Abstract
Aquatic photosynthetic eukaryotes represent highly diverse groups (green, red, and chromalveolate algae) derived from multiple endosymbiosis events, covering a wide spectrum of the tree of life. They are responsible for about 50% of the global photosynthesis and serve as the foundation for oceanic and fresh water food webs. Although the ecophysiology and molecular ecology of some algal species are extensively studied, some basic aspects of algal cell biology are still underexplored. The recent wealth of genomic resources from algae has opened new frontiers to decipher the role of cell signaling pathways and their function in an ecological and biotechnological context. Here, we took a bioinformatic approach to explore the distribution and conservation of TOR and autophagy-related (ATG) proteins (Atg in yeast) in diverse algal groups. Our genomic analysis demonstrates conservation of TOR and ATG proteins in green algae. In contrast, in all 5 available red algal genomes, we could not detect the sequences that encode for any of the 17 core ATG proteins examined, albeit TOR and its interacting proteins are conserved. This intriguing data suggests that the autophagy pathway is not conserved in red algae as it is in the entire eukaryote domain. In contrast, chromalveolates, despite being derived from the red-plastid lineage, retain and express ATG genes, which raises a fundamental question regarding the acquisition of ATG genes during algal evolution. Among chromalveolates, Emiliania huxleyi (Haptophyta), a bloom-forming coccolithophore, possesses the most complete set of ATG genes, and may serve as a model organism to study autophagy in marine protists with great ecological significance.
Keywords: algae, ATG8, ATG9, autophagy, blooms, chromalveolata, phylogenetics, phytoplankton, rhodophyta, stress
Abbreviations
- ATG
autophagy related
- DUF
domain of unknown function
- EhV
Emiliania huxleyi virus
- EST
expressed sequence tag
- GABARAP
GABA(A) receptor-associated protein
- PtdIns3K
phosphatidylinositol 3-kinase
- RPTOR
regulatory associated protein of MTOR, complex 1
- TOR
target of rapamycin
- TORC
target of rapamycin complex
- Ubl
ubiquitin-like
- Vps
vacuolar protein sorting
Introduction
Almost 50% of the organic carbon on our planet is fixed by a highly diverse, polyphyletic group of algae.1 Marine photosynthetic organisms consist of cyanobacteria, eukaryotic unicellular phytoplankton (microalgae), and eukaryotic multicellular algae (macroalgae), which are at the base of marine food webs.2,3 The fate of each individual algal cell is determined by an array of abiotic conditions such as nutrient availability, temperature, and light intensity,4,5 as well as biotic interactions with grazers and viruses.6,7 When favorable conditions for growth occur, algae can proliferate rapidly to form vast oceanic blooms.8,9 Phytoplankton blooms shape the ecology of the marine environment, as they play key roles in the global cycling of carbon, nitrogen, phosphate and sulfur, with widescale geological and climatic effects.10-16 Hence, elucidating the basic cellular machinery and signaling pathways that mediate phytoplankton acclimation to abiotic and biotic stress is highly important to our understanding of marine biogeochemical cycles.
The first oxygenic photosynthetic eukaryote is suggested to result from an endosymbiosis event involving the engulfment of a cyanobacterium by a heterotrophic eukaryote, a process estimated to have occurred ∼1.5 billion y ago.2 The resulting unicellular alga was the origin of 3 distinct clades, all formed by primary endosymbiosis: the viridiplantae, from which all green algae and land plants evolved (streptophyta, embryophyta), also known as the green plastid lineage; the rhodophyta, which includes an ancient group of marine red microalgae and seaweeds, also known as the red plastid lineage; and the glaucophyta, which comprise a small group of freshwater microalgae. A second endosymbiosis is hypothesized to have occurred ∼1 billion y ago, in which a second heterotroph engulfed and retained a member of one of these clades. Both chlorophytes and rhodophytes gave rise during secondary endosymbiosis events to dominant lineages of algae, such as stramenopiles, dinoflagellates, cryptophytes, and haptophytes.17,18 The immense diversity of algal groups, as derived from their unique evolutionary history (Fig. 1), gives an interesting opportunity to examine key cellular metabolic pathways across different branches of the eukaryotic tree of life. In contemporary oceans, the most abundant, diverse, and ecologically important eukaryotic algae are the ones descended from the red plastid lineage.2 These successful phytoplankton communities belong to the chromalveolata clade, including coccolithophores, diatoms, and dinoflagellates, which originated ∼250 million y ago and compose massive blooms in the ocean.19
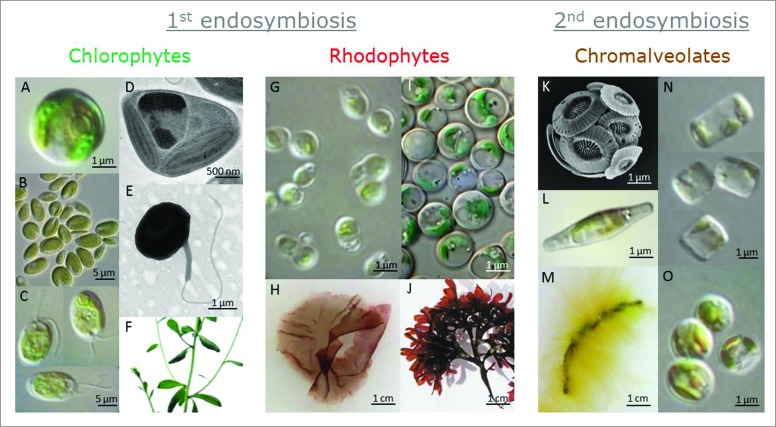
Figure 1.
A variety of fully sequenced algal species. Chlorophytes, land plants, and rhodophytes evolved through primary endosymbiosis, and chromalveolates evolved through secondary endosymbiosis. Representative species that were studied in the current work: (A) Chlorella. (B) Coccomyxa (Image courtesy of Thomas Proeschold). (C) Chlamydomonas. (D) Ostreococcus (Image courtesy of Martin F. Hohmann-Marriott). (E) Micromonas (Image courtesy of Daphné Grulois, Sophie Lepanse, and Nathalie Simon). (F) Arabidopsis thaliana was used as a representative terrestrial viridiplantae. (G) Cyanidioschyzon merolae. (H) Pyropia sp. (I) Galdieria sulphuraria (Image courtesy of Gerald Schoenknecht). (J) Chondrus crispus (Image courtesy of Inga Kjersti Sjøtun). (K) Emiliania huxleyi (Image courtesy of Daniella Schatz). (L) Phaeodactylum tricornutum. (M) Ectocarpus sp. (N) Thalassiosira pseudonana. (O) Nannochloropsis sp.
Although the ecophysiology and molecular ecology of some algal species are extensively studied, some basic aspects of algal cell biology are still underexplored. Here we wish to elucidate the genomic potential for a well-studied and pivotal cellular pathway, known as autophagy, in marine and freshwater algae. Autophagy is a catabolic process that degrades a variety of intracellular substrates, such as long-lived or defective proteins, protein aggregates, and aging organelles. Basal autophagy is important in balancing anabolic and catabolic pathways, and is therefore required to maintain the general cell homeostasis.20 In addition, autophagy is an effective defense mechanism against pathogens and recycling machinery for macromolecules that helps the cell to cope with nutrient scarcity.20-22 Along with its survival role, autophagy can lead to programmed cell death upon severe stress.23,24 Autophagy is negatively regulated by the target of rapamycin complex 1 (TORC1).25 TORC1 includes the kinase TOR as the catalytic subunit, the scaffold protein RPTOR (regulatory associated protein of MTOR, complex 1; Kog1 in yeast), as well as MLST8 (MTOR associated protein, LST8 homolog [S. cerevisiae]; Lst8 in yeast), which stabilizes the MTOR-RPTOR interaction.26-27 Nonphotosynthetic organisms contain a second TOR complex (TORC2) which consist of TOR, LST8, and additional members, and controls cytoskeleton organization.28 The autophagic process itself is mediated by more than 30 autophagy-related (ATG) proteins (Atg in yeast nomenclature). The current study will focus on ATG proteins which are obligatory for autophagosome biogenesis, which constitute 3 core functional units: The Atg9 cycling system, the class III phosphatidylinositol 3-kinase (PtdIns3K) complex and the ubiquitin-like (Ubl) conjugation system.29,30 Atg9 and its cycling machinery (Atg1, Atg2, Atg13, Atg18, Atg27) are negatively regulated by TORC1 and mediate the formation of autophagosomes, a unique double-membrane vesicles.25,31,32 Recruitment of additional autophagic factors to the maturing autophagosome is mediated by the PtdIns3K complex, which is composed of Vps34, Vps15, Vps30/Atg6 (BECN1 in mammals), Atg14, and Atg38.33 The maturation and fusion of the autophagosome with the lytic organelle (lysosome or vacuole) requires the activation of the Ubl conjugation machinery (Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16).34-37
The molecular machinery described above is extensively investigated in yeast, plants, and animal models. Accumulating evidence in plant research demonstrates that autophagy is triggered upon carbon and nitrogen starvation, specific developmental stages, senescence, pathogen infection, hypersensitive response, and also during normal metabolism.38-42 Whereas yeast serves as a common model organism to study autophagy, the role of ATG proteins has recently been studied in a few other protists.43-45 However, very little information is available regarding the conservation and origin of autophagy across the diverse aquatic phototrophs and about the importance of autophagy during acclimation to environmental stress conditions. Since the origin of these microorganisms spans divergent branches in the evolutionary tree of life which are largely underexplored, deciphering the conservation of genes, pathways, and cellular hallmarks related to autophagy will shed new light on its origin and diverse function.
Results
Genomic analysis of the autophagy pathway in key phytopl-ankton species
Recently published genomes of algae enable us to explore autophagy and other fundamental cellular processes at the molecular level. In order to identify orthologs of ATG genes from yeast and mammalian models, we conducted extensive genomic analysis and in-silico predictions on a total of 17 phylogenetically diverse algal species. Among these, 7 sequenced representatives of the green plastid lineage were chosen. These ”green algae” are unicellular chlorophytes, including Chlorella variabilis, Chlamydomonas reinhardtii, Coccomyxa subellipsoidea, Volvox carteri, Micromonas (strains pusilla CCMP 1545 and sp. RCC299) and Ostreococcus lucimarinus.46-51 In addition, the genome of the model land plant Arabidopsis thaliana was used as a representative terrestrial viridiplantae.52 We also analyzed the genomes of 5 rhodophytes: the microalgae Porphyridium purpureum, Galdieria sulphuraria, and Cyanidioschyzon merolae, and the macroalgae Pyropia yezoensis and Chondrus crispus.53-57 Furthermore, 5 available genomes of chromalveolates were analyzed; the coccolithophore Emiliania huxleyi, the diatoms Thalassiosira pseudonana, and Phaeodactylum tricornutum, the marine Nannochloropsis gaditana and the filamentous algae Ectocarpus siliculosus (Fig. 1).58-62
We searched algal genomes for ATG protein orthologs using the translated Basic Local Alignment Search Tool (TBLASTN), Phytozome v9.1, and the Joint Genome Institute (JGI) portal for most of the genomes (see Methods). Tables 1 to 4 summarize the distribution of TORC1 proteins and 17 essential ATG proteins (designated as protein IDs), which are encoded by the algal genomes. Identification of the ATG genes was based on the similarity of the total protein sequence to the yeast, plant and mammalian orthologs, and on the presence of conserved domains that were previously characterized based on the conserved domain database (CDD) at NCBI.63 Moreover, we manually defined sequences that were missing or poorly annotated in the current genome annotation (Fig. S1). For example, we constructed the sequences of ATG5 and ATG12 in V. carteri; ATG1, ATG2, and ATG5 in T. pseudonana; TOR, RPTOR, and LST8 in P. purpureum; TOR and RPTOR in P. yezoensis; TOR and LST8 in C. crispus; and all TORC1-encoding and ATG genes in E. huxleyi. The expression of the predicted genes in chromalveolates was verified by meta-analysis of recent publically available expression datasets (Fig. 2, S2 to 4). In addition, the expression of TOR1, TOR2, ATG2, ATG3, ATG5, VPS30/ATG6, ATG7, ATG8a, ATG8b, ATG12, and VPS34 in E. huxleyi was verified by real-time qPCR (Fig. S2B).
Table 1.
Distribution of TOR and related proteins in algae
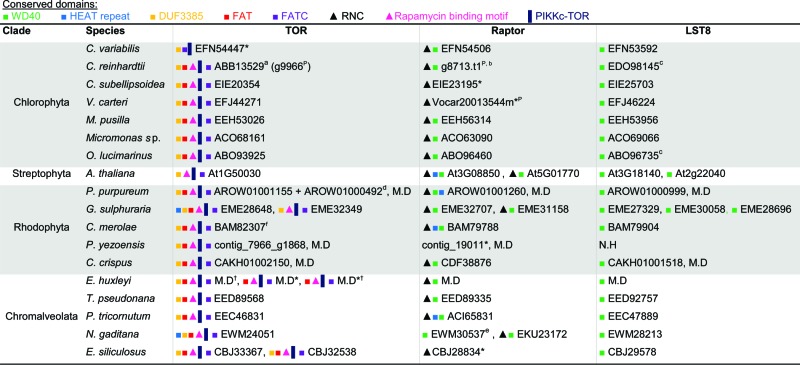 |
ID numbers refer to the NCBI database. N.H, no homolog identified. M.D, manually defined sequences, which are available in Figure S1 (DNA ID numbers are indicated).
P. yezoensis protein and DNA IDs refer to the P. yezoensis genome database; Arabidopsis homologs refer to the TAIR database (see Methods) and were previously described.67
Domain Accession numbers: WD40 cd00200/cl19087; HEAT pfam13646; DUF3385 pfam11865; FAT pfam02259; FATC pfam02260; RNC pfam14538; Rapamycin binding pfam08771; PIKKc-TOR cd05169.
Phytozome database (see Methods).110
According to genome version 1.65 In brackets is the same genomic locus in the updated version of the genome (no.5).
Reference: Díaz-Troya et al., 2008.67
Reference: Díaz-Troya et al., 2008.64
IDs represent genomic DNA contigs that were concatenated in order to build the gene.
Strain b-31. Only the middle of the protein was identified. EKU23172 refers to strain CCMP526.
Reference: Imamura et al., 2013.66
Only partial sequence is available.
Validated by real-time qPCR (Fig. S2B).
Table 2.
Distribution of ATG proteins that are involved in the ATG9 cycling system in algae
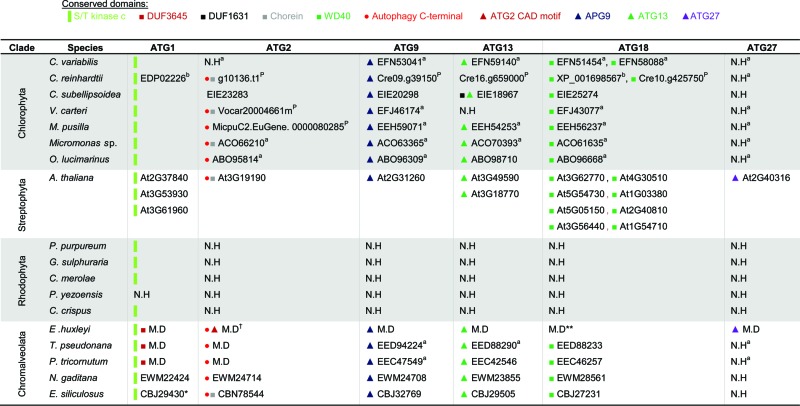 |
ID numbers refer to the NCBI database. N.H, no homolog identified. M.D, manually defined sequences, which are available in Figure S1.
Arabidopsis homologs refer to the TAIR database (see Methods), and were previously described.67, 100
Domain Accession numbers: S/T kinase c smart00220; DUF3645 cl13493/pfam12063; DUF1631 pfam07793; Chorein pfam12624; WD40 cd00200; Autophagy C-terminal pfam09333; ATG2 CAD motif pfam13329; APG9 pfam04109; ATG13 pfam10033; ATG27 pfam09451.
Phytozome database (see Methods).110
Reference: Jiang et al., 2012.99
References: Díaz-Troya et al., 2008; Avin-Wittenberg et al., 2012.67, 100
According to CDD version 3.11, the DUF3645 domain is not conserved. Yet, according to pfam, DUF3645 domain was detected, but below the default cutoff.
According to CDD version 3.11, the WD40 domain is not conserved. Yet, according to pfam, WD40 domain was detected, but below the default cutoff.
Validated by real-time qPCR (Fig. S2B).
Table 3.
Distribution of ATG proteins that are involved in the PtdIns3K complex in algae
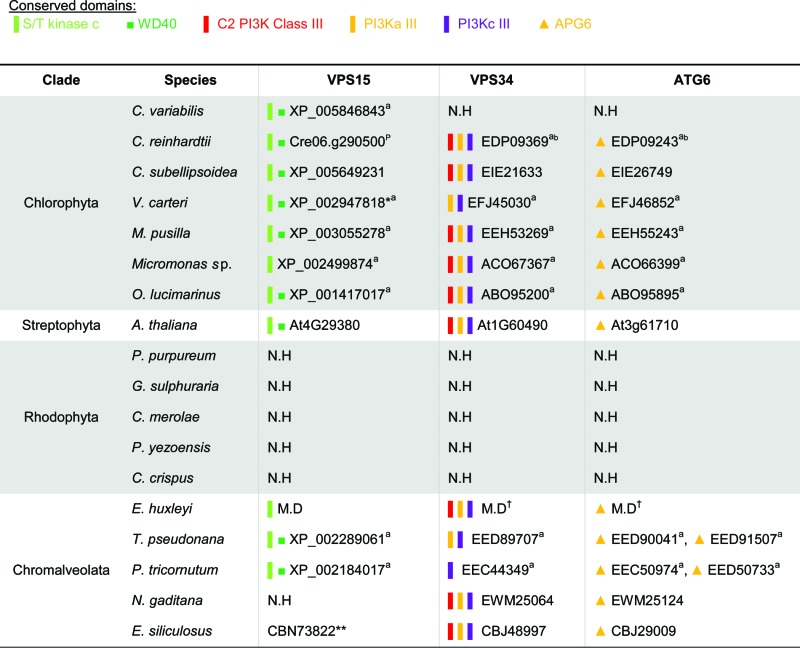 |
ID numbers refer to the NCBI database. N.H, no homolog identified. M.D, manually defined sequences, which are available in Figure S1.
Arabidopsis homologs refer to the TAIR database (see Methods), and were previously described.67, 100
Domain Accession numbers: S/T kinase c smart00220; WD40 cl02567; C2 cl14603/pfam00792; PI3Ka_III cl00271/cd00870; PI3Kc_III cd00896; APG6 pfam04111.
Phytozome database.110
Reference: Jiang et al., 2012.99
Reference: Avin-Wittenberg et al., 2012.100
From position 880, as the first half of the sequence is a mistaken fusion of unrelated RNA binding protein.
Missing the kinase domain, but can be found by BLAST directly upstream, predicted wrong.
Validated by real-time qPCR (Fig. S2B).
Table 4.
Distribution of ATG proteins that are involved in the Ubl-like conjugation system in algae
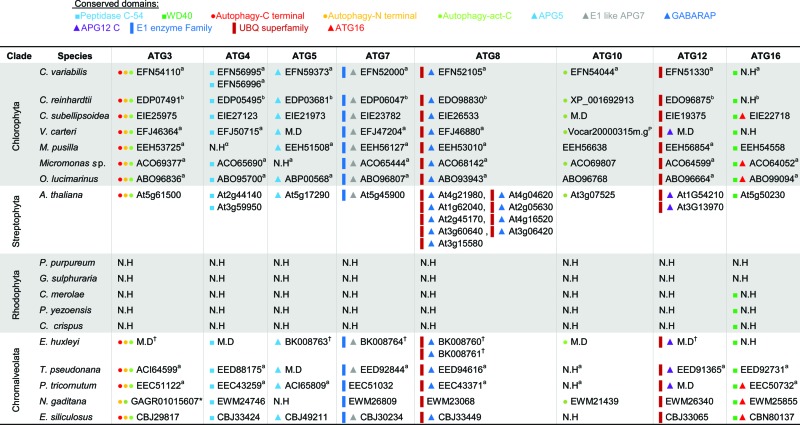 |
ID numbers refer to the NCBI database. N.H, no homolog identified. M.D, manually defined sequences, which are available in Figure S1.
Arabidopsis homologs refer to the TAIR database (see Methods) and were previously described.67, 100
Domain Accession numbers: Peptidase C-54 pfam03416; WD40 cd00200; Autophagy-C terminal pfam10381; Autophagy-N terminal pfam03986; Autophagy-act-C pfam03987; APG5 pfam04106; E1 like APG7 TIGR01381; GABARAP cd01611; APG12 C cd01612; E1 enzyme family cl17196; UBQ superfamily cl00155; ATG16 pfam08614.
Phytozome database.110
Reference: Jiang et al., 2012.99
References: Díaz-Troya et al., 2008; Avin-Wittenberg et al., 2012.67, 100
No protein ID was found. NCBI Transcriptome shotgun assembly (TSA) accession number is indicated.
Validated by real-time qPCR (Fig. S2).
Figure 2.
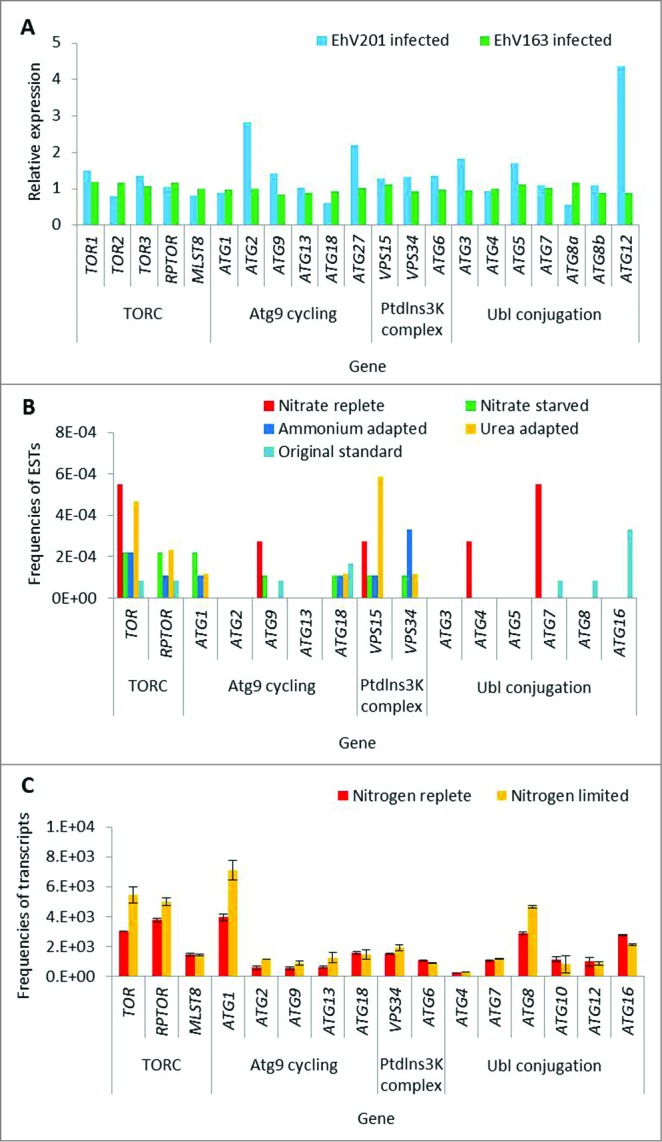
Expression of TOR and autophagy-related genes in chromalveolates. (A) Expression level of ATG and TOR genes in transcriptome of E. huxleyi cells during infection with EhV (24 h post infection). Two different viral strains were used (lytic, EhV201; non-lytic, EhV163).72 The expression level of each gene in the infected cells was normalized to the level of expression in the uninfected control cells, at the same time point. RNA-seq-based gene expression profiles are presented as RPM values. (B) Expression levels in EST libraries derived from P. tricornutum cells grown under different nitrogen regimes. Level of expression is presented as frequencies of ESTs in each library.76 (C) Transcriptome analysis of N. gaditana cells grown under nitrogen limitation. Error bars represents biological duplicates.61
Conservation of autophagy genes in the red vs. green plastid lineage
The conservation and functionality of TORC1 components has been described previously in C. reinhardtii and C. merolae.64-67 We identified TORC1 proteins (TOR, RPTOR, and LST8) as gene products in additional algal genomes. The sequence coding for Tco89, a TOR-interacting protein that is described as part of the yeast TORC1, is not detected in the algal genomes.68 However, TOR, RPTOR, and LST8 homologs are highly conserved in all species of interest (Table 1). The N-terminus of yeast and mammalian TOR contains a HEAT repeat (named after the proteins in which it was found, Huntingtin, EF3, regulatory subunit A of PP2A, and TOR1. pfam13646) involved in proper localization,69 which is missing from most of the algal homologs. Conversely, most algae maintain accessory domains in addition to the catalytic kinase domain (cd05169), such as the FAT (named after the proteins in which it was identified: FRAP, ATM, TRRAP, pfam02259) and FATC (pfam02260) domains, which are important for protein-protein interaction, and shared by all PtdIns3K-related kinases.69,70 The rapamycin-binding motif (pfam08771) is conserved, and may be a putative binding site for the FKBP12-rapamycin complex to specifically inhibit TOR.69 G. sulphuraria and E. siliculosus encode 2 TOR proteins, and E. huxleyi encodes 3 different TORs. In yeast, 2 TOR proteins are active in 2 distinct complexes and account for different functionality.26 However, our homology-based and domain conservation analysis is not sufficient to determine if the algal homologs we identified are TOR1- or TOR2-like proteins. Regarding RPTOR, all algal orthologs maintain the unique RAPTOR N-terminal (RNC) domain (pfam14538) and the C-terminal WD40 repeats (cl19087).71 However, as in the TOR homologs, the HEAT repeats are not as conserved in the algal RPTOR orthologs as they are conserved in other eukaryotes. Downstream of TOR signaling, the autophagy machinery is highly conserved in the green plastid lineage and in chromalveolates, with minor exceptions. In order to strengthen our functional annotations, we applied meta-analysis on available gene expression databases from chromalveolates and confirmed that the predicted TOR and ATG genes are expressed (Fig. 2, S2 to 4). In E. huxleyi, all 3 TOR isoforms, the TORC1-encoding genes and ATG genes were generally expressed (Fig. 2A, S2).72 Notably, infection with E. huxleyi-specific virus (EhV), an important pathogenic driver of E. huxleyi bloom demise at sea,73 induced specific ATG genes (Fig. 2A and ref. 74). Differential expression of ATG genes was also detected in expressed sequence tag (EST) libraries derived from different E. huxleyi strains, which differ in their viral susceptibility.75 The E. huxleyi strain RCC1216 is a diploid, viral-sensitive host, which expressed higher level of ATG genes, when compared with strain RCC1217, a haploid, viral-resistant cell (Fig. S2A). This is consistent with the observation that diploid cells exhibit greater transcriptome richness which may support their versatility and wide distribution, as compared to haploid cells.75 In P. tricornutum, ATG- and TOR-related transcripts were detected in 16 EST libraries derived from different growth conditions of ecological relevance, including stress conditions (Fig. 2B, S4).76 In N. gaditana, an important model algae to study biofuel production, ATG- and TOR-related transcripts were detected in transcriptome profiling during nitrogen replete and limited growth, and higher levels of TOR, RPTOR, ATG1, and ATG8 transcripts were reported upon nitrogen deficiency. Expression of ATG3 cannot be detected since ATG3 is not defined in the Nannochloropsis gene database, even though it is present in the genome sequence, as can be seen by translated searches directly against the genome (Fig. 2C, Table 4).61 In the diatom T. pseudonana, an important ecological model organism to study algal physiology and acclimation to stress, ATG- and TOR-related transcripts were expressed in EST libraries derived from phosphate-replete and -limited growth, yet without obvious differences in expression level between the 2 libraries (Fig. S3).77 In stark contrast, none of the 17 ATG genes from the 3 functional units (ATG9 cycling system, PtdIns3K complex and Ubl conjugation machinery) were identified in rhodophytes (Tables 2 to 4). Queries from different genomes such as E. huxleyi, E. siliculosus, A. thaliana, S. cerevisiae, and a variety of algae were used against the rhodophyte genomes, yet no comparable sequence was detected. ATG genes are probably not conserved in rhodophytes as in other eukaryotes. This raises a fundamental question regarding the evolutionary origin and function of algal ATG proteins during acclimation to environmental stress conditions in aquatic ecosystems.
Composition and conservation of the ATG9 cycling system
Upon induction of autophagy, Atg9 delivers lipids to the developing sequestering vesicles. Atg9 trafficking in the yeast cytoplasm is mediated by Atg1, Atg2, Atg11, Atg13, Atg18, and Atg27.32,78-80 We found that ATG9 is highly conserved in the green lineage and chromalveolate genomes (Table 2). However, the proteins associated with the ATG9 cycling system are partially conserved. ATG1 orthologs in chromalveolates maintain a serine/threonine protein kinase domain in the carboxy terminal (smart00220), and a DUF3543 domain (pfam12063), whose function is unclear, in the N-terminal. In chlorophytes, the DUF3543 domain is completely missing, and only the kinase domain is present. Moreover, the putative kinase domain of ATG1 is the only available match in rhodophytes, which lack any other autophagic components, suggesting that this match is not ATG1-specific. ATG18 is conserved in the green lineage and in chromalveolates, including the characteristic WD40 domain (except in E. huxleyi, where there is a poorer conservation of the WD40 domain). ATG2 is conserved in the green lineage (except for C. variabilis) and in chromalveolates, including an autophagy-related domain in the carboxy-terminus (pfam09333). Several algal ATG2 orthologs also maintain an N-terminal region of VPS13A/chorein (pfam12624), which presumably participates in protein trafficking between membrane compartments in the cytoplasm.63,81 E. huxleyi ATG2 is the only ortholog in which a CAD motif (pfam13329) is conserved, yet the function of this region is unknown. ATG13 is conserved in the green lineage (except for V. carteri) and in chromalveolates. Finally, ATG27 is the least conserved protein among the ATG9 cycling members, as it was identified only in the genomes of E. huxleyi and A. thaliana.
Composition and conservation of the PtdIns3K complex
The PtdIns3K complex is integral for autophagosome formation and maturation.33,82 The lipid kinase Vps34 generates phosphatidylinositol 3-phosphate, a central molecule involved in cellular membrane trafficking. Vps15 acts as a Vps34 regulator,83 and Vps30/Atg6 interacts with other proteins which are involved in different cellular processes, converging signals, and balancing between autophagy and other signaling cascades.33,84 The PtdIns3K complex members are well conserved in the algal genomes, with the exception of rhodophytes and VPS30/ATG6/BECN1 in C. variabilis (Table 3). None of the algal VPS30/ATG6/BECN1 orthologs contain a BCL2 homology 3 (BH3) domain, that has been characterized in mammals.85 This is not surprising in light of the major loss and divergence of canonical apoptosis machinery in protists and plants, which lack the BCL2 and TP53 components.86,87
Composition and conservation of the Ubl conjugation machinery
Synthesis and elongation of the autophagic membrane require 2 distinct Ubl conjugation reactions. Atg12 is a ubiquitin-like protein whose coupling to Atg5 is mediated by Atg7, an E1-like enzyme, and Atg10, an E2-like enzyme.36 The Atg12-Atg5 conjugate associates with Atg16, and the resulting complex probably serves as an E3 ligase in a subsequent Ubl conjugation reaction.37 In the second reaction, the cysteine protease Atg4 cleaves the nascent form of Atg8, which is also structurally similar to ubiquitin,88 in order to expose a glycine residue at the carboxy terminus of Atg8.35,89 Atg7 initiates the conjugation of the processed Atg8, while Atg3, an E2-like enzyme, catalyzes the formation of an amide bond between phosphatidylethanolamine and the exposed glycine of Atg8.89 Excluding rhodophytes, almost all the Ubl conjugation machinery components are encoded in the algal genomes (Table 4). Notably, ATG3 and ATG4 are highly conserved, including the typical peptidase C-54 domain (pfam03416) in ATG4. Likewise, 3 conserved regions are maintained in the predicted Atg3 homologs: an N-terminal autophagy domain (pfam03986); an active site (pfam03987) that includes a Cys234 residue within an HPC motif, which is probably important for ATG5 recognition; and a carboxy terminal autophagy domain (pfam10381), that includes the FLKF sequence motif, which is likely to be a distinct binding region for the stabilization of the autophagosome complex.90 ATG10 is only partially conserved in algae, as it is missing in T. pseudonana, P. tricornutum, and E. siliculosus. The ATG16 protein sequence is problematic to predict accurately, as it contains a WD40 domain. WD40 is shared by a wide superfamily of proteins that covers a variety of functions,91 and therefore cannot guarantee the specific conservation of ATG16. Yet the presence of an ATG16 domain (pfam08614), together with the WD40 domain, suggests that ATG16 is conserved in specific algal genomes (Table 4). A single ATG8 copy was identified in each of the algal genomes, with the exception of 2 putative ATG8 homologs in E. huxleyi.
Conservation of putative functional residues within central autophagy proteins
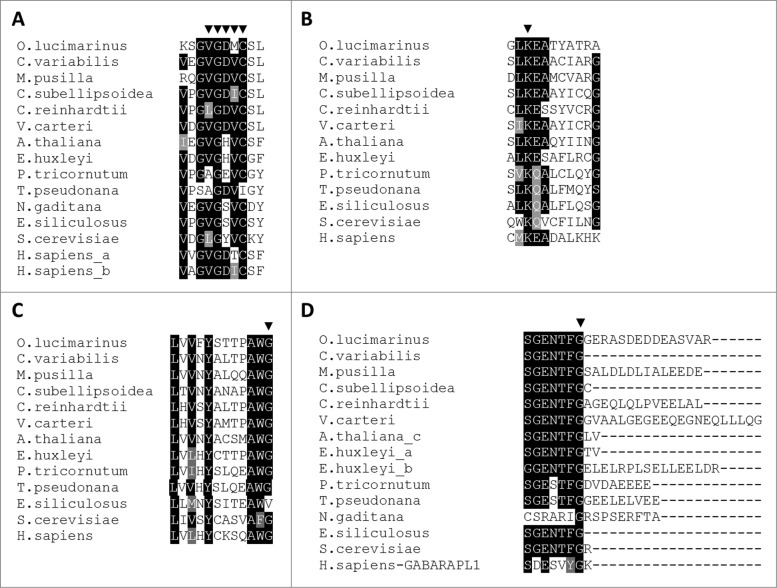
In order to examine the putative functionality of the predicted ATG homologs, we investigated specific residues, which were reported in earlier biochemical studies to be critical for the specific activity and function of ATG5, ATG8, ATG9, and ATG12. In yeast, Atg9 self-interaction via the conserved LGYVC motif is essential for the function of Atg9 as a lipid carrier to the forming autophagosome membrane (positions 766 to 770, according to the yeast Atg9 sequence).92 We identified the LGYVC motif in all algal ATG9 orthologs, in which the Gly and Cys are the most conserved, and the Tyr is the least canonical residue, in agreement with previous reports from yeast and mammals (Fig. 3A).92 Next, we confirmed that the algal ATG5 and ATG12 orthologs maintain the amino acids that form an isopeptide bond during ATG12-ATG5 conjugation, including the conserved Lys in ATG5 and Gly in ATG12 (Fig. 3B to C). The E. siliculosus ATG12 is the only ortholog that lacks the carboxy terminal glycine, and instead encodes a valine residue (Fig. 3C). Finally, during the Atg8 lipidation reaction, the carboxy terminal glycine is coupled to the phospholipid phosphatidylethanolamine (position 116, according to the yeast sequence).35,89 This glycine is conserved in the algal ATG8 homologs. Notably, the ATG8 proteins of O. lucimarinus, M. pusilla, V. carteri, and E. huxleyi (ATG8b), contain an unusually long (8 to 21 amino acids) and divergent peptide at the carboxy terminus (Fig. 3D). A similar extension was reported in the ATG8 sequence of C. reinhardtii.93 However, the function of this extension and the peptide that results from the cleavage of ATG8 at the carboxy terminal glycine is unknown.
Figure 3.
Conservation of amino acid residues that are essential for ATG protein interaction and function. Amino acid sequence alignments of ATG5, ATG8, ATG9, and ATG12, from green algae, chromalveolates, plants, yeast, and human. (A) ATG9 C-terminal sequence. The arrowheads indicate residues 763 to 772 (according to the yeast sequence), which are essential for ATG9 self-interaction. (B) ATG5 N-terminal sequence. The arrowhead indicates the lysine residue that is required for the isopeptide bond with ATG12. (C) ATG12 C-terminal sequence. The arrowhead indicates the glycine residue that is required for conjugation with ATG5. (D) ATG8 C-terminal sequence. The arrowhead indicates the processing site by ATG4. Identical and similar residues are shaded in black and gray, respectively. Multiple sequence alignment was performed with ClustalW version 2.1 using the default parameters.
Phylogeny of ATG8 and ATG9
Phylogenetic analysis of 76 homologs of yeast Atg8 across eukaryotes shows that ATG8 in green algae forms a separate clade, which is related to land plants (Fig. 4A, S5). Based on the phylogenetic distribution, ATG8 homologs in green algae comprise 3 main branches: prasinophyceae (Ostreococcus, Micromonas), trebouxiophyceae (Chlorella, Coccomyxa), and volvocaceae (Chlamydomonas, Volvox). The ATG8 protein of the glaucophyte Cyanophora paradoxa was clustered together with the prasinophytes. ATG8 homologs in chromalveolates emerged in a separate branch, which is related to amoeba and fungi. Notably, the E. huxleyi ATG8b homolog diverged away from chromalveolates, yet with low bootstrap values. The ATG8 sequence of N. gaditana was not aligned properly with the other sequences, because the sequence in the database has extensions and may not have been constructed properly, and so it was omitted from the current analysis.
Figure 4.
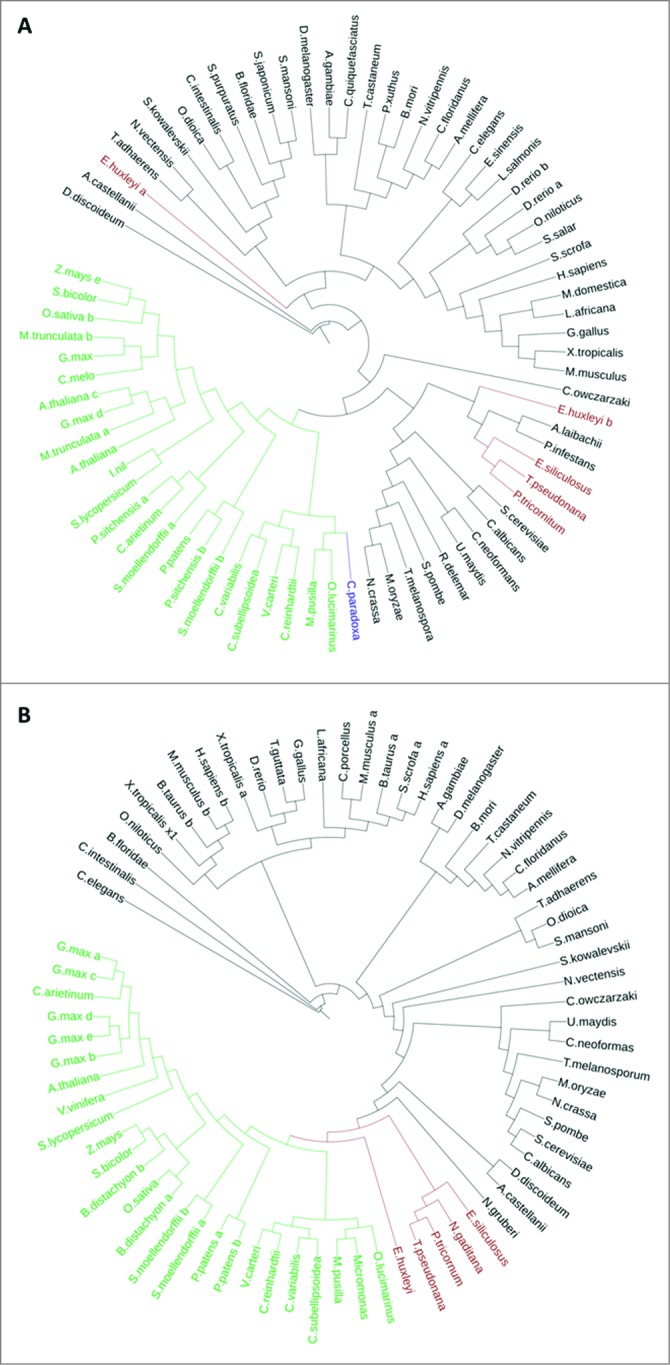
Phylogenetic trees of ATG8 and ATG9 proteins. (A) ATG8 Protein sequences were aligned using Muscle version 3.8.31. (B) ATG9 domain (cl04403) sequences were aligned using ClustalW version 2.1.113 Maximum Likelihood trees (Phylip ProML) are shown. Highlighted in green are chlorophytes and land plants, in brown are chromalveolates, and in blue is the glaucophyte.
A similar phylogenetic analysis was conducted with 72 ATG9 orthologs (Fig. 4B, S5). The phylogeny of ATG9 in green algae resembles their evolutionary history, as was demonstrated by the ATG8 phylogeny. ATG9 orthologs in chromalveolates diverged as a separate clade between green algae and amoeba, while the ATG9 ortholog of E. huxleyi emerged in a closely related, but separate, branch.
Discussion
Autophagy genes are not conserved in the genomes of red algae
The current bioinformatics study argues that ATG orthologs are not conserved in unicellular and multicellular red algae as in green algae, chromalveolates, and other eukaryotes. To our knowledge, this is the first study to report such phenomena, which revisits the common notion that ATG genes are generally conserved in all eukaryotes. If red algae functionally maintain autophagy, the proteins that activate this putative machinery are considerably different from their yeast, plants, and mammalian counterparts. The absence of ATG genes in the red algal genomes stands in contrast to the conserved TORC1-encoding genes. Hypothetically, the putative TORC1 may control catabolic processes in red algae, which may functionally compensate for the autophagic machinery.
How did specific red algae adapt to diverse environmental niches without autophagy, which serves as an innate survival response in eukaryotes?94,95 Are there alternative survival mechanisms for autophagy in red algae? Such questions remain elusive and intriguing, and demand experimental evidence, especially in light of the high stress tolerance to extreme environmental conditions of high temperature and acidity which are attributed to some red algae.54,96
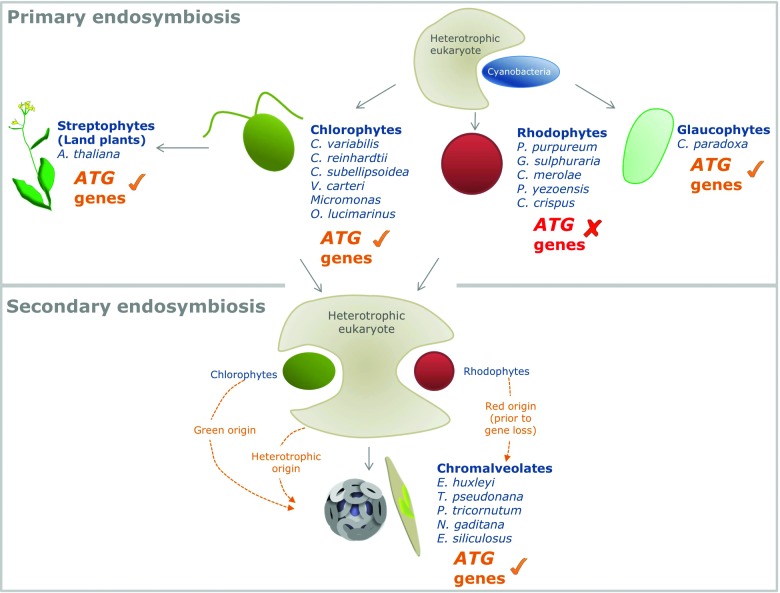
As chlorophytes and glaucophytes derived from primary endosymbiosis, theoretically, their autophagy pathway could have been acquired from the photosynthetic cyanobacterium or the eukaryotic heterotroph that engulfed it. We searched for ATG genes in the genomes of 31 cyanobacteria strains. No bacterial ATG gene was identified, suggesting that the autophagy pathway that we identified probably has a heterotrophic origin, and perhaps was acquired multiple times through evolution. Theoretically, rhodophyte genomes might have once encoded an autophagy pathway, but lost it. The reduced genome content in essential metabolic pathways in C. crispus (105 Mbp) and C. merolae (16.5 Mbp) supports this theory.55,57 Genome size is hypothesized to be reduced by strong selection pressure, and facilitates the invasion of C. crispus into extreme habitats.57 However, reduction in genome size alone cannot account for ATG gene loss in red algae, because some non-red algal genomes are also relatively small (Nannochloropsis, ~28Mbp; Ostreococcus, ~13Mbp), and notwithstanding contain a variety of ATG genes.61,51
Red algal genomes contributed via secondary endosymbiosis to several chromalveolate lineages, including stramenopiles and haptophytes.17 Since canonical ATG genes are missing in red algae, the conservation of ATG genes in chromalveolates is puzzling (Fig. 5). Considering red algae once encoded ATG genes, the origin of chromalveolates may have taken place prior to, or post-ATG gene loss in the red algal genomes. In the former scenario, chromoalveolate ATG genes may be of red algal origin. In the second hypothetical scenario, the origin of chromoalveolate ATG genes may be attributed to the heterotroph that engulfed a green or red alga during secondary endosymbiosis. Another possibility is that chromoalveolate ATG genes are of green algal origin, and were transferred by cryptic plastid endosymbiosis and/or by horizontal gene transfer.18
Figure 5.
A possible loss of the autophagy pathway from red alga (Rhodophyta) genomes. Primary endosymbiosis gave rise to the green algae and glaucophytes, which possess ATG genes, and to red algae, whose genomes lack ATG genes. Red algae possibly lost their autophagy machinery before or after secondary endosymbiosis, which gave rise to chromalveolates. The origin of ATG genes in chromalveolates may be attributed to the heterotrophic host that engulfed a red or green alga, to a red alga (considering that ATG gene loss in red genomes occurred after the secondary endosymbiosis event), or to a green algae (orange arrows). The distribution of ATG genes in algae raises fundamental questions regarding the origin of autophagy and the inheritance of ATG genes during the evolution of photosynthetic organisms.
Autophagy is conserved in green algae and chromalveolates
Here, we demonstrate the remarkable conservation of the ATG genes in green algae and chromalveolates, in agreement with previous evidence for functional autophagy in these algae in response to the TOR-inhibitor rapamycin, nitrogen and carbon starvation, aging, oxidative stress, carotenoid deficiency, and heterotrophic growth conditions.67,93,97-100 By manually curating genes, we were able to detect essential homologs that were previously suggested to be missing, and are available as Figure S1. In addition, we were able to define at least 5 key autophagy genes (ATG1, ATG3, ATG7, ATG8, and VPS15) in C. paradoxa, the only glaucophyte whose genome was sequenced (Fig. 4A, S1).101 The presence of a single ATG8 gene in algae (except for E. huxleyi, where there are 2) corroborates the notion that the increase in genome complexity of higher metazoans includes extended subfamilies of ATG8 as compared to protists and invertebrates.88 The A. thaliana ATG8 gene family comprises 9 isoforms, with different spatial and temporal expression patterns.38,41,102 The major expansion of the ATG8 family in plants, as compared to algae, suggests late duplication events of this pivotal protein in autophagy. This may also be true for animals, where 3 ATG8-subfamilies are defined: MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), GABARAP (GABA[A] receptor-associated protein) and GABARAPL2/GATE-16.88 Sequence homology suggests that E. huxleyi ATG8 homologs, as all other algal ATG8, belong to the GABARAP subfamily. Unlike ATG8, proteins that contain general kinase or WD40 domains are very difficult to predict because they are shared among numerous protein families.91 For example, the Atg1 kinase, which directly phosphorylates Atg9,79 cannot be predicted accurately based on the presence of the kinase domain alone. In contrast, some ATG orthologs are clearly missing in specific algal genomes. In all chromalveolates but E. huxleyi, ATG10 is not found, although other ATG genes are highly conserved. In addition, ATG2, VPS34, VPS30/ATG6, ATG16, and ATG27 are not conserved in the genome of C. variabilis.99 The absence of specific ATG genes, together with the relatively high conservation of ATG8 and other ATG genes, suggests that some ATG proteins may be dispensable in certain models. However, in A. thaliana, ATG10 mutants are hypersensitive to nitrogen and carbon starvation, and undergo early aging and cell death, and VPS30/ATG6 mutants are defective in pollen germination.103,104 Therefore, it is plausible that in certain algae like C. variabilis, autophagy (or a specific functional unit within the autophagic machinery), does not operate in the same manner as in higher photosynthetic organisms. The absence of specific ATG genes from the genomes of green algae and chromalveolates may either be because the genomes are not sequenced to completion, leading to ‘missing’ genes, or because unknown alternative components substitute for the missing proteins during autophagy.
E. huxleyi, an ecologically important model to study autophagy in marine protists
E. huxleyi, a cosmopolitan bloom-forming coccolithophore, whose genome encodes for the most complete repertoire of ATG proteins, may be an attractive model to study autophagy in unicellular algae. E. huxleyi derived from secondary endosymbiosis and is one the most abundant algae in modern oceans.2 E. huxleyi blooms play prominent roles in marine biogeochemical cycles of carbon and sulfur, and create a massive flux of calcium carbonate to marine sediment, as well as dimethylsulfide flux to the atmosphere.8,14-16 Infection by E. huxleyi virus (EhV, phycodnaviridae), a specific large DNA virus, is a major factor that routinely terminates E. huxleyi blooms.7,105 Recent work demonstrates that autophagy hallmarks are induced in EhV-infected cells (Fig. 2A and ref. 74). Viral infection is accompanied by the presence of double-membrane vesicles in the cytosol. Furthermore, elevated transcription of ATG5, ATG7, and ATG8 and lipidation of ATG8 suggest that specific autophagy components are induced in response to viral attack. Moreover, the host ATG8 protein is detected in the membranes of highly purified virions.74 This intriguing data, together with the observation that the virus is composed of several membrane layers,106 suggests that EhV may benefit from enhanced formation and remobilization of membranes during host autophagy, as a source for new viral membranes.74
The fact that autophagy is highly conserved in coccolithophores, as well as in diatoms, raises fundamental questions regarding the role that autophagy may play during acclimation response to environmental stress conditions, and contribute to their ecological success. The current genomic analysis may serve as a valuable resource for future laboratory-based exploration, and illuminate the role of autophagy on the cellular and population-level fitness during bloom succession in the ocean.
Methods
Gene definition
The ATG and TORC1-related gene sequences in E. huxleyi were manually defined and aligned, and phylogenetic trees were built in order to prove family relationships as described in detail by Feldmesser et al.107 In brief, a target gene was chosen, and the protein sequences from human, Arabidopsis and yeast were taken from the National Center for Biotechnology Information (NCBI). The protein sequences were blasted directly against the genome sequence of E. huxleyi at the Joint Genome Institute (JGI). All hits were analyzed to see if there was any transcript evidence (ESTs) or gene models. A gene model was then manually built based on the existing transcripts, models, and the BLAST results. If no hits were found with the sequences taken from the initial 3 species, BLAST searches were run at NCBI to find the target gene in potentially more closely related species, and then these were used as input sequences. When the transcriptome sequences of E. huxleyi became available,72 they were used to improve the manual gene models. After the gene in E. huxleyi was defined and translated, the protein sequence that was used for searches against the protein collections of NCBI to ensure that the gene constructed was the homolog of the desired target. The protein sequences were analyzed for the presence of the proper domains, related sequences were collected, and phylogenetic trees were built. For the other better-studied species, the initial analysis was BlastP at NCBI.108 In addition, the same approach taken for E. huxleyi was used for all the other species, with searches against the genome databases, to ensure the genes were defined correctly. The E. huxleyi sequence was also used as a target input for other algae, and in the case of rhodophyta, all the sequences we were able to identify or define as ATG genes in this study were used as additional input target sequences. In several cases in species other than E. huxleyi, the sequences were manually defined. In addition to searching the algal genomes, their protein collections were also used, at NCBI, the individual genome databases, Pico-Plaza (http://bioinformatics.psb.ugent.be/plaza/versions/pico-plaza/),109 and RNA collections available either at the genome databases or the NCBI TSA (Transcript Shotgun Assembly) database. All genes identified from all species were translated and domain analysis was performed. The final sequences were also blasted at NCBI to ensure that the proper target sequence were the best hits. The manually defined genes from E. huxleyi were deposited in GenBank, (accession numbers pending), and the predicted genes from E. huxleyi and other genomes are attached as Figure S1. Data sources for the various algal genomes:
JGI: http://genome.jgi-psf.org/: Emiliania huxleyi v1; Phaeodactylum tricornutum v2; Thalassiosira pseudonana v3; Chlorella variabilis v1.
Phytozome110: http://www.phytozome.org/: Chlamydomonas reinhardtii JGI v5; Coccomyxa subellipsoidea JGI v2; Micromonas pusilla CCMP1545 JGI v3; Micromonas sp RCC299 JGI v3; Ostreococcus lucimarinus JGI v2; Volvox carteri JGI v2.
Species-specific databases:
Ectocarpus siliculosus: http://bioinformatics.psb.ugent.be/blast/moderated/?project=orcae_Ectsi
Porphyridium purpureum: http://cyanophora.rutgers.edu/porphyridium/
Cyanidioschyzon merolae: http://merolae.biol.s.u-tokyo.ac.jp/blast/blast.html
Galdieria sulphuraria: http://genomics.msu.edu/cgi-bin/galdieria/blast.cgi
BLAST at NCBI vs. whole genome shotgun contigs: Chondrus Crispus (taxid:2769)
Arabidopsis: TAIR http://www.arabidopsis.org/
Downloaded data sets, local BLAT running against the genome,111 predicted genes, predicted proteins:
Nannochloropsis gaditana B-31 (December 8, 2012): http://www.nannochloropsis.org/page/ftp
Cyanophora paradoxa (11/2010): http://cyanophora.rutgers.edu/cyanophora/blast.php
Pyropia yezoensis (Nori) V1: http://nrifs.fra.affrc.go.jp/ResearchCenter/5_AG/genomes/nori/
Multiple alignment and phylogenetic analysis
Multiple alignments were performed with both ClustalW version 2.1112 and Muscle version 3.8.31,113 and the better alignment was chosen for phylogenetic analysis (ATG8, Muscle; ATG9, ClustalW). In cases where only one region was properly aligned, the alignment was cut manually (ATG9, according to the domain definition of CDD,63 cl04403: APG9 superfamily). Phylogenetic analysis was performed with Neighbor Joining in ClustalW and ProML (Maximum Likelihood) in Phylip (version 3.69),114 with the following parameters: 100 datasets, seed=9, jumble=3. The trees had similar topologies, and the maximum likelihood trees are shown. Visualization of the trees was done using iTol: http://itol.embl.de/.115
Gene expression
Gene expression analysis was performed using publicly available expression data sets. For E. huxleyi, our transcriptome data was used,72 and for the haploid (RCC1217)/ diploid (RCC1216) strains, EST data from von Dassow et al., 2009, was mined at NCBI.75 P. tricornutum gene expression data was taken from Maheswari et al., 2010.76 T. pseudonana gene expression was taken from Dyhrman et al., 2012.77 N. gaditana RNASeq expression data was taken from Carpinelli et al., 2014.61
Real-time qRT-PCR was performed as follows: RNA was isolated from cells with the RNeasy Plant Mini kit (Qiagen, 74904) according to the manufacturer's instructions, followed by DNAse treatment with Turbo DNase (Ambion, AM1907). Equal amounts of RNA were used for cDNA synthesis with the ThermoScript RT-PCR system (Invitrogen, 11146-016). For transcript abundance analysis, Platinum SYBER Green qPCR SuperMix-UDG with ROX (Invitrogen, 11744-500) was used as instructed by the manufacturer. Reactions were performed on StepOnePlus real-time PCR Systems (Applied Biosystems California, USA) as follows: 50°C for 2 min, 95°C for 2 min, 40 cycles of 95°C for 15 sec, 60°C for 30 sec. Transcript abundance was calculated by normalizing the results to the expression level of tubulin in each sample at a given time point. A list of primers used in this study is added as Table S1.
Credits for images of algae presented in Figure 1
The following micrographs were kindly provided to this study: 1B, Thomas Proeschold; 1D, Martin F. Hohmann-Marriott; 1E, Daphné Grulois, Sophie Lepanse and Nathalie Simon; 1I, Gerald Schoenknecht; 1J, Inga Kjersti Sjøtun; 1K, Daniella Schatz. Alvaro Israel kindly contributed the samples of Pyropia and Ectocarpus. Nathan Nelson kindly provided the samples of C. merolae.
Acknowledgments
We thank Daniella Schatz and Shilo Rosenwasser for fruitful discussions and commenting on the manuscript, and Orr Shapiro for assisting with the microscopy imaging.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the European Research Council (ERC) StG (INFOTROPHIC grant #280991) and the generous support of Edith and Nathan Goldenberg Career Development Chair both awarded to A.V.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski PG. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 1998; 281: 237-40; PMID:9657713; http://dx.doi.org/ 10.1126/science.281.5374.237 [DOI] [PubMed] [Google Scholar]
- 2.Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR. The evolution of modern eukaryotic phytoplankton. Science 2004; 305: 354-60; PMID:15256663; http://dx.doi.org/ 10.1126/science.1095964 [DOI] [PubMed] [Google Scholar]
- 3.Falkowski PG, Raven JA. Aquatic photosynthesis 2nd ed New Jersey: Taylor & Francis; 2007. 484 p. [Google Scholar]
- 4.Tyrrell T, Merico A. Emiliania huxleyi: bloom observations and the conditions that induce them In: Thierstein HR, Young JR, eds. Coccolithophores. From molecular processes to global impact. New York: Taylor & Francis, 2004; 75-97 [Google Scholar]
- 5.Falkowski PG. Primary productivity in the sea. New York: Taylor & Francis, 1980. 531 p. [Google Scholar]
- 6.Jackson GA. Phytoplankton growth and zooplankton grazing in oligotrophic oceans. Nature 1980; 284: 439-41; http://dx.doi.org/ 10.1038/284439a0 [DOI] [Google Scholar]
- 7.Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser 1993; 93: 39-48; http://dx.doi.org/ 10.3354/meps093039 [DOI] [Google Scholar]
- 8.Holligan PM, Viollier M, Harbour DS, Camus P, Champagne-Philippe M. Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature 1983; 304: 339-42; http://dx.doi.org/ 10.1038/304339a0 [DOI] [Google Scholar]
- 9.Pingree RD, Pugh PR, Holligan PI, Forster GR. Summer phytoplankton blooms and red tides along tidal fronts in the approaches to the English Channel. Nature 1975; 258: 672-7; http://dx.doi.org/ 10.1038/258672a0 [DOI] [Google Scholar]
- 10.Winter A, Siesser WG. Coccolithophores. Cambridge UK: Taylor & Francis, 1994; p 252. [Google Scholar]
- 11.Capone DG, Bronk DA, Mulholland MR, Carpenter EJ. Nitrogen in the marine environment. 2nd ed Taylor & Francis, 2008; 1729 p. [Google Scholar]
- 12.Dyhrman ST, Ammerman JW, Van Mooy BAS. Microbes and the marine phosphorus cycle. Oceanography 2007; 20: 110-6; http://dx.doi.org/ 10.5670/oceanog.2007.54 [DOI] [Google Scholar]
- 13.Lovelock JE, Maggs RJ, Rasmussen RA. Atmospheric dimethyl sulphide and the natural sulphur cycle. Nature 1972; 237: 452-3; http://dx.doi.org/ 10.1038/237452a0 [DOI] [Google Scholar]
- 14.de Vargas C, Aubry MP, Probert I, Young J. Origin and evolution of coccolithophores: From coastal hunters to oceanic farmers In: Falkowski PG, Knoll AH, eds. Evolution of primary producers in the sea. USA: Taylor & Francis, 2007; 251-85. [Google Scholar]
- 15.Iglesias‐Rodríguez MD, Brown CW, Doney SC, Kleypas J, Kolber D, Kolber Z, Hayes P, Falkowski PG. Representing key phytoplankton functional groups in ocean carbon cycle models: Coccolithophorids. Global Biogeochem Cy 2002; 16: 1-47; http://dx.doi.org/ 10.1029/2001GB001398 [DOI] [Google Scholar]
- 16.Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature 1987; 326: 655-61; http://dx.doi.org/ 10.1038/326655a0 [DOI] [Google Scholar]
- 17.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 2004; 21: 809-18; PMID:14963099; http://dx.doi.org/ 10.1093/molbev/msh075 [DOI] [PubMed] [Google Scholar]
- 18.Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 2009; 324: 1724-6; PMID:19556510; http://dx.doi.org/ 10.1126/science.1172983 [DOI] [PubMed] [Google Scholar]
- 19.Falkowski PG, Knoll AH, eds. Evolution of primary producers in the sea. USA: Taylor & Francis, 2007; 441 p. [Google Scholar]
- 20.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: 463-77; PMID:15068787; http://dx.doi.org/ 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 21.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120: 159-62; PMID:15680321 [DOI] [PubMed] [Google Scholar]
- 22.de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28: 435-92; PMID:5322983; http://dx.doi.org/ 10.1146/annurev.ph.28.030166.002251 [DOI] [PubMed] [Google Scholar]
- 23.Bursch W, Ellinger A, Gerner C, Schulte-Hermann R. Autophagocytosis and programmed cell death In: Klionsky DJ, ed. Autophagy. USA: Taylor & Francis, 2003: 290-306. [Google Scholar]
- 24.Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ 2005; 12: 1528-34; PMID:16247500; http://dx.doi.org/ 10.1038/sj.cdd.4401777 [DOI] [PubMed] [Google Scholar]
- 25.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30: 1049-58; PMID:19995911; http://dx.doi.org/ 10.1128/MCB.01344-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002; 10: 457-68; PMID:12408816; http://dx.doi.org/ 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 2003; 11: 895-904; PMID:12718876; http://dx.doi.org/ 10.1016/S1097-2765(03)00114-X [DOI] [PubMed] [Google Scholar]
- 28.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature Cell Biol 2004; 6: 1122-8; PMID:15467718; http://dx.doi.org/ 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- 29.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell 2002; 1: 11-21; PMID:12455967; http://dx.doi.org/ 10.1128/EC.01.1.11-21.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118: 7-18; PMID:15615779; http://dx.doi.org/ 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2002; 148: 465-80; http://dx.doi.org/ 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6: 79-90; PMID:14723849; http://dx.doi.org/ 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 33.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152: 519-30; PMID:11157979; http://dx.doi.org/ 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature 1998; 395: 395-8; PMID:9759731; http://dx.doi.org/ 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- 35.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151: 263-76; PMID:11038174; http://dx.doi.org/ 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 1998; 273: 33889-92; PMID:9852036; http://dx.doi.org/ 10.1074/jbc.273.51.33889 [DOI] [PubMed] [Google Scholar]
- 37.Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy 2013; 9: 424-5; PMID:23321721; http://dx.doi.org/ 10.4161/auto.22931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose TL, Bonneau L, Der C, Marty‐Mazars D, Marty F. Starvation‐induced expression of autophagy‐related genes in Arabidopsis. Biol Cell 2006; 98: 53-67; PMID:16354162; http://dx.doi.org/ 10.1042/BC20040516 [DOI] [PubMed] [Google Scholar]
- 39.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 2002; 129: 1181-93; PMID:12114572; http://dx.doi.org/ 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant cell physiol 2006; 47: 1641-52; PMID:17085765; http://dx.doi.org/ 10.1093/pcp/pcl031 [DOI] [PubMed] [Google Scholar]
- 41.Sláviková S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot 2005; 56: 2839-49 [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005; 121: 567-77; PMID:15907470; http://dx.doi.org/ 10.1016/j.cell.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 43.Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, Golstein P, Escalante R. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 2010; 6: 686-701; PMID:20603609; http://dx.doi.org/ 10.4161/auto.6.6.12513 [DOI] [PubMed] [Google Scholar]
- 44.Picazarri K, Nakada‐Tsukui K, Sato D, Nozaki T. Analysis of autophagy in the enteric protozoan parasite Entamoeba. Methods Enzymol 2008; 451: 359-71; PMID:19185732; http://dx.doi.org/ 10.1016/S0076-6879(08)03224-2 [DOI] [PubMed] [Google Scholar]
- 45.Alvarez VE, Kosec G, Sant'Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem 2008; 283: 3454-64; PMID:18039653; http://dx.doi.org/ 10.1074/jbc.M708474200 [DOI] [PubMed] [Google Scholar]
- 46.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindguist E, Lucas S, Pangilinan J, Polle J, et al.. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010; 22: 2943-55; PMID:20852019; http://dx.doi.org/ 10.1105/tpc.110.076406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al.. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007; 318: 245-50; PMID:17932292; http://dx.doi.org/ 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanc G, Agarkova I, Grimwood J, Kuo A, Brueggeman A, Dunigan DD, Gurnon J, Ladunga I, Lindguist E, Lucas S, et al.. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol 2012; 13; R39; PMID:22630137; http://dx.doi.org/ 10.1186/gb-2012-13-5-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK et al.. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 2010; 329: 223-6; PMID:20616280; http://dx.doi.org/ 10.1126/science.1188800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Cuvelier E, Derelle ML, Everett MV, Foulon E, et al.. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009; 324: 268-72; PMID:19359590; http://dx.doi.org/ 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- 51.Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Darelle E, Rombauts S, et al.. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci U S A 2007; 104: 7705-10; PMID:17460045; http://dx.doi.org/ 10.1073/pnas.0611046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000; 408: 796-815; PMID:11130711; http://dx.doi.org/ 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya D, Price DC, Chan CX, Qiu H, Rose N, Ball S, Weber APM, Arias MC, Henrissat B, Coutinho PM et al.. Genome of the red alga Porphyridium purpureum. Nat Commun 2013;4: 1941; PMID: 23770768; http://dx.doi.org/ 10.1038/ncomms2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schönknecht G, Chen W, Tarens CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Benfield JF, Garavito RM, et al.. Gene transfer from bacteria and archaea facilitated evolution of anextremophilic eukaryote. Science 2013; 339: 1207-10; PMID:23471408; http://dx.doi.org/ 10.1126/science.1231707 [DOI] [PubMed] [Google Scholar]
- 55.Matsuzaki M, Misumi O, Shin-i T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al.. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2004; 428: 653-7; PMID:15071595; http://dx.doi.org/ 10.1038/nature02398 [DOI] [PubMed] [Google Scholar]
- 56.Nakamura Y, Sasaki N, Kobayashi M, Ojima N, Yasuike M, Shigenobu Y, Satomi M, Fukuma Y, Shiwaku K, Tsujimoto A, et al.. The first symbiont-free genome sequence of marine red alga, susabi-nori (Pyropia yezoensis). PLoS One 2013; 8: e57122; PMID:23536760; http://dx.doi.org/ 10.1371/journal.pone.0057122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collén J, Porcel B, Carré W, Ball SG, Chaparro C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, et al.. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc Natl Acad Sci U S A 2013; 110: 5247-52; http://dx.doi.org/ 10.1073/pnas.1221259110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, Mayer C, Miller J, Monier A, Salamov A, et al.. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 2013; 499: 209-13; PMID:23760476; http://dx.doi.org/ 10.1038/nature12221 [DOI] [PubMed] [Google Scholar]
- 59.Armbrust EV, Bergres JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al.. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 2004; 306: 79-86; PMID:15459382; http://dx.doi.org/ 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- 60.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al.. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008; 456: 239-44; PMID:18923393; http://dx.doi.org/ 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- 61.Carpinelli EC, Telatin A, Vitulo N, Forcato C, D'Angelo M, Schiavon R, Vezzi A, Giacometti GM, Morosinotto T, Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol Plant 2014; 7: 323-35; PMID:23966634; http://dx.doi.org/ 10.1093/mp/sst120 [DOI] [PubMed] [Google Scholar]
- 62.Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al.. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010; 465: 617-21; PMID:20520714; http://dx.doi.org/ 10.1038/nature09016 [DOI] [PubMed] [Google Scholar]
- 63.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al.. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013; 41: D348-52; PMID:23197659; http://dx.doi.org/ 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Díaz-Troya S, Florencio FJ, Crespo JL. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell 2008; 7: 212-22; http://dx.doi.org/ 10.1128/EC.00361-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crespo JL, Díaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol 2005; 139: 1736-49; PMID:16299168; http://dx.doi.org/ 10.1104/pp.105.070847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imamura S, Ishiwata A, Watanabe S, Yoshikawa H, Tanaka K. Expression of budding yeast FKBP12 confers rapamycin susceptibility to the unicellular red alga Cyanidioschyzon merolae. Biochem Biophys Res Commun 2013; 439: 264-9; PMID:23973485; http://dx.doi.org/ 10.1016/j.bbrc.2013.08.045 [DOI] [PubMed] [Google Scholar]
- 67.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 2008; 4: 851-865; http://dx.doi.org/ 10.4161/auto.6555 [DOI] [PubMed] [Google Scholar]
- 68.Reinke A, Anderson S, McCaffery JM, Yates J 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 2004; 279: 14752-62; PMID:14736892; http://dx.doi.org/ 10.1074/jbc.M313062200 [DOI] [PubMed] [Google Scholar]
- 69.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell 2000; 103: 253-62; PMID:11057898; http://dx.doi.org/ 10.1016/S0092-8674(00)00117-3 [DOI] [PubMed] [Google Scholar]
- 70.Dames SA, Mulet JM, Rathgeb-Szabo K, Hall MN, Grzesiek S. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J Biol Chem 2005; 280: 20558-64; PMID:15772072; http://dx.doi.org/ 10.1074/jbc.M501116200 [DOI] [PubMed] [Google Scholar]
- 71.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110: 177-89; PMID:12150926; http://dx.doi.org/ 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- 72.Rosenwasser S, Mausz MA, Schatz D, Sheyn U, Malitsky S, Aharoni A, Weinstock E, Tzfadia O, Ben-Dor S, Feldmesser E, et al.. Rewiring host lipid metabolism by large vruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell 2014; 26: 2689-707; PMID:24920329; http://dx.doi.org/ 10.1105/tpc.114.125641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lehahn Y, Koren I, Schatz D, Frada M, Sheyn U, Boss E, Efrati S, Rudich Y, Trainic M, Sharoni S, et al.. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr Biol 2014; 24: 2041-6; PMID: 25155511; http://dx.doi.org/ 10.1016/j.cub.2014.07.046 [DOI] [PubMed] [Google Scholar]
- 74.Schatz D, Shemi A, Rosenwasser S, Sabanay H, Wolf S, Ben-Dor S, Vardi A. Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol 2014; In press; PMID:25195618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Dassow P, Ogata H, Probert I, Wincker P, Da Silva C, Audic S, Claverie JM, de Vargas C. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol 2009; 10: R114; PMID:19832986; http://dx.doi.org/ 10.1186/gb-2009-10-10-r114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maheswari U, Jabbari K, Petit JL, Porcel BM, Allen AE, Cadoret JP, De Martino A, Heijde M, Kaas R, La Roche J, et al.. Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol 2010; 11: R85; PMID:20738856; http://dx.doi.org/ 10.1186/gb-2010-11-8-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, Whitney LP, Drzewianowski A, Bulygin VV, Bertrand EM, et al.. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One 2012; 7: e33768; PMID:22479440; http://dx.doi.org/ 10.1371/journal.pone.0033768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem 2001; 276: 30452-60; PMID:11382761; http://dx.doi.org/ 10.1074/jbc.M102346200 [DOI] [PubMed] [Google Scholar]
- 79.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al.. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 Kinase. Mol Cell 2014; 53: 471-83; PMID:24440502; http://dx.doi.org/ 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell 2007; 18: 581-93; PMID:17135291; http://dx.doi.org/ 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velayos-Baeza A, Lévecque C, Dobson-Stone C, Monaco AP. The function of chorein In: Walker RH, Saiki S, Danek A, eds. Neuroacanthocytosis syndromes II. Taylor & Francis, 2008;87-105 [Google Scholar]
- 82.Fry MJ. Structure, regulation and function of phosphoinositide 3-kinases. Biochim Biophys Acta 1994; 1226: 237-68; PMID:8054357; http://dx.doi.org/ 10.1016/0925-4439(94)90036-1 [DOI] [PubMed] [Google Scholar]
- 83.Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol 1995; 129: 321-34; PMID:7721937; http://dx.doi.org/ 10.1083/jcb.129.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571-80; PMID:21311563; http://dx.doi.org/ 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl−2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell 2005; 122: 927-39; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 86.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 2002; 9: 394-404; PMID:11965492; http://dx.doi.org/ 10.1038/sj.cdd.4400991 [DOI] [PubMed] [Google Scholar]
- 87.Lam E. Controlled cell death, plant survival and development. Nat Rev Mol Cell Bio 2004; 5: 305-15; http://dx.doi.org/ 10.1038/nrm1358 [DOI] [PubMed] [Google Scholar]
- 88.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12: 226; PMID:21867568; http://dx.doi.org/ 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al.. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408: 488-92; PMID:11100732; http://dx.doi.org/ 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 90.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al.. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 2011; 39: D225-9; PMID:21109532; http://dx.doi.org/ 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994; 371: 297-300; PMID:8090199; http://dx.doi.org/ 10.1038/371297a0 [DOI] [PubMed] [Google Scholar]
- 92.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell 2008; 19: 5506-16; PMID:18829864; http://dx.doi.org/ 10.1091/mbc.E08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 2010; 152: 1874-88; http://dx.doi.org/ 10.1104/pp.109.152520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy 2006; 2: 2-11; PMID:16874030; http://dx.doi.org/ 10.4161/auto.2092 [DOI] [PubMed] [Google Scholar]
- 95.Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol 2014; 12: 101-14; PMID:24384599; http://dx.doi.org/ 10.1038/nrmicro3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blouin NA, Brodie JA, Grossman AC, Xu P, Brawley SH. Porphyra: a marine crop shaped by stress. Trends Plant Sci 2011; 16: 29-37. [DOI] [PubMed] [Google Scholar]
- 97.Inwood W, Yoshihara C, Zalpuri R, Kim KS, Kustu S. The ultrastructure of a Chlamydomonas reinhardtii mutant strain lacking phytoene synthase resembles that of a colorless alga. Mol Plant 2008; 1: 925-37; PMID:19825593; http://dx.doi.org/ 10.1093/mp/ssn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pérez-Pérez ME, Couso I, Crespo JL. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 2012; 8: 376-388; http://dx.doi.org/ 10.4161/auto.18864 [DOI] [PubMed] [Google Scholar]
- 99.Jiang Q, Zhao L, Dai J, Wu Q. Analysis of autophagy genes in microalgae: Chlorella as a potential model to study mechanism of autophagy. PLoS One 2012; 7: e41826; PMID:22848622; http://dx.doi.org/journal.pone.0041826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Avin-Wittenberg T, Honig A, Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 2012; 249: 285-99; PMID:21660427; http://dx.doi.org/ 10.1007/s00709-011-0296-z [DOI] [PubMed] [Google Scholar]
- 101.Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, et al.. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 2012; 335: 843-7; PMID:22344442; http://dx.doi.org/ 10.1126/science.1213561 [DOI] [PubMed] [Google Scholar]
- 102.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002; 277: 33105-14; PMID:12070171; http://dx.doi.org/ 10.1074/jbc.M204630200 [DOI] [PubMed] [Google Scholar]
- 103.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008; 178: 1339-53; PMID:18245858; http://dx.doi.org/ 10.1534/genetics.107.086199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harrison-Lowe NJ, Olsen LJ. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 2008; 4: 339-48; PMID:18227644; http://dx.doi.org/ 10.4161/auto.5629 [DOI] [PubMed] [Google Scholar]
- 105.Vardi A, Van Mooy BA, Fredricks HF, Popendorf KJ, Ossolinski JE, Haramaty L, Bidle KD. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 2009; 326: 861-5; PMID:19892986; http://dx.doi.org/ 10.1126/science.1177322 [DOI] [PubMed] [Google Scholar]
- 106.Mackinder LC, Worthy CA, Biggi G, Hall M, Ryan KP, Varsani A, Harper GM, Wilson WH, Brownlee C, Schroeder DC. A unicellular algal virus, Emiliania huxleyi virus 86, exploits an animal-like infection strategy. J Gen Virol 2009; 90: 2306-16; PMID:19474246; http://dx.doi.org/ 10.1099/vir.0.011635-0 [DOI] [PubMed] [Google Scholar]
- 107.Feldmesser E, Rosenwasser S, Vardi A, Ben-Dor S. Improving transcriptome construction in non-model organisms: integrating manual and automated gene definition in Emiliania huxleyi. BMC Genomics 2014; 15: 148; PMID:24559402; http://dx.doi.org/ 10.1186/1471-2164-15-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389-402; PMID:9254694; http://dx.doi.org/ 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vandepoele K, Van Bel M, Richard G, Van Landeghem S, Verhelst B, Moreau H, Van de Peer Y, Grimsley N, Piganeau G. Pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ Microbiol 2013; 15: 2147-53; PMID:23826978; http://dx.doi.org/ 10.1111/1462-2920.12174 [DOI] [PubMed] [Google Scholar]
- 110.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 2012; 40: D1178-86; PMID:22110026; http://dx.doi.org/ 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kent WJ. BLAT - the BLAST-like alignment tool. Genome Res 2002; 12: 656-64; PMID:11932250; http://dx.doi.org/ 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al.. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947-8; PMID:17846036; http://dx.doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 113.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32: 1792-7; PMID:15034147; http://dx.doi.org/ 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Taylor & Francis, Seattle: 2005 [Google Scholar]
- 115.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011; 39: W475-8; PMID:21470960; http://dx.doi.org/ 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.