
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity α)
- CXCR2
chemokine (C-X-C motif) receptor 2
- DRAM2
DNA-damage regulated autophagy modulator 2
- GFP-LC3B
green fluorescent protein-LC3B
- IL12B
interleukin 12B
- IL13
interleukin 13
- IFNG
interferon, gamma
- IPF
idiopathic pulmonary fibrosis
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- RELA
v-rel reticuloendotheliosis viral oncogene homolog A
- SQSTM1
sequestosome 1
- TGFB1
transforming growth factor, β 1
- TGFBR2
transforming growth factor, β receptor II (70/80kDa)
- TNF
tumor necrosis factor
- TUBB4
tubulin, β 4, class IV
- WT
wild type
Introduction
Lung fibrosis is the final result of a large and heterogeneous group of lung disorders, known as interstitial lung diseases.1 One of the most common and the most aggressive interstitial lung disease is idiopathic pulmonary fibrosis (IPF), a chronic, progressive, irreversible, and usually lethal lung disease of unknown etiology.2 Evidence suggests that IPF is the result of an aberrant wound healing process, characterized by epithelial cell injury and activation followed by the expansion of fibroblasts/myofibroblasts population and an aberrant deposit of extracellular matrix in lung parenchyma.3,4
IPF is an aging-associated disease, and actually most patients are older than 60 y at the time of diagnosis.5 However, the mechanisms that link IPF with aging are uncertain. Aging is characterized by the time-dependent accumulation of cellular damage. There is evidence that autophagy and the proteasome proteolytic systems decline with aging, resulting in accumulation of damaged proteins and organelles.6 Interestingly, decreased autophagic activity in lung tissue from IPF compared with chronic obstructive pulmonary disease and healthy controls has been reported.7 Additionally, studies in vitro have provided evidence that autophagy inhibition accelerates epithelial cell senescence and induces fibroblast to myofibroblast differentiation, 2 key processes in the pathogenesis of lung fibrosis.8 Collectively, these findings suggest that impaired autophagy may contribute to pulmonary fibrosis. However the specific role of autophagy in IPF is still undefined.
Autophagy is a major intracellular proteolytic system in which portions of cytoplasm are engulfed within double-membrane vesicles called autophagosomes that are delivered to and degraded in the lysosome.9 This degradative process is executed by a complex molecular machinery that includes more than 30 proteins, encoded by the evolutionarily conserved ATG genes.10 Among the autophagic proteins required for autophagosome formation is ATG4, a cysteine proteinase that proteolytically activates the uncleaved and unlipidated yeast Atg8 (also known as pro-Atg8) or its 6 currently identified mammalian orthologs and paralogs. A process that is strictly required for the subsequent conjugation of yeast Atg8 (or its mammalian orthologs and paralogs) with membrane-bound phosphatidylethanolamine (PE), essential for autophagosome completion. ATG4 is also required for deconjugating PE from the C-terminal Gly of Atg8–PE.11 There is one single member in yeast, and deletion of ATG4 arrests the autophagy process. Four mammalian orthologs of the yeast proteinase gene ATG4, named Atg4a/autophagin-2, Atg4b/autophagin-1, Atg4c/autophagin-3, and Atg4d/autophagin-4 have been identified in mice.12 Recent kinetics analysis of all 4 ATG4 orthologs has indicated that ATG4B had the broadest substrate spectrum with similar affinity and catalytic efficiency toward each of the 6 mammalian orthologs and paralogs of yeast Atg8 (of which, the most widely characterized group is the MAP1LC3/LC3 family), suggesting that ATG4B is the main ATG4 protease for autophagy in mammalian cells.13 Recently, we have generated mutant mice deficient in Atg4b and demonstrated that these animals exhibit a clear reduction of basal- and starvation-induced autophagic flux in all tissues, and that the 3 other Atg4 isoforms (Atg4a/c/d) do not compensate for the lack of Atg4b.14 Atg4b-deficient mice reach adulthood and are fertile, enabling to study in vivo the effect of autophagy impairment in lung fibrosis.
In this work, we provide evidence that autophagic activity and ATG4B expression increase during experimental bleomycin-induced lung fibrosis, and importantly, that Atg4b deficiency causes more severe inflammatory and fibrotic responses indicating that ATG4B plays a protective role in lung fibrosis.
Results
Autophagy is activated after bleomycin-induced pulmonary fibrosis
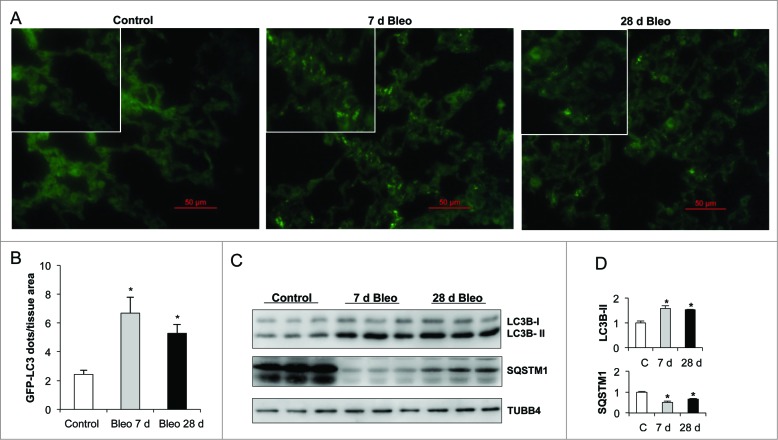
Autophagic activity is constitutively active at low basal levels and is enhanced in response to a wide variety of intracellular and extracellular stress conditions, including nutrient or growth factor deprivation, hypoxia, reactive oxygen species, DNA damage, protein aggregates, damaged organelles, or intracellular pathogens.15 To investigate whether autophagy is induced during pulmonary fibrosis in vivo, GFP-LC3 transgenic mice were treated with bleomycin and autophagosome formation was monitored by fluorescence microscopy analysis in lung tissue cryosections as previously described.16 In lungs from saline-treated control mice, the GFP-LC3 signal was detected diffusely in the cytoplasm with no or few punctate dots. The number of GFP-LC3-positive dots per tissue area was significantly increased in lungs at 7 (inflammatory phase) and 28 (fibrotic phase) d post-bleomycin treatment, compared to uninjured saline control lung (Fig. 1A and B). Next, we followed the autophagic flux through conversion of cytosolic LC3B-I to lipidated membrane-bound LC3B-II in lung tissue extracts. Time course analysis of LC3B demonstrated that LC3B-II levels were remarkably increased at 7 and 28 d post-bleomycin compared to saline controls. Consistent with these results, a decrease in SQSTM1/p62 level was also revealed (Fig. 1C and D).
Figure 1.
Autophagic activity increases after lung inflammation and fibrosis. Bleomycin (Bleo) or saline were administered to GFP-LC3 transgenic mice by endotracheal instillation and lungs were harvested after indicated time points. (A) Representative fluorescent microphotographs of lung tissue from saline control and bleomycin-treated GFP-LC3 transgenic mice. (B) Quantification of LC3B dots per mm2 of lung tissue from GFP-LC3 transgenic mice. (C) Representative immunoblots of endogenous LC3B-I/II and SQSTM1 in lung tissue. TUBB4 was used as a loading control. (D) Densitometry. Protein levels were normalized to saline control. Results represent mean ± SD. Statistical significance was determined by one-way ANOVA (*P < 0.05).
Additionally, we evaluated in vitro the autophagic activity in mouse alveolar epithelial (MLE 12) cells after bleomycin injury. Very few autophagosomes were observed in MLE 12 cells in basal conditions. However, after 24 h of bleomycin treatment the number of autophagosomes per cell increased significantly (Fig. 2A). To further analyze the autophagic flux in MLE 12 cells, the turnover of the autophagosomal markers LC3B-II and SQSTM1 was measured after bleomycin challenge in presence of lysosomal inhibitors. Bleomycin treatment induced autophagy activation, as indicated by increased LC3B-II and decreased SQSTM1 protein level (Fig. 2B). Chloroquine and leupeptin treatment led to accumulation of LC3B-II in both control and bleomycin-treated cells, however, LC3B-II accumulation was significantly higher in bleomycin-treated cells compared to control cells (Fig. 2B and C). Both inhibitors also led to greater accumulation of SQSTM1 in bleomycin-treated compared to control cells (Fig. 2B and C). These results reinforce the notion that autophagic activity is induced after alveolar epithelial cell injury.
Figure 2.
Autophagic activity in MLE 12 cells increases after bleomycin challenge. (A) Representative fluorescent microphotographs of endogenous LC3B dots in MLE 12 cells under basal or bleomycin-treated condition, and LC3B dots per cell area (μm2 × 100). (B) Representative immunoblots of endogenous LC3B-I/-II and SQSTM1 in MLE 12 cells after 0 or 100 mU/ml of bleomycin alone, or combined with 10 μM chloroquine or 10 μM leupeptin for 24 h (left), and densitometry normalized to the basal control condition (right). TUBB4 was used as loading control. (C) LC3B-II and SQSTM1 ratio (protein level in the presence of inhibitor vs protein level in the absence of inhibitor) in bleomycin-treated compared to control cells. Results represent mean ± SD. Statistical significance was determined by one-way ANOVA (*P < 0.05, ** P < 0.01).
To evaluate the molecular mechanism underlying the increased autophagic activity during bleomycin-induced pulmonary fibrosis, we performed a mouse autophagy RT-PCR array to analyze the expression level of 84 key genes involved in autophagy, including genes that encode proteases and components of the autophagosomal machinery, signaling, and apoptosis. Bleomycin-induced lung injury resulted in the upregulation of Ulk1, Bax, Atg5, Atg4b, Fadd, Atg9b, Hsp90aa1, and Ifna2 gene expression (Fig. 3A; Table S1). Conversely, Eva1, Gabarapl2, and Dram2 were downregulated (Fig. 3A; Table S2). Additionally, protein levels of ATG3, ATG4B, ATG5, and ATG7 were determined by immunoblot in lung tissue extracts from bleomycin-treated and saline control mice. Consistent with gene expression results, we found that ATG4B and ATG5 significantly increased at 7 and 28 d of bleomycin instillation compared to saline controls (Fig. 3B and C), suggesting that these autophagic proteins could play a functional role in the pathogenic process of pulmonary fibrosis.
Figure 3.
Gene expression changes after bleomycin-induced lung fibrosis. (A) Gene expression infogram for differentially expressed genes at 28 d after bleomycin exposure performed by qPCR. Every row represents a gene and every column a mouse lung sample (control vs fibrosis, n = 3 for each group). (B) Representative immunoblots of ATG3, ATG4B, ATG5, and ATG7 in lung tissue extracts from control and bleomycin-treated mice. TUBB4 was used as loading control. (C) Densitometry. Protein levels were normalized to saline control. Results represent mean ± SD. Statistical significance was determined by one-way ANOVA (*P < 0.05).
ATG4B immunolocalization in bleomycin-induced pulmonary fibrosis and IPF lungs
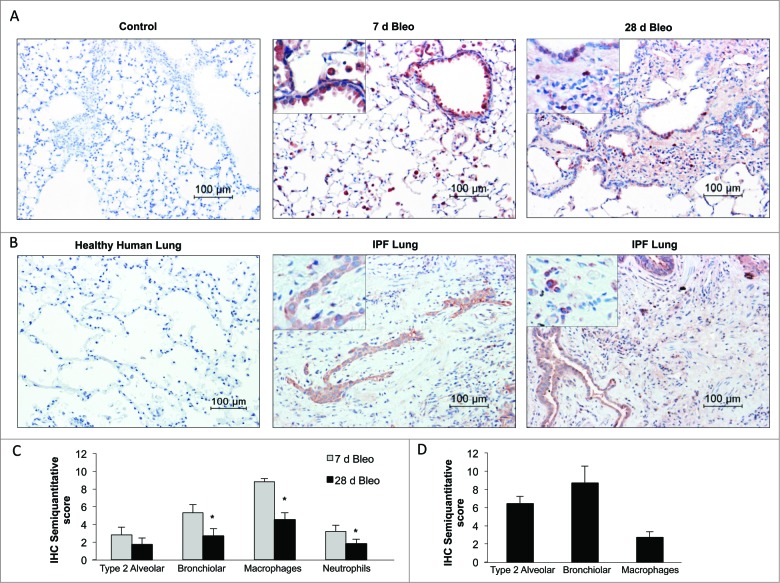
The cellular source of the protease ATG4B was examined by immunohistochemistry in both the bleomycin mouse model as well as IPF lung tissues. In the mouse model, lung staining for ATG4B was negative in saline controls (Fig. 4A). At 7 d postbleomycin instillation, ATG4B was strongly expressed in alveolar macrophages and bronchiolar epithelial cells, with moderate staining of type 2 alveolar epithelial cells and neutrophils, as illustrated in Figure 4A and C. At 28 d post-treatment, ATG4B was also expressed in alveolar macrophages and neutrophils however, the percentage of positive stained inflammatory cells was significantly decreased compared to 7 d (Fig. 4A and C). Also, while at 7 d about all bronchiolar epithelium was strongly positive for ATG4B, at 28 d post-treatment only a few bronchiolar epithelial cells were positive stained. Moderate staining was observed in type 2 alveolar epithelial cells and putative nuclear staining in scarce interstitial cells in fibrotic areas (Fig. 4A and C). Regarding human tissues, ATG4B was undetectable in all the healthy lungs examined (Fig. 4B). Interestingly, in IPF lungs ATG4B was mainly localized in hyperplastic and hypertrophic type 2 alveolar epithelial cells usually overlying the fibroblastic foci, and in hyperplastic epithelial cells with metaplasia, nonciliated columnar cells, bronchiolar epithelial cells, and few interstitial inflammatory cells while it was negative in fibroblasts (Fig. 4B and D).
Figure 4.
ATG4B immunolocalization in bleomycin-induced pulmonary fibrosis and IPF lung. (A) Representative photomicrographs of immunohistochemical staining for ATG4B in saline control and bleomycin-treated mouse lung at 7 and 28 d. (B) Healthy and IPF human lung tissue sections. Positive staining was observed in red. All sections were counterstained with hematoxylin. (C) Semiquantitative score of mouse and (D) human lung tissue sections. Results represent mean ± SD. Statistical significance was determined by one-way ANOVA (*P < 0.05).
Atg4b deficiency results in autophagic activity disruption in lung
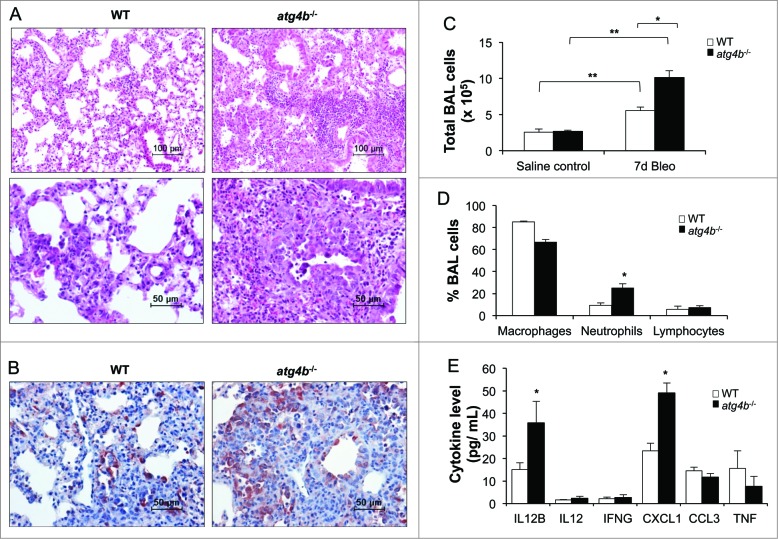
In order to evaluate the role of the protease ATG4B and autophagy in lung homeostasis, Atg4b-deficient mice were used as an experimental model. ATG4B proteolytically activates pro-LC3B to render possible its lipidation and subsequent incorporation into phagophore membranes. We and others have previously demonstrated, through the generation and characterization of atg4b−/− mice, that the absence of ATG4B results in deficient proteolytic processing of LC3B and other murine orthologs of yeast Atg8, leading to decreased autophagic activity in most atg4b−/− mice tissues.14 Here, we first tested whether Atg4b disruption leads to a reduced autophagic activity specifically in the lungs, examining LC3BI/II and SQSTM1 in lung tissue extracts from untreated mice. As shown in Figure 5A, immunoblot analysis confirmed that ATG4B was absent in the lungs from atg4b−/− mice. The LC3B-I form accumulated while the LC3B-II form was absent in the lungs from atg4b−/− mice as compared with their corresponding wild-type (WT) controls. We also observed SQSTM1 accumulation in lungs from untreated atg4b−/− mice as compared with their corresponding controls (Fig. 5A). These results demonstrated a dramatic impairment of autophagic activity in the lungs from atg4b−/− mice, under basal conditions. Comparative morphological analysis of lung tissue between atg4b−/− and WT mice under basal conditions did not reveal any abnormalities. Thus, at least at 8 to 10 wk old, Atg4b deletion did not appear to impact lung architecture (Fig. 5D). Then, we analyzed the autophagic flux in the lungs from atg4b−/− mice after bleomycin-induced inflammation and fibrosis. As shown in Figure 5B and C, an accumulation of LC3B-I, impaired conversion to LC3B-II and a marked accumulationof SQSTM1 were found in lung tissue from atg4b−/− at 7 and 28 d after bleomycin treatment compared to WT mice. These results indicate that autophagic activity is impaired in the lungs of atg4b−/− mice, even under stress conditions, such as bleomycin treatment, and suggest that the protease ATG4B is required for induction of autophagy after lung stress and injury.
Figure 5.
Reduced autophagic flux in lung tissue from atg4b−/− mice. Representative immunoblots of ATG4B, LC3B and SQSTM1 in lung tissue extracts from (A) control (B) 7 and (C) 28 d after bleomycin treatment from age-matched WT and mutant mice. Lung tissue extracts from 3 mice per genotype are shown. TUBB4 was used as loading control. (D) Representative light microscopy images of H&E stained lung tissue sections from WT and atg4b−/− control mice. (E) Percentage of body weight changes over a 28-day period for atg4b−/− and WT mice after bleomycin instillation. (F) Percentage of survival were plotted for atg4b−/− (n = 18) and WT (n = 18) mice for 28 d after challenge with bleomycin. Statistical significance was determined by log-rank test or Student t test (*P < 0.05).
Increased inflammation after bleomycin treatment in atg4b−/− mice is linked to alterations in proinflammatory cytokine profiles
To determine the role of ATG4B in pulmonary fibrosis, atg4b−/− mice were treated with bleomycin, and the inflammatory and fibrotic responses were analyzed. Mice from both genotypes experienced progressive weight loss during the first week after bleomycin instillation. However, atg4b−/− mice showed a more pronounced weight loss and never recovered the initial weight (Fig. 5E). Consistently, Atg4b deficiency resulted in reduced survival rate after bleomycin instillation compared to WT (Fig. 5F). Histopathological analysis of lungs from mice sacrificed at 7 d post-bleomycin revealed multifocal interstitial and intra-alveolar inflammation in both atg4b−/− and WT mice. However, increased inflammatory response was observed in atg4b−/− mice, mainly characterized by enhanced neutrophilic infiltration (Fig. 6A). We also found an increased RELA/NFκB p65 immunostaining in type 2 alveolar epithelial cells and macrophages in lungs from atg4b−/− compared to WT bleomycin-treated mice (Fig. 6B). Lung histology correlated with BALF cell profile, where a significant increase of total inflammatory cells and in the percentage of neutrophils at 7 d after bleomycin instillation was observed in atg4b−/− compared with WT mice (Fig. 6C and D). To address mechanisms that may contribute to the increased neutrophil accumulation in atg4b−/− mice, we performed a multiplex immunoassay to evaluate the pattern of cytokine expression in BALF. We found a significant increase of CXCL1/KC (chemokine [C-X-C motif] ligand 1 [melanoma growth stimulating activity, α]), a strong chemotactic mediator for neutrophils,17 and of IL12B levels in BALF from atg4b−/− compared with WT mice at 7 d post-bleomycin (Fig. 6E). These results suggest that impaired autophagy and deregulated CXCL1 expression could contribute to the exacerbated neutrophilic inflammation observed in the lungs of atg4b−/− bleomycin-treated mice. Regarding IL12B, recent evidence suggested that this cytokine can act as a profibrotic and proinflammatory cytokine, since il12b−/− mice were protected against silica-induced fibrosis.18
Figure 6.
Increased lung inflammatory response in atg4b−/− mice. (A) Representative light microscopy images of H&E stained lung tissue sections from WT and atg4b−/− mice at 7 d after bleomycin instillation. (B) Immunohistochemical staining performed with specific antibody against RELA/NFκB p65 in lung tissue sections from WT and atg4b−/− mice at 7 d post-bleomycin. (C) Total and (D) differential cell count in BALF. (E) Protein level of proinflammatory cytokines in BALF from atg4b−/− and WT bleomycin-treated mice. Results are shown as mean ± SD. Statistical significance was determined by one-way ANOVA (C) or Student t test (D and E) (*P < 0.05).
Atg4b deficiency exacerbates bleomycin-induced lung fibrosis
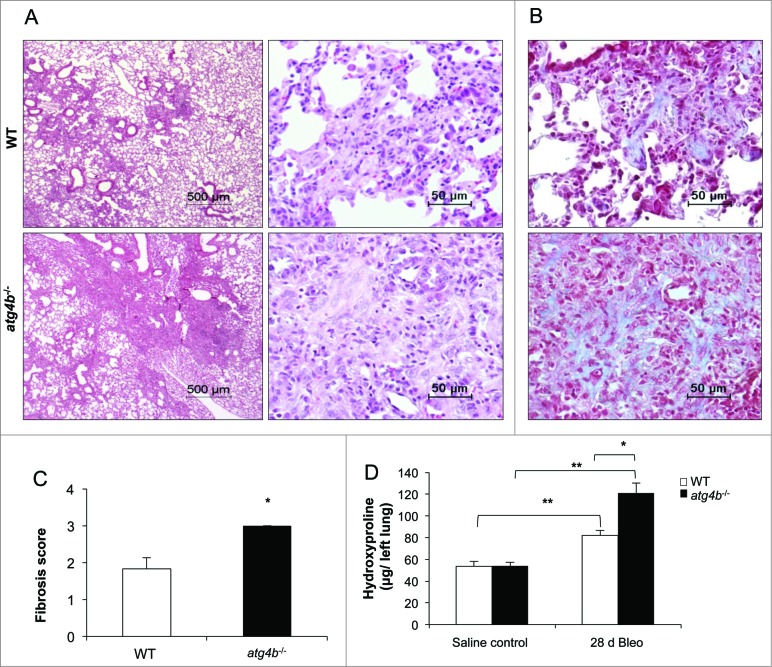
Mice from both genotypes developed fibrosis at 28 d after bleomycin instillation. However, Atg4b-deficient mice exhibited more extensive and severe fibrosis (Fig. 7A). As illustrated in Figure 7B, lungs from atg4b−/− mice had large areas of peribronchiolar and parenchymal fibrosis than their corresponding WT littermates, and increased collagen deposition assessed by Masson trichrome staining. Consequently, both semiquantitative fibrosis scoring and hydroxyproline lung content were significantly higher in atg4b−/− mice relative to WT mice (Fig. 7C and D). Additionally, the myofibroblast population and the protein levels of the myofibroblast biomarker ACTA2/α-SMA were markedly increased in the lungs from atg4b−/− mice, consistent with more severe lung fibrosis phenotype (Fig. 8A and B). To better understand the molecular mechanisms associated with the phenotype observed in atg4b−/− mice, we performed a mouse fibrosis RT-PCR array to analyze the gene expression level of key growth and transcription factors that regulate extracellular matrix synthesis and remodeling, extracellular matrix components, such as collagens, matrix metalloproteinases, and their specific inhibitors. This analysis revealed the upregulation of Il13ra2, Col1a2, Timp3, Col3a1, Cxcr4, Smad3, Mmp13, Tgfbr2, and Tgfb3 in Atg4b−/−compared to WT mice at 28 d post-bleomycin injury (Fig. 8C; Table S3). Among downregulated genes we found Cav1, Il4, Smad7, Hgf, and Ccl11 (Fig. 8C; Table S4). Taken together, these results indicate that the absence of ATG4B exacerbates the lung fibrotic reaction to bleomycin, suggesting a protective role for this protease and autophagy in lung fibrosis.
Figure 7.
Atg4b deficiency sensitizes mice to experimental lung fibrosis. (A) Representative light microscopy images of H&E stained lung tissue sections from WT and atg4b−/− mice at 28 d after bleomycin instillation. (B) Masson Trichrome staining of lung sections from the same experimental groups. (C) Fibrosis score for grading lung histopathological changes. (D) Hydroxyproline content in lung at 28 d after bleomycin. Results are shown as mean ±SD . Statistical significance was determined by Student t test (C) or one-way ANOVA (D) (*P < 0.05).
Figure 8.
Deregulated expression of profibrotic mediators in atg4b−/− mice. (A) Representative light microscopy images of stained lung tissue sections performed with specific antibody against ACTA2 from WT and atg4b−/− mice at 28 d after bleomycin instillation. (B) Representative immunoblot of ACTA2 in lung tissue extracts from WT and atg4b−/− mice at 28 d after bleomycin instillation and densitometry analysis. TUBB4 was used as loading control. Results are shown as mean ± SD. Statistical significance was determined by Student t test (*P < 0.05). (C) Gene expression infogram for differentially expressed genes at 28 d after bleomycin instillation performed by qPCR. Every row represents a gene and every column a mouse lung sample (WT vs atg4b−/− mice, n = 3 for each group).
Atg4b deficiency increases lung apoptosis following bleomycin injury
Alveolar epithelial cell death is probably an initiating mechanism in IPF and experimental lung fibrosis. Bleomycin produces DNA damage, senescence, oxidative stress and apoptosis of the alveolar epithelium.19,20 Since atg4b−/− mice are highly susceptible to bleomycin-induced pulmonary fibrosis we explored lung apoptosis by TUNEL assay and anti-active CASP3 immunohistochemistry in WT and atg4b−/− lungs. The number of TUNEL-positive cells was significantly increased in atg4b−/− mice compared with WT mice as early as 7 d after bleomycin exposure (Fig. 9A and D). Moreover, we observed that in lungs from both WT and atg4b−/− mice, bleomycin exposure was associated with active CASP3 positive staining at 7 d post-treatment. However, the percentage of positive stained cells and staining intensity in lungs from atg4b−/− was significantly higher compared to WT mice. Thus, WT mice exhibited a moderate staining of alveolar macrophages and bronchiolar and type 2 alveolar epithelial cells. While, atg4b−/− lungs showed strong active CASP3 immunostaining primarily observed in alveolar macrophages and in bronchiolar and type 2 alveolar epithelial cells located in areas of high inflammatory cell infiltration (Fig. 9C and E). Western blot analysis of lung tissue homogenates corroborated the marked increase of cleaved CASP3 levels in atg4b−/− compared with WT mice at 7 d after bleomycin (Fig. 9B and D). These data indicate that ATG4B deficiency resulted in increased cell death after bleomycin challenge.
Figure 9.
Mice with Atg4b deficiency display increased apoptosis after bleomycin challenge. (A) Lung tissue sections labeled with TUNEL assay and counterstained with methyl green. Apoptotic DAB-positive nuclei are brown, normal nuclei are green. (B) Representative immunoblots for pro- and active-CASP3 in lung tissue homogenates from WT and atg4b−/− mice at 7 d after bleomycin. (C) Immunohistochemistry for active CASP3. (D) Percentage of TUNEL-positive cells in lung tissue sections and immunoblot densitometry. TUBB4 was used as loading control. (E) Semiquantitative score. Results are shown as mean ± SD. Statistical significance was determined by one-way ANOVA (*P < 0.05).
Discussion
Defects in autophagy have been implicated in a wide range of human diseases, including lung disorders such as cystic fibrosis, chronic obstructive pulmonary disease, asthma, pulmonary hypertension, lung cancer, and lung infectious diseases.21-24 The first study to address the role of autophagy in IPF suggested that this process is not induced in this disease despite the upregulation of potent inducers of autophagy such as increased levels of ER stress and oxidative stress.7 However, IPF is an age-related disease and it is widely assumed that the autophagic activity decreases with age and probably contributes to the accumulation of damaged macromolecules and organelles in lung.6 Therefore, how autophagy deficiency may contribute to lung fibrosis development has not been explored in vivo.
We used the Atg4b-deficient mouse, which could phenotypically mimic the impairment of autophagic activity found in IPF, to explore the involvement of insufficient autophagy in experimental lung fibrosis. The present study has discovered ATG4B as a novel protein that regulates lung homeostasis and plays a crucial role in the control of the inflammatory and fibrotic response occurring during bleomycin-induced lung injury.
We first sought to determine if there were changes in the lung autophagic flux after bleomycin lung injury and to investigate this, we monitored autophagosome formation in the lungs from GFP-LC3 transgenic mice. We found marked increase in autophagic activity as revealed by the accumulation of GFP-LC3 puncta, a marked increase in LC3B-II, and a decrease in SQSTM1 protein in lungs from bleomycin-treated mice compared with saline control mice at 7 and 28 d post-treatment. Mi et al.25 have found that instillation of bleomycin significantly increases autophagy activation in fibrotic tissues, and demonstrate that blocking IL17A with neutralizing antibodies further enhances autophagy, leading to less lung fibrosis. Likewise, Yang et al.26 have demonstrated that autophagic activity is increased in the bleomycin-injured lung tissues and that TLR4 (toll-like receptor 4) deficiency attenuates autophagy resulting in higher collagen accumulation. This evidence suggests that both IL17A and TLR4 regulate lung fibrosis at least partially by modulating autophagy. These results and our findings are in apparent discrepancy with the study by Patel et al.,7 which reports absence of autophagic activity based on the phosphorylation status of RPS6/S6 protein in bleomycin-induced experimental lung fibrosis. However, in this study the autophagic flux was not directly evaluated. Although MTORC1 is a key regulator of autophagy, recent research findings have unveiled that there are also MTORC1-independent regulatory pathways of this process.27–29
We also explored the gene expression profile associated with increase autophagic flux in bleomycin induced lung inflammation and fibrosis using an autophagy array. We found that Ulk1, Bax, Atg5, Atg4b, Fadd, Atg9b, Hsp90aa1, and Infa2 gene expression were upregulated, while Eva1, Gabarapl2, and Dram2 gene expression were downregulated. Additionally, we demonstrated the increase of ATG4B at the protein level concomitantly with autophagy induction during experimental lung inflammation and fibrosis.
ATG4B is a cysteine protease that cleaves C-terminal Gly residues from LC3B and together with ATG7 and ATG3 mediates the lipidation of all LC3 paralogs.30 ATG4B is also required to recycle LC3B-I from the autophagosome outer membrane, therefore the ATG4-LC3B conjugation system has an essential role during elongation and closure of the phagophore membrane.13,14 This is supported by recent evidence showing that overexpression of an inactive mutant of ATG4B inhibits the lipidation of LC3B-I, which led to a defect in autophagosome expansion and closure.31
In the present study we evaluated the effect of Atg4b deficiency in lung homeostasis. Under basal conditions, lung morphology of atg4b−/− mice did not disclose any abnormalities. However, lungs from atg4b−/− mice displayed a markedly reduction in LC3B-II conversion together with SQSTM1 accumulation, indicating impaired autophagic activity. After 7 and 28 d post-bleomycin treatment, we observed that while autophagic flux was significantly increased in lung from WT mice, autophagy was not induced in lung from atg4b−/− mice despite cytotoxic and stressful conditions. This reduced autophagic flux, was associated with an extensive inflammation, characterized by increased neutrophilic infiltration in atg4b−/− lungs at 7 d after bleomycin.
On the one hand, the increased inflammation observed in lung from atg4b−/− mice correlated with higher BALF levels of CXCL1, a strong inducer of the activation and recruitment of neutrophils.32 In mice, CXCR2 is the receptor for this chemokine, and it has been shown that blocking tissue neutrophil recruitment with a CXCR2 antagonist diminished neutrophil transmigration into airway space and consequently the collagen deposition in the bleomycin-induced lung fibrosis model.33,34 On the other hand, we also found a high level of IL12B in BALF from atg4b−/− mice at 7 d after bleomycin treatment. Recent evidence indicates that IL12B participates in lung inflammation and fibrosis after injury.35 Thus, the use of il12b−/− mice or the administration of anti-IL12B antibody reduces the inflammation and fibrosis induced by silica or bleomycin.36,37 This evidence indicates that high IL12B level could promote a worse inflammatory and fibrotic response in Atg4b-deficient mice.
ATG4B deficiency resulted in an extensive and enhanced lung fibrosis compared with their WT littermates at 28 d after bleomycin, characterized by increased numbers of myofibroblasts and collagen accumulation. This exaggerated fibrotic response was associated with upregulated expression of key TGFB pathway members such as Tgfb3, Tgfbr2, and Smad3, that regulate extracellular matrix turnover.38 Signaling through SMAD3 is essential for TGFB1-mediated fibrosis, as evidenced in smad3-deficient mice that are resistant to develop fibrosis after bleomycin instillation.39,40 Additionally, Il13ra2 was upregulated in fibrotic lung from Atg4b-deficient mice compared to WT. IL13 signals through IL13RA2 and activates fibrogenesis via TGFB1-dependent and independent mechanisms.41,42 In this context, excessive signaling through TGBR2 and IL13RA2 could be associated with Col1a2, Col3a1, and Timp3 overexpression found in the fibrotic lungs from Atg4b-deficient mice. CXCR4, a critical chemokine receptor expressed by fibrocytes, was also upregulated in atg4b−/− fibrotic lungs. Increased circulating fibrocytes as well as CXCR4-expressing fibrocytes have been reported in the lungs of patients with IPF.43-45
Among downregulated genes we found Cav1, Il14, Smad7, Hgf, and Ccl11. A marked reduction of CAV1 has been reported in IPF and experimental bleomycin-induced lung fibrosis.46,47 CAV1 is an integral membrane protein essential for the formation of caveole that plays a major role in signal transduction because it is able to bind to a variety of kinase members and coordinates the compartmentalization of specific signaling cascades.47 Interestingly, cav1-deficient mice have profound abnormalities in lung morphology and develop lung fibrosis spontaneously.48 Evidence suggest that downregulation of Cav1 increases gene expression of TGFB1-signaling components. This is consistent with our data, given that Tgfb3, Tgfbr2, and Smad3 were upregulated in atg4b−/− fibrotic lung.49 This negative correlation between the TGFB1 and CAV1 pathways in Atg4b-deficient lung could be associated with the increased collagen deposition after bleomycin injury.
On the one hand, autophagy is an adaptive prosurvival response, and depletion of autophagy genes in vivo and in vitro leads to failure in the protection against several stressors and apoptosis inducers.15,50,51 On the other hand, apoptosis of alveolar epithelial cells is a key early initiating event in the pathogenesis of pulmonary fibrosis.3,4,52-54 In this context we hypothesized that ATG4B deficiency may result in greater epithelial cell apoptosis leading to increased inflammation and consequently, extensive fibrotic areas. We show that ATG4B deficiency resulted in higher apoptosis predominantly observed in type 2 alveolar and bronchiolar epithelium and increased inflammation after bleomycin-induced lung injury.
Alveolar and bronchiolar epithelial cell injury and apoptosis modulate inflammatory cell recruitment into the lungs promoting the release of chemotactic factors that attract neutrophils and monocytes to the interstitium and alveolar space. In turn, neutrophils release reactive oxygen species, metalloproteinases, and proinflammatory cytokines that may perpetuate epithelial apoptosis and lung inflammation creating a positive feedback loop between apoptosis and inflammation.55-57 Moreover, autophagy could contribute to the clearance of dead cells, thus influencing the inflammatory response, resolution of inflammation, and tissue repair.58,59 In fact, it has been shown that macrophages lacking autophagy do not efficiently degrade their engulfed cargo and produce increased amounts of proinflammatory cytokines.60 We can propose that an increased autophagic activity after bleomycin injury, as we found in WT lungs, may contribute to interrupt this positive feedback loop by reducing the propensity of cells to undergo apoptosis and enhancing the clearance of dead cells.
Conversely, in lungs from WT mice, ATG4B was mainly expressed by bronchiolar and type 2 alveolar epithelial cells at 7 and 28 d after bleomycin treatment suggesting that expression of ATG4B by the injured epithelium could be necessary for survival. At 7 d, macrophages and neutrophils were also stained, suggesting that ATG4B expression could regulate the inflammatory response. Similarly, in IPF lungs we found that ATG4B positive stained cells were mostly bronchiolar and alveolar epithelial cells with altered phenotypes, while mesenchymal cells in dense fibroblastic foci such as fibroblasts were negative. ATG4B was undetectable in all the healthy human lungs examined. Recently, Chen et al.61 have quantified by western blot, several autophagy-related proteins in healthy human lung, and have found that ATG4B is almost undetectable and significantly lower compared to ATG5 or ATG7. This observation is in agreement with our results.
In summary, our findings point out that the ATG4B protease and autophagy play a crucial role protecting epithelial cells against bleomycin-induced stress and apoptosis, and in the regulation of the inflammatory and fibrotic responses.
Materials and Methods
Animals and bleomycin treatment
The generation of atg4b−/− mice has been previously described.14 In all experiments, homozygous atg4b−/− mice and their corresponding wild-type (WT) littermates were derived from interbreeding of heterozygous mice. Mice genotypes were determined by PCR analysis of tail DNA. Transgenic mice expressing the fluorescent autophagosome marker GFP-LC3 have been previously described. GFP-LC3 mice were maintained as heterozygous and were genotyped by PCR as previously described.16 Mice were bred under specific pathogen-free conditions. All experiments were performed with 8- to 10-wk-old mice and were approved by the Committee on Animal Experimentation of the National Institute of Respiratory Diseases of Mexico (INER).
Lung fibrosis was induced by intratracheal instillation of bleomycin sulfate (Sigma-Aldrich, B8416) at a single dose of 0.05U on day 0 in 50 μl of sterile saline. Control groups received the same volume of sterile saline. Mice were sacrificed at 7 and 28 d after bleomycin or saline treatment. For tissue harvesting, the lungs were perfused with sterile saline from right to left ventricle of the heart. Lungs were removed for fixation overnight in paraformaldehyde or snap frozen in liquid nitrogen followed by storage at –80°C.
Analysis of autophagosome accumulation in GFP-LC3 lungs
Lungs from mice expressing GFP-LC3 were perfused with 4% paraformaldehyde in 1× phosphate-buffered saline (1× PBS diluted from a 10× PBS stock; Life Technologies, 70011–044), pH 7.4. Lungs were harvested and fixed with the same fixative solution for at least 4 h, followed by treatment with 15% sucrose (Sigma-Aldrich, S0389) in 1× PBS for 4 h, and then with 30% sucrose solution at 4°C overnight. Tissue samples were embedded in Tissue-Tek OCT compound (Fisher Scientific, 14–373–65) and stored at –80°C. Samples were sectioned at 5-μm thickness with a cryostat (Leica, CM3050 S, Leica Biosystems, Buffalo Grove, IL), air-dried for 1 h, washed in 1× PBS for 5 min, dried at room temperature for 30 min, and mounted with a conventional antifading medium. GFP- LC3 dots were observed and counted in 3 independent visual fields from at least 6 independent mice under fluorescent microscope.
Analysis of autophagosome accumulation in MLE 12 cells
Mouse lung epithelial cell line MLE 12 was purchased from American Type Culture Collection (ATCC, CRL2110). MLE 12 cells were cultured in a CO2 incubator (5% CO2–95% air) at 37°C in Dulbecco's modified Eagle's medium, Nutrient Mixture F-12 (DMEM/F-12; Life Technologies, 11330–057), supplemented with 2% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 1% L-glutamine, 1% HEPES (Sigma-Aldrich, H9136), 1% insulin/transferrin/sodium selenite (Sigma-Aldrich, I-3146), 0.01% β-estradiol (Sigma-Aldrich, E2257), and 0.01% hydrocortisone (Sigma-Aldrich, I1882). MLE 12 cells were exposed to 0 or 100 mU/ml of bleomycin (Sigma-Aldrich, B8416) for 24 h. For immunofluorescence analyses, MLE 12 cells were grown on coverslips, washed in 1× PBS, and fixed in 4% paraformaldehyde in 1× PBS at 4°C during 10 min. The coverslips were blocked in 5% BSA (Sigma-Aldrich, A9418) for 10 min, incubated with anti-LC3B (Novus Biologicals, NB 100–2220) primary antibody for 1 h at 37°C, washed in PBS 1×, incubated for 40 min with secondary antibody, thoroughly washed in water, and mounted in Vectashield containing DAPI for nuclear staining (Vector Laboratories, H-1200).
Quantitative real-time RT-PCR
Total RNA of lung tissue from saline control and bleomycin-treated WT and atg4b−/− mice was isolated using TRIzol reagent (Invitrogen Life Technologies, 15596–026) following the manufacturer's instructions. Additional cleanup of total RNA was carried out using the RNeasy Mini kit (SABiosciences, 74104). Total RNA was used as template for double-stranded cDNA synthesis using the RT2 First Strand Kit according to the manufacturer's instructions (SABiosciences, 330401). cDNA from saline control and bleomycin-treated WT lungs was added to qPCR master mix (SABiosciences, 330512) and Mouse Autophagy RT2 Profiler™ PCR Array (SABiosciences, PAMM-084A) was performed using CFX 96 Real-Time PCR Detection System (BioRad, Hercules CA) to quantify the expression of 84 key genes involved in autophagy. To compare differential gene expression in fibrotic lung from WT and atg4b−/−, Mouse Fibrosis RT2 Profiler™ PCR Array was performed according to the manufacturer's instructions (SABiosciences, PAMM-120).
PCR array data were analyzed by a web software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Data analysis web portal selects an optimal set of internal housekeeping genes for normalization and analyzes Ct values to calculate changes in gene expression. The fold change was calculated by the equation 2(ΔΔCt).
Immunoblotting
MLE 12 cells were exposed to 0 or 100 mU/ml of bleomycin (Sigma-Aldrich, B8416) alone, or combined with 10 μM chloroquine (Sigma-Aldrich, C6628) or 10 μM leupeptin (Sigma-Aldrich, L2884) for 24 h. MLE 12 cells or lung tissue were homogenized in a 20 mM Tris buffer pH 7.4, containing 150 mM NaCl, 1% Triton X-100 (Sigma-Aldrich, T8787), 10 mM EDTA and Complete→ protease inhibitor cocktail (Roche Applied Science, 05–892–791–001). Then, tissue or cells extracts were centrifuged at 15,000 × g at 4°C and supernatant fractions were collected. Protein concentration was quantified by bicinchoninic acid technique (BCA protein assay kit, Pierce Biotechnology, 23225). A total of 25 µg of protein was loaded on either 8% or 13% SDS-polyacrylamide gels. After electrophoresis, gels were electrotransferred onto polyvinylidenedifluoride membranes (PVDF, Millipore, IPV H00010), and then membranes were blocked with 5% nonfat dried milk in PBT (phosphate-buffered saline with 0.05% Tween 20 [Sigma-Aldrich, P9416]) and incubated overnight at 4°C with the following primary antibodies diluted in 3% nonfat dried milk in PBT: anti-ATG4B (Sigma-Aldrich, A2981), anti-SQSTM1 (Sigma-Aldrich, P0068), anti-LC3B (Sigma-Aldrich, L7543), anti-TUBB4 (Santa Cruz Biotechnology, sc-9104), anti-ATG3 (Sigma-Aldrich, A3231), anti-ATG5 (Sigma-Aldrich, A0731), anti-ATG7 (Sigma-Aldrich, A2856), anti-ACTA2 (Abcam, AB7817) and anti-pro/active-CASP3/caspase 3 (Abcam, ab13585). After 3 washes with PBT, filters were incubated with the corresponding secondary antibody at 1:10,000 dilution in 1.5% milk in PBT, and developed with Immobilon Western Chemiluminescent HRP substrate (Millipore, WBKLS0500).
Immunohistochemistry
The tissue sections were deparaffinized and were then rehydrated and blocked with 3% H2O2 in methanol followed by antigen retrieval in a microwave in 10 mM citrate buffer, pH 6.0. Tissue sections were treated with universal blocking solution (BioGenex, HK085–5K) for 10 min, and then incubated overnight at 4°C with the following primary antibodies: anti-ATG4B (Sigma-Aldrich, A2981), anti-RELA/NFκB p65 (Abcam, ab7970), anti-ACTA2 (Abcam, AB7817) or anti-active CASP3 (Abcam, ab2302). A secondary biotinylated anti-immunoglobulin followed by horseradish peroxidase-conjugated streptavidin (BioGenex, HK330–5K) was used according to the manufacturer's instructions. 3-amino-9-ethyl-carbazole (BioGenex, HK092–5K) in acetate buffer containing 0.05% H2O2 was used as the substrate. The sections were counterstained with hematoxylin. The primary antibody was replaced by nonimmune serum for negative control slides.
For semi-quantitative score assessment, 10 fields were randomly chosen at × 400 magnification in each slide and the staining intensity was scored as no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3. The extent of positive cells was scored as 0 = no positive cells, 1 = comprising <25 %, 2 = 25 to 50%, 3 = 50 to 75% and 4 = 75 to 100% (the percentage of labeled cells, was determined according to the following equation: % = number of labeled cells/total counted cells ×100). The final score was determined by multiplying the intensity scores with the extent of positivity scores of stained cells, with the minimum score of 0 and a maximum score of 12.
Human lung specimens
Paraffin-embedded lung sections were obtained from surgical lung biopsies from patients diagnosed with IPF. The diagnosis of IPF was based on clinical, radiological, and functional findings and was corroborated by the characteristic morphology of UIP and made in accordance with American Thoracic Society/European Respiratory Society (ATS/ERS) Consensus Statements. Control lung tissue samples were obtained from autopsies of patients who died from nonlung-related causes.
Bronchoalveolar lavage fluid (BALF)
A 20-gauge intravenous catheter (BD insyte autoguard, 381834) was inserted into the trachea, and BALF was performed twice with 0.5 ml of sterile saline. The recovered BALF was centrifuged at 2,000 × g for 10 min at 4°C, and the supernatant fraction was stored at –80°C for subsequent biochemical analyses. Cell counts were performed by manual counting under light microscopy with a standard hemocytometer. Differential cell counts were obtained using Wright-Giemsa stain.
Multiplex immunoassay
Cytokine/chemokine levels in BALF were determined using a multiplex assay. BAL supernatants were normalized to 1 mg/ml of protein. A magnetic bead-based multiplex assay, containing fluorescent dyed microspheres conjugated with a monoclonal antibody specific for IL12B, IL12 (the p70 IL12 heterodimer), CCL3/MIP1α (chemokine [C-C motif] ligand 3), IFNG, CXCL1/KC or TNF, was used according to the manufacturer's instructions (Bio-Rad, 171–304070M and 171–150001), with a Bio-Rad Luminex Bio-Plex 200 System (Bio-Rad, Hercules, CA). The analyte concentration was calculated using software provided by the manufacturer (Bio-Plex Manager Software, Hercules, CA) and expressed as pg/mL.
Morphology
Lungs were removed and fixed by inflation with 4% paraformaldehyde in 1× PBS at continuous pressure of 25 cm H2O and were embedded in paraffin. Sections were stained with hematoxylin-eosin and Masson trichrome stain, and were scored blindly for severity and extent of lung lesions.
Hydroxyproline assay
Left lungs were hydrolyzed in 6 N HCl for 24 h at 110°C. Aliquots of 5 μL were then assayed by adding chloramine T (Sigma-Aldrich, 857319) solution for 20 min followed by development with Erlich reagent at 65°C for 15 min as previously described.17 Absorbance was measured at 550 nm, and the amount of hydroxyproline was determined against a standard curve generated using known concentrations of hydroxyproline standard (Sigma-Aldrich, H54409). Each sample was tested in triplicate. Data are expressed as μg of hydroxyproline/ left lung.
In situ apoptosis staining (TUNEL) assay
Paraffin-embedded lung sections were deparaffinized in xylene, and rehydrated with decreasing concentrations of ethanol. Apoptosis was detected by in situ TUNEL (terminal deoxynucleotidyltransferase biotin-dUTP nick end labeling) using the TACS2 TdT DAB in situ apoptosis detection kit (Trevigen, 4810–30-K). In brief, sections were treated with 50 μL of a proteinase K solution for 20 min and quenched in freshly prepared 3% hydrogen peroxide in methanol for 5 min at room temperature. Then, sections were washed in PBS 1× and labeled with the TdT reaction mix at 37°C for 1 h in a humidified chamber. Chromogenic detection was developed with streptavidin-HRP solution for 10 min, followed by incubation with DAB solution for 5 min at room temperature. Tissues were counterstaining with 1% methyl green and mounted with Clarion mounting medium (Sigma-Aldrich, C0487).
Statistics
All experimental data are reported as mean ± SD. Statistical analyses were performed by the 2-tailed Student t test and survival differences between groups were analyzed by Log-rank test. In experiments with more than 2 groups, differences were analyzed by multifactorial one-way analysis of variance (ANOVA) followed by Tukey post hoc test. using Graphpad Prism software Version 4.0 (Graphpad Software Inc.., San Diego CA) and P values lower than 0.05 were considered significant.
Acknowledgments
The authors thank Alberto Pizaña, Remedios Ramírez, Erika Monterrubio, and Guadalupe Hiriart for their technical assistance. We also thank N. Mizushima for GFP-LC3 transgenic mice.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a grant from PAPIIT program (project IA202613) from Universidad Nacional Autónoma de México. Mariana Maciel was supported by a fellowship from PAPIIT program (project IA202613).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Selman M, King TE, Pardo A. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001; 134:136–51; PMID:11177318; http://dx.doi.org/ 10.7326/0003-4819-134-2-200101160-00015 [DOI] [PubMed] [Google Scholar]
- 2.King TE, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet 2011; 378:1949–61; PMID:21719092; http://dx.doi.org/ 10.1016/S0140-6736(11)60052-4 [DOI] [PubMed] [Google Scholar]
- 3.Selman M, Pardo A. Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res 2002; 3:3; PMID:11806838; http://dx.doi.org/ 10.1186/rr175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selman M, Pardo A. Alveolar epithelial cell disintegrity and subsequent activation: a key process in pulmonary fibrosis. Am J RespirCrit Care Med 2012; 186:119–21; http://dx.doi.org/ 10.1164/rccm.201206-0997ED [DOI] [PubMed] [Google Scholar]
- 5.Selman M, Rojas M, Mora AL, Pardo A. Aging and interstitial lung diseases: unraveling an old forgotten player in the pathogenesis of lung fibrosis. SeminRespirCrit Care Med 2010; 31:607–17 [DOI] [PubMed] [Google Scholar]
- 6.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 2013; 153:1194–217; http://dx.doi.org/ 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, Rosas IO, Morse D. Autophagy in idiopathic pulmonary fibrosis. PLoS One 2012; 7: e41394; PMID:22815997; http://dx.doi.org/ 10.1371/journal.pone.0041394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araya J, Kojima J, Takasaka N, Ito S, Fujii S, Hara H, Yanagisawa H, Kobayashi K, Tsurushige C, Kawaishi M, et al.. Insufficient autophagy idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2013; 304: L56–69; PMID:23087019; http://dx.doi.org/ 10.1152/ajplung.00213.2012 [DOI] [PubMed] [Google Scholar]
- 9.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell 2014; 157:300–11; PMID:24725401; http://dx.doi.org/ 10.1016/j.cell.2014.01.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells 2010; 15:923–33; PMID:20670274; http://dx.doi.org/ 10.1111/j.1365-2443.2010.01433.x [DOI] [PubMed] [Google Scholar]
- 11.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, et al.. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488–92; PMID:11100732; http://dx.doi.org/ 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 12.Fernández AF, López-Otín C. The functional and pathologic relevance of autophagy proteases. J Clin Invest 2015; 125:33–41; PMID:2117786525654548; http://dx.doi.org/ 10.1172/JCI73940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem 2011; 286:7327–38; PMID:21177865; http://dx.doi.org/ 10.1074/jbc.M110.199059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, Salvador-Montoliu N, Vega JA, Germanà A, Fueyo A, et al.. Autophagy is essential for mouse sense of balance. J Clin Invest 2010; 120:2331–44; http://dx.doi.org/ 10.1172/JCI42601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280–93; PMID:20965422; http://dx.doi.org/ 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol 2009; 452:13–23; PMID:19200873; http://dx.doi.org/ 10.1016/S0076-6879(08)03602-1 [DOI] [PubMed] [Google Scholar]
- 17.Russo RC, Guabiraba R, Garcia CC, Barcelos LS, Roffê E, Souza AL, Amaral FA, Cisalpino D, Cassali GD, Doni A, et al.. Role of the chemokine receptor CXCR2 in bleomycin-induced pulmonary inflammation and fibrosis. Am J Respir Cell Mol Biol 2009; 40:410–21; PMID:18836137; http://dx.doi.org/ 10.1165/rcmb.2007-0364OC [DOI] [PubMed] [Google Scholar]
- 18.Huaux F, Arras M, Tomasi D, Barbarin V, Delos M, Coutelier JP, Vink A, Phan SH, Renauld JC, Lison D. A profibrotic function of IL12p40 in experimental pulmonary fibrosis. J Immunol 2002; 169:2653–61; PMID:12193738; http://dx.doi.org/ 10.4049/jimmunol.169.5.2653 [DOI] [PubMed] [Google Scholar]
- 19.Plataki M, Koutsopoulos AV, Darivianaki K, Delides G, Siafakas NM, Bouros D. Expression of apoptotic and antiapoptotic markers in epithelial cells in idiopathic pulmonary fibrosis. Chest 2005; 127:266–74; PMID:15653994; http://dx.doi.org/ 10.1378/chest.127.1.266 [DOI] [PubMed] [Google Scholar]
- 20.Wallach-Dayan SB, Izbicki G, Cohen PY, Gerstl-Golan R, Fine A, Breuer R. Bleomycin initiates apoptosis of lung epithelial cells by ROS but not by Fas/Fas L pathway. Am J Physiol Lung Cell Mol Physiol 2006; 290: L790–L796; PMID:16306138; http://dx.doi.org/ 10.1152/ajplung.00300.2004 [DOI] [PubMed] [Google Scholar]
- 21.Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol 2012; 74:377–401; PMID:22035347; http://dx.doi.org/ 10.1146/annurev-physiol-020911-153348 [DOI] [PubMed] [Google Scholar]
- 22.Ryter SW, Choi AM. Autophagy in the lung. Proc Am Thorac Soc 2010; 7:13–21; PMID:20160144; http://dx.doi.org/ 10.1513/pats.200909-101JS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon A, Eidelman D, Laprise C, Hamid Q. ATG5, autophagy and lung function in asthma. Autophagy 2012; 8:694–5; PMID:22498476; http://dx.doi.org/ 10.4161/auto.19315 [DOI] [PubMed] [Google Scholar]
- 24.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina DL, Settembre C, Gavina M, Raia V, Ballabio A, Maiuri L. Cystic fibrosis: a disorder with defective autophagy. Autophagy 2011; 7:104–6; PMID:21048426; http://dx.doi.org/ 10.4161/auto.7.1.13987 [DOI] [PubMed] [Google Scholar]
- 25.Mi S, Li Z, Yang HZ, Liu H, Wang JP, Ma YG, Wang XX, Liu HZ, Sun W, Hu ZW. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Immunol 2011; 187:3003–14; PMID:21841134; http://dx.doi.org/ 10.4049/jimmunol.1004081 [DOI] [PubMed] [Google Scholar]
- 26.Yang HZ, Wang JP, Mi S, Liu HZ, Cui B, Yan HM, Yan J, Li Z, Liu H, Hua F, et al.. TLR4 activity is required in the resolution of pulmonary inflammation and fibrosis after acute and chronic lung injury. Am J Pathol 2012; 180:275–92; PMID:22062220; http://dx.doi.org/ 10.1016/j.ajpath.2011.09.019 [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S, Rubinsztein DC. Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations. Autophagy 2006; 2(2):132–4; http://dx.doi.org/ 10.4161/auto.2387 [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto A, Mizushima N, Tsukamoto S. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol Reprod 2014; 91(1):7; http://dx.doi.org/ 10.1095/biolreprod.113.115816 [DOI] [PubMed] [Google Scholar]
- 29.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, Xavier RJ, Yuan J. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 2010; 18(6):1041–52; http://dx.doi.org/ 10.1016/j.devcel.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 2004; 279:36268–76; PMID:15187094; http://dx.doi.org/ 10.1074/jbc.M401461200 [DOI] [PubMed] [Google Scholar]
- 31.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol BiolCell 2008; 19:4651–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizutani N, Nabe T, Yoshino S. IL-17A promotes the exacerbation of IL-33-induced airway hyperresponsiveness by enhancing neutrophilicinflammation via CXCR2 signaling in mice. J Immunol 2014; 192:1372–84; PMID:24446518; http://dx.doi.org/ 10.4049/jimmunol.1301538 [DOI] [PubMed] [Google Scholar]
- 33.Redente EF, Jacobsen KM, Solomon JJ, Lara AR, Faubel S, Keith RC, Henson PM, Downey GP, Riches DW. Age and sex dimorphisms contribute to the severity of bleomycin-induced lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol 2011; 301:L510–8; PMID:21743030; http://dx.doi.org/ 10.1152/ajplung.00122.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo RC, Guabiraba R, Garcia CC, Barcelos LS, Roffê E, Souza AL, Amaral FA, Cisalpino D, Cassali GD, Doni A, et al.. Role of the chemokine receptor CXCR2 in bleomycin-induced pulmonary inflammation and fibrosis. Am J Respir Cell Mol Biol 2009; 40:410–21; PMID:18836137; http://dx.doi.org/ 10.1165/rcmb.2007-0364OC [DOI] [PubMed] [Google Scholar]
- 35.Walter MJ, Kajiwara N, Karanja P, Castro M, Holtzman MJ. Interleukin 12 p40 production by barrier epithelial cells during airway inflammation. J Exp Med 2001; 193:339–51; PMID:11157054; http://dx.doi.org/ 10.1084/jem.193.3.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeyama T, Kuwano K, Kawasaki M, Kunitake R, Hagimoto N, Hara N. Attenuation of bleomycininduced pneumopathy in mice by monoclonal antibody to interleukin-12. Am J Physiol Lung Cell Mol Physiol 2001; 280: L1128–37; PMID:11350791 [DOI] [PubMed] [Google Scholar]
- 37.Huaux F, Arras M, Tomasi D, Barbarin V, Delos M, Coutelier JP, Vink A, Phan SH, Renauld JC, et al.. A profibrotic function of IL-12p40 in experimental pulmonary fibrosis. J Immunol 2002; 169:2653–61; PMID:12193738; http://dx.doi.org/ 10.4049/jimmunol.169.5.2653 [DOI] [PubMed] [Google Scholar]
- 38.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003; 425:577–84; PMID:14534577; http://dx.doi.org/ 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Shi W, Wang YL, Chen H, Bringas P Jr, Datto MB, Frederick JP, Wang XF, Warburton D. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol 2002; 282: L585–93; PMID:11839555; http://dx.doi.org/ 10.1152/ajplung.00151.2001 [DOI] [PubMed] [Google Scholar]
- 40.Bonniaud P, Kolb M, Galt T, Robertson J, Robbins C, Stampfli M, Lavery C, Margetts PJ, Roberts AB, Gauldie J. Smad3 null mice develop airspace enlargement and are resistant to TGF-beta-mediated pulmonary fibrosis. J Immunol 2004; 173:2099–108; PMID:15265946; http://dx.doi.org/ 10.4049/jimmunol.173.3.2099 [DOI] [PubMed] [Google Scholar]
- 41.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006; 12:99–106; PMID:16327802; http://dx.doi.org/ 10.1038/nm1332 [DOI] [PubMed] [Google Scholar]
- 42.Kaviratne M, Hesse M, Leusink M, Cheever AW, Davies SJ, McKerrow JH, Wakefield LM, Letterio JJ, Wynn TA. IL-13 activates a mechanism of tissue fibrosis that is completely TGF-beta independent. J Immunol 2004; 173:4020–9; PMID:15356151; http://dx.doi.org/ 10.4049/jimmunol.173.6.4020 [DOI] [PubMed] [Google Scholar]
- 43.Andersson-Sjöland A, de Alba CG, Nihlberg K, Becerril C, Ramírez R, Pardo A, Westergren-Thorsson G, Selman M. Fibrocytes are a potential source of lung fibroblasts in idiopathic pulmonary fibrosis. Int J Biochem Cell Biol 2008; 40:2129–40; PMID:18374622; http://dx.doi.org/ 10.1016/j.biocel.2008.02.012 [DOI] [PubMed] [Google Scholar]
- 44.García-de-Alba C, Becerril C, Ruiz V, González Y, Reyes S, García-Alvarez J, Selman M, Pardo A. Expression of matrix metalloproteases by fibrocytes: possible role in migration and homing. Am J Respir Crit Care Med 2010; 182:1144–52; http://dx.doi.org/ 10.1164/rccm.201001-0028OC [DOI] [PubMed] [Google Scholar]
- 45.Makino H, Aono Y, Azuma M, Kishi M, Yokota Y, Kinoshita K, Takezaki A, Kishi J, Kawano H, Ogawa H, et al.. Antifibrotic effects of CXCR4 antagonist in bleomycin-induced pulmonary fibrosis in mice. J Med Invest 2013; 60:127–37; PMID:23614921 [DOI] [PubMed] [Google Scholar]
- 46.Gvaramia D, Blaauboer ME, Hanemaaijer R, Everts V. Role of caveolin-1 in fibrotic diseases. Matrix Biol 2013; 32:307–15; PMID:23583521; http://dx.doi.org/ 10.1016/j.matbio.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 47.Le Saux O, Teeters K, Miyasato S, Choi J, Nakamatsu G, Richardson JA, Starcher B, Davis EC, Tam EK, et al.. The role of caveolin-1 in pulmonary matrix remodeling and mechanical properties. Am J Physiol Lung Cell Mol Physiol 2008; 295: L1007–17; PMID:18849439; http://dx.doi.org/ 10.1152/ajplung.90207.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al.. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 2001; 293:2449–52; PMID:11498544; http://dx.doi.org/ 10.1126/science.1062688 [DOI] [PubMed] [Google Scholar]
- 49.Lee EK, Lee YS, Han IO, Park SH. Expression of Caveolin-1 reduces cellular responses to TGF-beta1 through down-regulating the expression of TGF-beta type II receptor gene in NIH3T3 fibroblast cells. Biochem Biophys Res Commun 2007; 359:385–90; PMID:17543885; http://dx.doi.org/ 10.1016/j.bbrc.2007.05.121 [DOI] [PubMed] [Google Scholar]
- 50.Ryter SW, Choi AM. Autophagy: an integral component of the mammalian stress response. J Biochem Pharmacol Res 2013; 1:176–88; PMID:24358454 [PMC free article] [PubMed] [Google Scholar]
- 51.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 2008; 19:797–806; PMID:18094039; http://dx.doi.org/ 10.1091/mbc.E07-10-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuwano K, Hagimoto N, Kawasaki M, Yatomi T, Nakamura N, Nagata S, Suda T, Kunitake R, Maeyama T, Miyazaki H, et al.. Essential roles of the Fas-Fas ligand pathway in the development of pulmonary fibrosis. J Clin Invest 1999; 104:13–9; PMID:10393694; http://dx.doi.org/ 10.1172/JCI5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee VY, Schroedl C, Brunelle JK, Buccellato LJ, Akinci OI, Kaneto H, Snyder C, Eisenbart J, Budinger GR, Chandel NS. Bleomycin induces alveolar epithelial cell death through JNK-dependent activation of the mitochondrial death pathway. Am J Physiol Lung Cell Mol Physiol 2005; 289: L521–8; PMID:16148050; http://dx.doi.org/ 10.1152/ajplung.00340.2004 [DOI] [PubMed] [Google Scholar]
- 54.Li X, Zhang H, Soledad-Conrad V, Zhuang J, Uhal BD. Bleomycin-induced apoptosis of alveolar epithelialcells requires angiotensin synthesis de novo. Am J Physiol Lung Cell Mol Physiol 2003; 284: L501–7; PMID:12573988; http://dx.doi.org/ 10.1152/ajplung.00273.2002 [DOI] [PubMed] [Google Scholar]
- 55.Keane MP, Belperio JA, Moore TA, Moore BB, Arenberg DA, Smith RE, Burdick MD, Kunkel SL, Strieter RM. Neutralization of the CXC chemokine, Macrophage Inflammatory Protein-2, attenuates bleomycin- induced pulmonary fobrosis. J Immunol 1999; 162:5511–8; PMID:10228032 [PubMed] [Google Scholar]
- 56.Chua F, Dunsmore SE, Clingen PH, Mutsaers SE, Shapiro SD, Segal AW, Roes J, Laurent GJ. Mice lacking neutrophil elastase are resistant to bleomycin-induced pulmonary fibrosis. Am J Pathol 2007; 170:65–74; PMID:17200183; http://dx.doi.org/ 10.2353/ajpath.2007.060352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan- 1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 2002; 111:635–46; PMID:12464176; http://dx.doi.org/ 10.1016/S0092-8674(02)01079-6 [DOI] [PubMed] [Google Scholar]
- 58.Zou W, Wang X, Vale RD, Ou G. Autophagy genes promote apoptotic cell corpse clearance. Autophagy 2012; 8(8):1267–8; http://dx.doi.org/ 10.4161/auto.20786 [DOI] [PubMed] [Google Scholar]
- 59.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15(2):81–94; http://dx.doi.org/ 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 2011; 108(42):17396–401; http://dx.doi.org/ 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, et al.. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One 2008; 3(10):e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.