Abstract
Impaired autophagy function and enhanced ARG2 (arginase 2)-MTOR (mechanistic target of rapamycin) crosstalk are implicated in vascular aging and atherosclerosis. We are interested in the role of ARG2 and the potential underlying mechanism(s) in modulation of endothelial autophagy. Using human nonsenescent “young” and replicative senescent endothelial cells as well as Apolipoprotein E-deficient (apoe−/−Arg2+/+) and Arg2-deficient apoe−/− (apoe−/−arg2−/−) mice fed a high-fat diet for 10 wk as the atherosclerotic animal model, we show here that overexpression of ARG2 in the young cells suppresses endothelial autophagy with concomitant enhanced expression of RICTOR, the essential component of the MTORC2 complex, leading to activation of the AKT-MTORC1-RPS6KB1/S6K1 (ribosomal protein S6 kinase, 70kDa, polypeptide 1) cascade and inhibition of PRKAA/AMPK (protein kinase, AMP-activated, α catalytic subunit). Expression of an inactive ARG2 mutant (H160F) had the same effect. Moreover, silencing RPS6KB1 or expression of a constitutively active PRKAA prevented autophagy suppression by ARG2 or H160F. In senescent cells, enhanced ARG2-RICTOR-AKT-MTORC1-RPS6KB1 and decreased PRKAA signaling and autophagy were observed, which was reversed by silencing ARG2 but not by arginase inhibitors. In line with the above observations, genetic ablation of Arg2 in apoe−/− mice reduced RPS6KB1, enhanced PRKAA signaling and endothelial autophagy in aortas, which was associated with reduced atherosclerosis lesion formation. Taken together, the results demonstrate that ARG2 impairs endothelial autophagy independently of the L-arginine ureahydrolase activity through activation of RPS6KB1 and inhibition of PRKAA, which is implicated in atherogenesis.
Keywords: ARGINASE, atherosclerosis, autophagy, endothelial cells, MTOR, PRKAA, senescence
Abbreviations: Three-MA, 3-methyladenine; AKT/PKB, v-akt murine thymoma viral oncogene homolog 1; PRKAA/AMPK, protein kinase, AMP-activated, α catalytic subunit; ANOVA, analysis of variance; apoe−/−, Apolipoprotein E-deficient; AR, aortic roots; ARG1, arginase 1; ARG2, arginase 2; arg2−/−, arginase type II deficient; Atg, autophagy-related; Baf A1, bafilomycin A1; BEC, S-12 bromoethyl-L-cystine-HCl; BECN1, Beclin 1, autophagy-related; CMV, cytomegalovirus; EC, endothelial cell; H160F, inactive mutant of mouse ARG2; HAEC, human aortic endothelial cells; HUVEC, human umbilical vein endothelial cells; LC3, microtubule-associated protein 1 light chain 3; LDL, low-density lipoprotein; MTOR, mechanistic target of rapamycin; nor-NOHA, Nω-hydroxy-nor-Arginine; NOS3/eNOS, nitric oxide synthase 3 (endothelial cell); PE, phosphatidylethanolamine; PtdIns3K, phosphatidylinositol 3-kinase; TP53/p53, tumor protein 53; RPS6, ribosomal protein S6; RPS6KB1/S6K1, ribosomal protein S6 kinase, 70kDa, polypeptide 1; shRNA, short hairpin RNA; SA-ß-gal, senescence-associated-β-gal; SMC, smooth muscle cells; SQSTM1/p62, sequestosome 1; ULK1, unc-51 like autophagy activating kinase 1; VWF, von Willebrand factor; WT, wild type
Introduction
Autophagy, the mechanism of lysosomal proteolysis aiming to remove harmful proteins in the cells, is implicated in maintaining the healthy status of an organism under stress conditions.1 Adequate functions of autophagy in a cell involve the formation of characteristic double-membrane vesicles, called autophagosomes, in which cytoplasmic material is sequestered.1 The autophagosomes fuse with lysosomes to form autolysosomes with degradative capacity. The dysfunctional cellular substances or organelles are degraded in the autolysosomes and the degraded components are then released to the cytosol for recycling or reuse for anabolic pathways, so that the cells can be protected from accumulation of nonfunctional or dysfunctional substances which may cause cellular damage.1 Numerous proteins are identified to participate in autophagosome formation and autophagy functions.2 Among them, the protein MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) undergoes changes upon autophagy induction, i.e., proteolytic processing and lipidation with phosphatidylethanolamine, resulting in formation of the phagophore membrane-bound form LC3-II, which leads to the elongation of the phagophore membrane.3 A portion of the LC3-II remains associated with the completed autophagosome and is therefore the most widely used autophagy marker.4 Since the ATG12–ATG5 conjugate is involved in LC3 lipidation (conversion of the proteolytically processed LC3-I form to LC3-II), it therefore serves as another marker for autophagosome formation.5 The receptor protein SQSTM1/p62 (sequestosome 1) targets ubiquitinated protein aggregates for lysosomal degradation and is selectively degraded via autophagy; it is thus used along with LC3 to monitor autophagic degradation/flux.6 Accumulation of SQSTM1 reflects impaired degradation in the autolysosome, whereas low SQSTM1 levels may indicate active degradation. Combinatory detection of LC3-II, ATG12–ATG5 conjugate, and SQSTM1 thus monitors both autophagosome formation and autophagic flux.
Endothelial senescence is involved in acceleration of the atherosclerotic process.7 Vascular aging exhibits decreased autophagy function, whereas autophagy induction in aging arteries is able to enhance endothelial function.8 Impairment of autophagy occurs in atherosclerosis and contributes to plaque development and vulnerability.9 Although the function of autophagy in atherosclerosis is not entirely clear, recent studies provide evidence suggesting that adequate induction of autophagy protects against cellular injury in endothelial and smooth muscle cells and formation of foam cells, resulting in anti-atherosclerotic effects.8-10
Autophagy is regulated by numerous signaling pathways such as those regulated by MTOR (mechanistic target of rapamycin), PRKAA, and TP53/p53 (tumor protein 53).1 Compelling evidence demonstrates that MTOR is a dominant signaling kinase inhibiting autophagy.11 Depending on its binding partners, MTOR forms 2 different protein complexes, namely MTORC1 and MTORC2,12 which are large complexes consisting of 6 and 7 known protein components, respectively. Among the multiprotein components, RPTOR (regulatory associated protein of MTOR, complex 1) and RICTOR (RPTOR independent companion of MTOR, complex 2) are characteristic components of MTORC1 and MTORC2, respectively.13 MTORC1 negatively regulates autophagy by directly phosphorylating ATG proteins, such as ULK1 (unc-51 like autophagy activating kinase 1) and ATG13.14 Many signaling pathways such as PRKAA and AKT that regulate autophagy converge on MTORC1.15,16 In line with the negative regulatory role of MTORC1 in autophagy, PRKAA which inhibits MTORC1 signaling, promotes autophagy,16 while AKT which is a downstream effector of MTORC2 and upstream activator of MTORC1,13 suppresses autophagy.14,17 In contrast to MTORC1, the function of its best-characterized downstream effector, RPS6KB1, is controversial. RPS6KB1 has been shown to stimulate autophagy in mammals,18,19 whereas, Sch9, a putative yeast homolog of RPS6KB1, inhibits autophagy in yeast.20
ARG2, an L-arginine ureahydrolase metabolizing L-arginine to urea and L-ornithine, has been implicated in NOS3/eNOS (nitric oxide synthase 3 [endothelial cell]) dysfunction in vascular aging and pathogenesis of atherosclerosis.21 Our recent studies demonstrate that ARG2 positively regulates RPS6KB1 signaling, which promotes endothelial cell senescence and inflammation,22 and causes vascular smooth muscle cell senescence and apoptosis independently of its enzymatic activity.23 ARG2 also promotes macrophage pro-inflammatory responses in atherosclerosis and type II diabetes.24 Given that impaired autophagy is implicated in vascular aging and atherosclerosis, we investigated here the role of ARG2 and the signaling mechanisms in regulation of endothelial autophagy and its contribution to the pathogenesis of atherosclerosis.
Results
ARG2 impairs autophagy independently of its enzymatic activity in endothelial cells
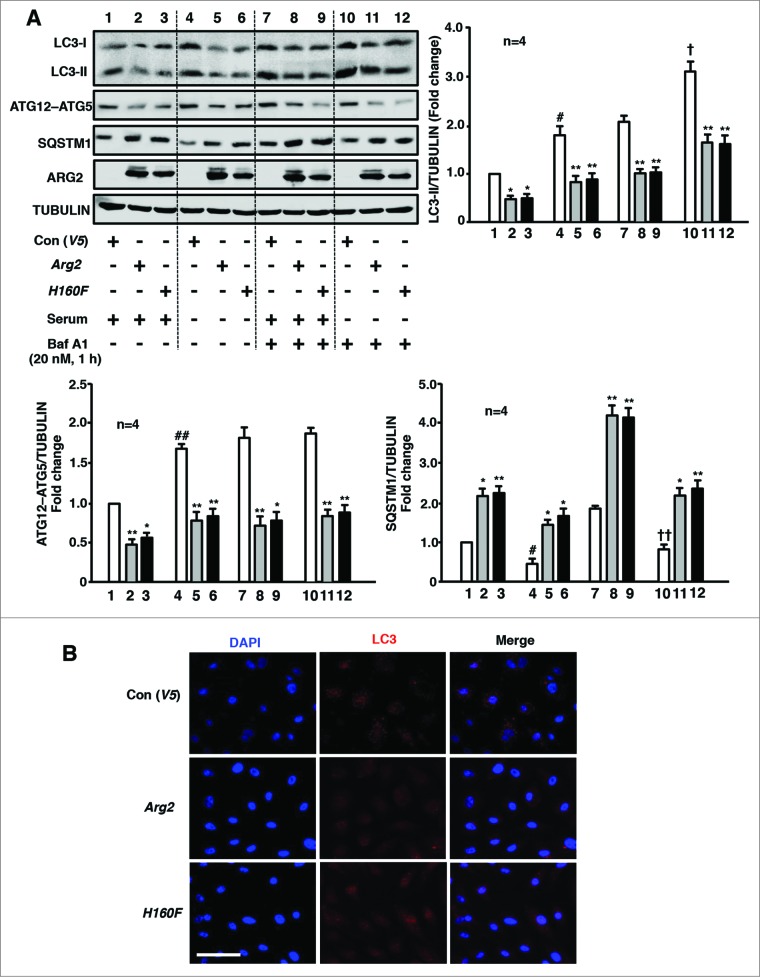
In the nonsenescent human umbilical vein endothelial cells (HUVECs), serum-starvation on its own increased LC3-II levels both in the absence (Fig. 1A, lane 4 vs. lane 1) and presence of bafilomycin A1 (Baf A1, 20 nmol/L, 60 min), a vacuolar H+-ATPase inhibitor that inhibits autophagosome fusion with lysosomes,25,26 (Fig. 1A, lane 10 vs. lane 7), demonstrating that the increase in LC3-II results from induced LC3-II/autophagosome formation, but not from reduced degradation of LC3-II via lysosomal turnover upon serum-starvation. Adenovirus-mediated overexpression of either wild-type (WT) or inactive ARG2 (H160F) decreased LC3-II levels both in the absence (Fig. 1A, lanes 2–3 vs. lane 1; lanes 5–6 vs. lane 4) and presence (Fig. 1A, lanes 8–9 vs. lane 7; lanes 11–12 vs. lane 10) of Baf A1, under both serum-growth (Fig. 1A, lanes 1–3 and 7–9) and serum-starvation (Fig. 1A, lanes 4–6 and 10–12) conditions. The lack of the L-arginine ureahydrolase activity of H160F and its inability to cause NOS3-uncoupling were demonstrated previously.22 Moreover, the alteration of another autophagosome formation marker, the ATG12–ATG5 conjugate, revealed the same pattern as that of LC3-II. In line with the inhibition of autophagosome formation as indicated by a decrease in LC3-II and ATG12–ATG5 conjugate formation, the SQSTM1 level was increased by WT or inactive ARG2 under all 4 of the experimental conditions, which is indicative of reduced autophagic flux (Fig. 1A). Furthermore, immunofluorescence confocal microscopy revealed a punctate pattern of LC3-I/-II staining in the cytosol of the control cells (Fig. 1B), which is indicative of LC3-I/-II accumulation in the lysosomes and an active autophagic response in the cells. Overexpression of ARG2 or the H160F mutant in the cells caused a diffuse distribution of LC3 throughout the cells (Fig. 1B), suggesting an inhibition of the autophagic response. Similar results were obtained in human aortic endothelial cells (HAEC; Fig. S1). These results provide firm evidence that ARG2 inhibits endothelial autophagy independently of its L-arginine ureahydrolase activity.
Figure 1.
ARG2 impairs autophagy independently of its enzymatic activity in endothelial cells. Young HUVECs were transduced with empty vector rAd/CMV-V5 as control (V5), rAd/CMV-Arg2 (Arg2) or rAd/CMV-Arg2-H160F (H160F, an inactive ARG2 mutant). Forty-8 h post transduction, cells were either further incubated in serum-growth medium or in serum-starvation medium for 16 h as indicated. During the last 1 h of incubation prior to the experiment, the cells were either untreated or treated with Baf A1 (20 nmol/L, 1 h) as indicated. Sixty-4 h post-transduction, the cells were subjected to (A) immunoblotting analysis for detection of ARG2, H160F, LC3-I/-II, ATG12–ATG5, and SQSTM1 protein levels. Tubulin served as the loading control. The bar graph presents the quantification of the signals (n = 4). *P < 0.05, **P < 0.01 vs. V5-control group within the corresponding experimental condition. #P < 0.05, ##P < 0.01 vs. lane 1. †P < 0.05, ††P < 0.01 vs. lane 7. To be clearer for the readers, the 4 different experimental conditions, i.e., serum-growth medium +/−Baf A1, and serum-starvation medium +/−Baf A1 were separated by dotted lines on the western blot, but all samples were run on the same gel. (B) Representative images showing the immunofluorescence staining of LC3-I/-II (red) in HUVECs followed by counterstaining with DAPI (blue). The merged images are also shown. The cells were transduced and treated as in (A) where cells were serum-starved during the last 16 h without treatment with Baf A1. Scale bar = 0.1 mm.
ARG2 enhances TP53, and MTOR signaling and impairs the PRKAA signaling pathway independently of its enzymatic activity
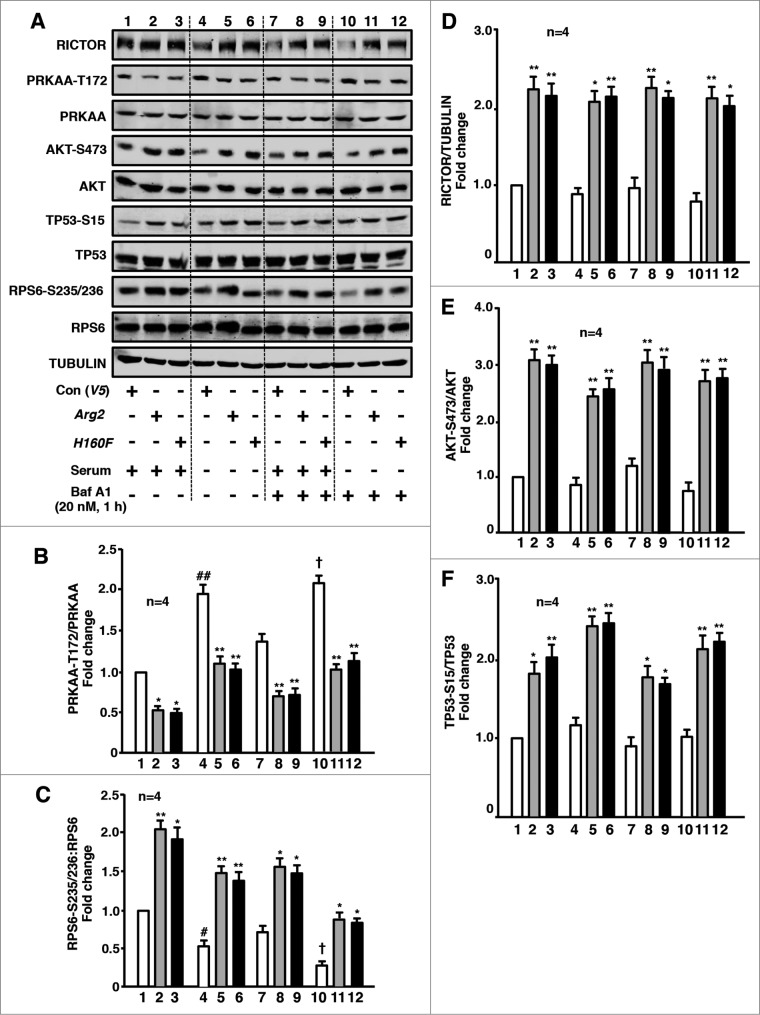
The signaling transduction mechanisms that are involved in the regulation of autophagy were further investigated in endothelial cells. Along with the assays of autophagy, we examined signaling events with 4 experimental conditions, i.e., serum-growth medium +/−Baf A1, and serum-starvation medium +/−Baf A1) for each of the parameters (control, ARG2 and H160F) with all conditions run on the same gel in order to make direct comparisons. Similar to the effect on autophagy, serum-starvation on its own increased basal levels of the PRKAA-T172/PRKAA ratio (Fig. 2A and B) while it decreased basal levels of the RPS6-S235/236:RPS6 ratio (Fig. 2A and C) in the absence (lane 4 vs. lane 1) and presence of Baf A1 (lane 10 vs. lane 7). The basal levels of RICTOR (Fig. 2A and D), AKT-S473/AKT (Fig. 2A and E) and TP53-S15/TP53 (Fig. 2A and F) were not significantly affected by serum-starvation. Although the basal levels of these readouts may be different under the 4 experimental conditions, the effects of ARG2 and H160F on these proteins revealed a similar pattern under all 4 conditions, i.e., overexpression of either WT or the inactive ARG2 reduced PRKAA-T172/PRKAA while it enhanced RICTOR, AKT-S473/AKT, RPS6-S235/236: RPS6 and TP53-S15/TP53 [Fig. 2A, the 2nd and 3rd lanes (2–3, 5–6, 8–9, 11–12) vs. the corresponding 1st lane (1, 4, 7, 10, respectively)] under each experimental condition. Similar results were obtained in HAEC (Fig. S1).
Figure 2.
ARG2 enhances TP53, and MTORC2-AKT-MTORC1-RPS6KB1 and impairs PRKAA signaling independently of its enzymatic activity. Young HUVECs were transduced as in Fig.1A. Sixty-4 h post transduction, cell lysates were prepared and subjected to (A) immunoblotting analysis of RICTOR, PRKAA-T172, and PRKAA, AKT-S473, and AKT, TP53-S15 and TP53, RPS6-S235/236 and RPS6, and tubulin. ((B)to F) Quantification of the signals is also shown (n = 4 as indicated in the figure). *P < 0.05, **P < 0.01, vs. V5-control group within the corresponding experimental condition. #P < 0.05, ##P < 0.01 vs. lane 1. †P < 0.05 vs. lane 7. To be clearer for readers, the 4 different experimental conditions were separated by dotted lines on the protein gel blot, but all conditions were run on the same gel.
The role of TP53, RPS6KB1, and PRKAA in ARG2-mediated inhibition of autophagy
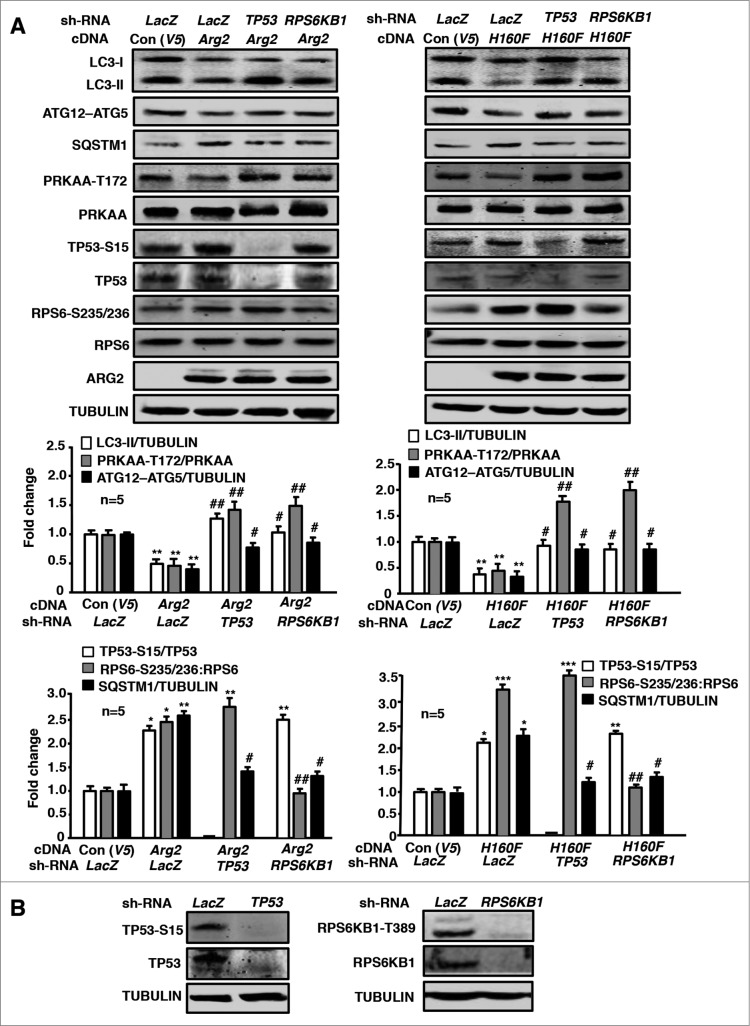
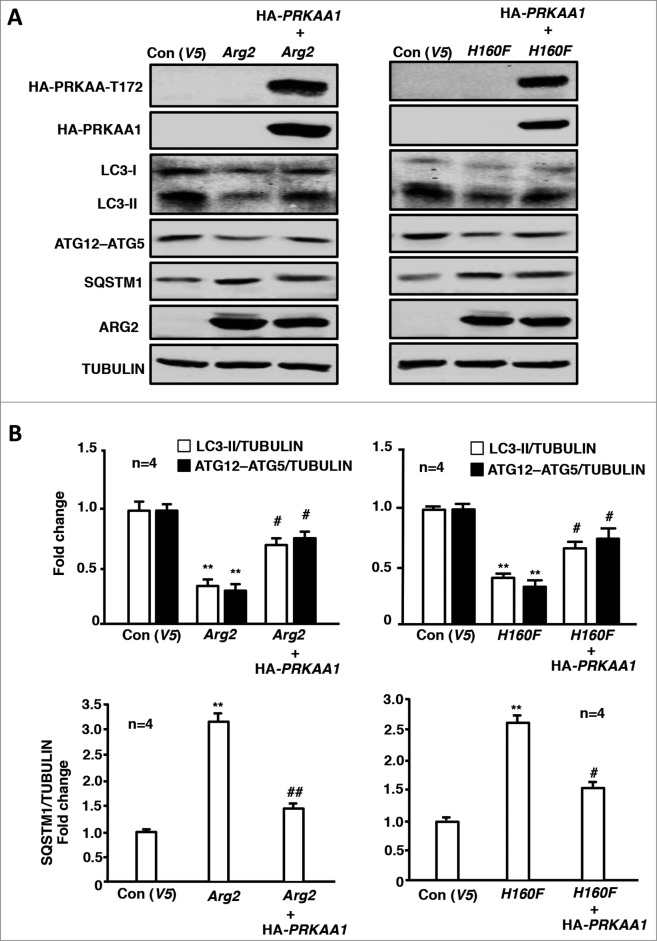
The role of TP53, RPS6KB1 and PRKAA in ARG2-induced inhibition of autophagy was further studied in the endothelial cells overexpressing ARG2 or H160F. In the young cells, overexpression of either ARG2 (Fig. 3A, left) or H160F (Fig. 3A, right) decreased LC3-II and ATG12–ATG5 conjugate levels while they increased SQSTM1 levels, reduced activation of PRKAA (PRKAA -T172/PRKAA ratio), and activated TP53 (increase TP53-S15:TP53 ratio), and enhanced RPS6KB1 activity (RPS6-S235/236:RPS6 ratio). Silencing TP53 or RPS6KB1 as confirmed by immunoblotting (Fig. 3B) independently prevented the effects of ARG2 or H160F on the decrease in LC3-II and ATG12–ATG5 conjugate levels, the increase in SQSTM1 levels, and the inhibition of PRKAA (Fig. 3A). Of note, silencing TP53 or RPS6KB1 did not affect the activity of each other (Fig. 3A), indicating that the 2 pathways are stimulated by ARG2 in parallel. Moreover, overexpression of a HA-tagged constitutively active PRKAA1 (HA-PRKAA1) prevented the suppressing effect of ARG2 or H160F on LC3-II and the ATG12–ATG5 conjugate as well as the accumulation of SQSTM1 (Fig. 4A and B). These results suggest that activation of TP53 and RPS6KB1 by ARG2 or H160F causes inhibition of PRKAA leading to suppression of autophagy.
Figure 3.
Silencing RPS6KB1 or TP53 in young HUVECs attenuates ARG2-induced inhibition of PRKAA signaling and autophagy suppression. Young HUVECs were first transduced either with rAd/U6-LacZshRNA as control, rAd/U6-RPS6KB1shRNA or rAd/U6- TP53shRNA. Twenty-4 h after the 1st transduction with the rAd/U6-shRNAs, the cells were then transduced either with rAd/CMV as control (Con), or with rAd/CMV-Arg2 or rAd/CMV-H160F to overexpress ARG2 or H160F, respectively. Experiments were performed 64 h (serum-free during the last 16 h) post the 2nd transduction. Shown are immunoblotting analyses of LC3-I/-II, ATG12–ATG5, SQSTM1, PRKAA-T172, and PRKAA, TP53-S15 and TP53, RPS6-S235/236 and RPS6, ARG2 and tubulin, which served as a loading control. Quantification of the signals is shown in the bar graphs below (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control; #P < 0.05, ##P < 0.01 vs. Arg2-cDNA + LacZshRNA.
Figure 4.
Overexpression of PRKAA1 in young HUVECs ameliorates ARG2-induced autophagy suppression. Young HUVECs were first transduced either with rAd/CMV as control or rAd/CMV-PRKAA1. Twenty-4 h after the 1st transduction, the cells were then transduced either with rAd/CMV as control (Con), rAd/CMV-Arg2 or rAd/CMV-H160F. Cell lysates were prepared 64 h (serum-free during the last 16 h) post the 2nd transduction for (A) immunoblotting analyses of PRKAA-T172 and PRKAA, LC3-I/-II, ATG12–ATG5, SQSTM1, and ARG2. Tubulin served as a loading control. (B) Quantification of the signals is shown in the bar graphs below (n = 4). **P < 0.01 vs. control; #P < 0.05, ##P < 0.01 vs. Arg2-cDNA + rAd/CMV.
Inhibition of autophagy promotes endothelial senescence
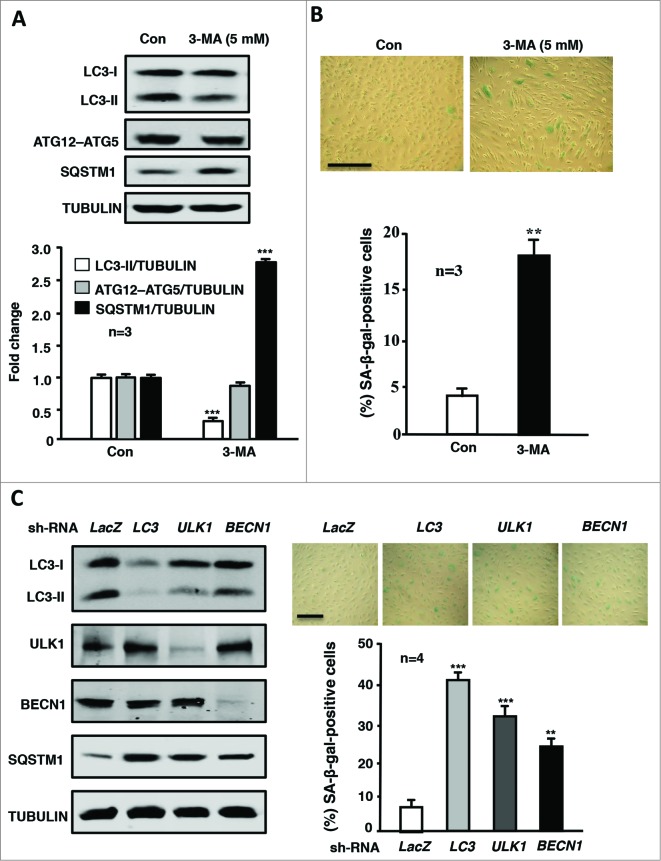
To evaluate the functional role of autophagy in endothelial cell fate or senescence, autophagy of the young endothelial cells was pharmacologically inhibited with an autophagy inhibitor, 3-methyladenine (3-MA; 5 mmol/L, 4 d), one of the most widely used inhibitors of autophagic sequestration of cytoplasmic materials that acts by blocking the class III phosphatidylinositol 3-kinase (PtdIns3K) that is critical during the early stage of autophagy.27,28 The inhibition of autophagy by 3-MA is revealed in Fig. 5A. Fig. 5B shows that inhibition of autophagy increased SA-β-gal-positive endothelial cells, implying that inhibition of autophagy promotes endothelial senescence. This conclusion was further confirmed by the experiments showing that genetic silencing of various autophagy-related genes such as LC3, ULK1, or BECN1 (the efficiency and specificity are shown in Fig. 5C, left) inhibited autophagy (reflected by increased SQSTM1 levels, Fig. 5C, left) and increased SA-β-gal-positive endothelial cells (Fig. 5C, right).
Figure 5.
Inhibition of autophagy promotes endothelial senescence. Young HUVECs were cultured in serum-growth medium in the absence (Con) or presence of the autophagy inhibitor 3-MA (5 mmol/L) for 5 d. The cells were then subjected to (A) immunoblotting analyses of LC3-I/-II, ATG12–ATG5, SQSTM1, and tubulin, which served as a loading control. (B) SA-β-gal staining. (C) The young HUVECs were transduced either with rAd/U6-LacZshRNA as control or rAd/U6-LC3shRNA or rAd/U6-ULK1shRNA or rAd/U6-BECN1shRNA. Five d post transduction, cells were subjected to immunoblotting analyses of LC3, ULK1, BECN1, SQSTM1, and tubulin (left) and senescence-associated (SA)-ß-gal staining (right). Quantification of the signals is shown in the bar graphs in the corresponding panels (n = 3 or n = 4 as indicated in the figures). **P < 0.01, *** < 0.001 vs. control (Con) or LacZshRNA.
Silencing ARG2 in senescent endothelial cells augments autophagy
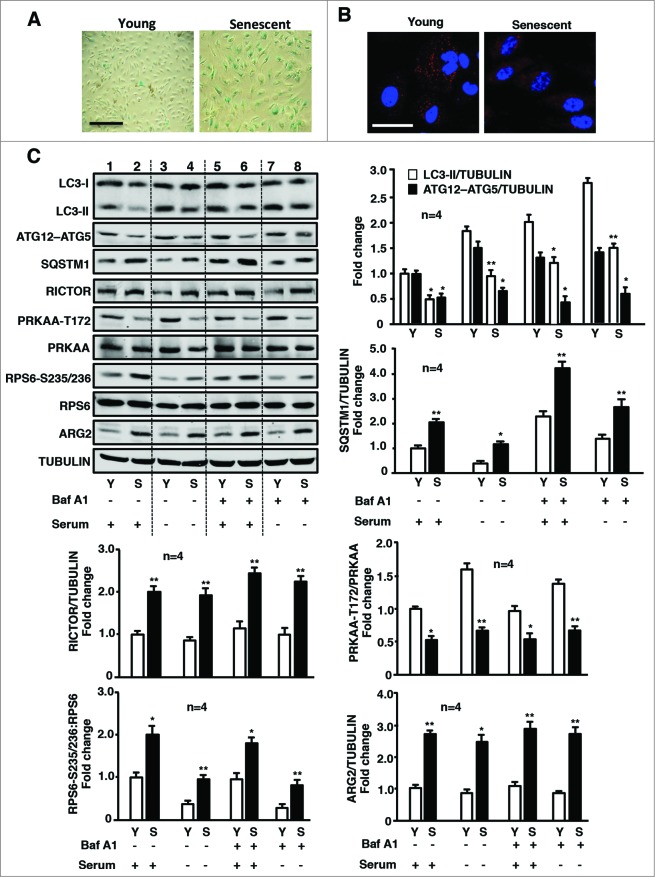
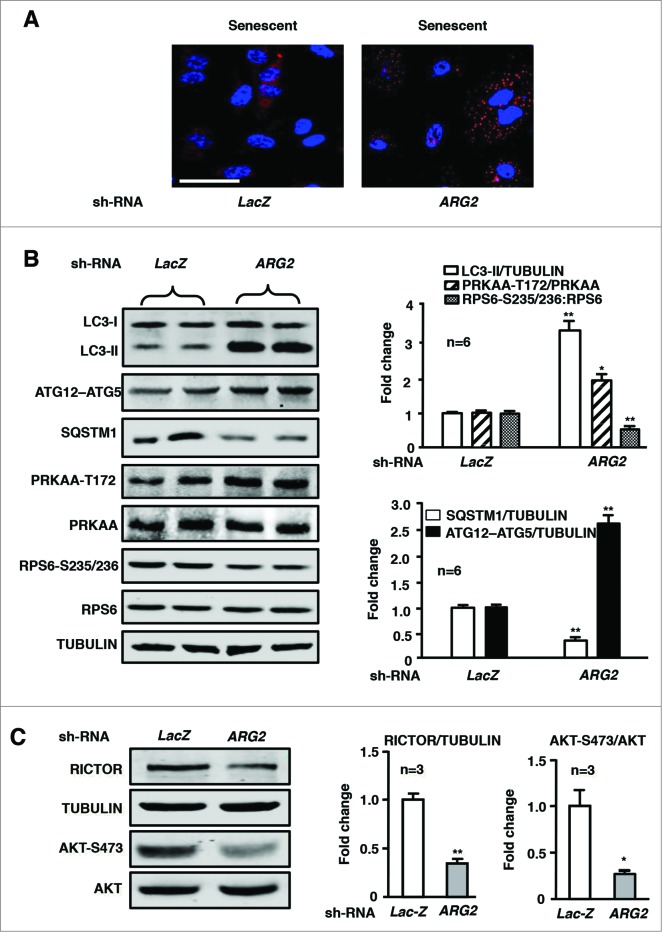
Our previous study showed a positive hyperactive crosstalk between ARG2 and RPS6KB1 in endothelial aging.22 Our present study further demonstrated an impaired autophagy in the senescent endothelial cells (Fig. 6). The cell senescence was confirmed by SA-β-gal staining (Fig. 6A). The decreased autophagosome formation in senescent cells was visualized by immunofluorescence confocal microscopy showing disappearing of the punctate pattern of LC3 staining (Fig. 6B). Moreover, impaired autophagy in senescent cells was also revealed by immunoblotting analysis showing decreased LC3-II protein and ATG12–ATG5 conjugate levels as well as enhanced SQSTM1 levels as compared to young cells under 4 experimental conditions, i.e., serum-growth medium +/− Baf A1, and serum-starvation medium +/− Baf A1) (Fig. 6C). Consistent with the previous study, elevated ARG2 protein levels and enhanced RPS6KB1 activity were detected in senescent endothelial cells (Fig. 6C), whereas ARG1 was not detectable in young nor in senescent HUVECs using human ARG1 expressed from plasmid pBudCE4.1-hARG129 as a positive control (Fig. S2). In addition, an elevated RICTOR protein level and decreased PRKAA activation were also observed in the senescent cells (Fig. 6C). Moreover, knocking down ARG2 in the senescent cells significantly enhanced autophagy as reflected by the increased punctate pattern of the immunostaining of LC3 (Fig. 7A), the augmented LC3-II and ATG12–ATG5 levels and reduced SQSTM1 levels (Fig. 7B), which was accompanied by enhanced PRKAA activation and decreased RPS6KB1 signaling (Fig. 7B). Furthermore, silencing ARG2 in the senescent endothelial cells reduced the MTORC2 component RICTOR levels and its downstream effector AKT activation (AKT-S473 levels, Fig. 7C). In contrast to silencing ARG2, treatment of senescent cells with the arginase inhibitor S-12 bromoethyl-L-cystine-HCl (BEC) or nor-NOHA did not affect autophagy markers or signaling (Fig. S3), demonstrating that ARG2 activates MTORC2-AKT-MTORC1-RPS6KB1 signaling, resulting in inactivation of PRKAA and suppression of autophagy in the senescent cells, which is independent of its L-arginine ureahydrolase activity and reinforce our finding with overexpression of the inactive ARG2 mutant H160F.
Figure 6.
Senescent HUVECs exhibit impaired autophagy, associated with elevated ARG2 and MTORC2-RPS6KB1, and decreased PRKAA1 signaling. Young and senescent HUVECs were subjected to (A) SA-β-gal staining to confirm the cell senescence. (B) The immunostaining for LC3-I/-II (red) and DAPI (blue). Shown are representative merged images from 6 independent experiments. Scale bar = 0.1 mm. (C) Young and senescent cells were cultured in serum-growth or serum-starvation medium for 16 h in the absence or presence of Baf A1 (20 nmol/L) for the last 1 h as indicated, followed by immunoblotting analysis of LC3-I/-II, ATG12–ATG5, SQSTM1, RICTOR, PRKAA-T172, and total PRKAA, RPS6-S235/236 and total RPS6, ARG2 levels and tubulin, which served as a loading control. Quantification of the signals is shown in the bar graphs (n = 4). * < 0.05, **P < 0.01 vs. young within each corresponding experimental condition.
Figure 7.
Silencing ARG2 in senescent HUVECs augments autophagy and PRKAA signaling while inhibiting RICTOR expression and activation of AKT and RPS6KB1. The senescent HUVECs were transduced either with rAd/U6-LacZshRNA as control or rAd/U6-ARG2shRNA. Eighty-8 h (serum-free during the last 16 h) post transduction, cells were subjected to (A) immunostaining of LC3-I/-II (red) followed by DAPI staining (blue). Shown are representative merged images from 6 independent experiments. Scale bar = 0.1 mm. ((B)and C) Immunoblotting analysis LC3-I/-II, ATG12–ATG5, SQSTM1, PRKAA-T172, and total PRKAA, RPS6-S235/236 and total RPS6. RICTOR and tubulin, AKT-S473 and total AKT. Quantification of the immunoblotting signals is shown in the bar graphs in the right panels (n = 3 to 6 as indicated in the figures). *P < 0.05, **P < 0.01 vs. LacZshRNA control.
Crosstalk between RICTOR and ARG2 in regulation of autophagy in senescent cells
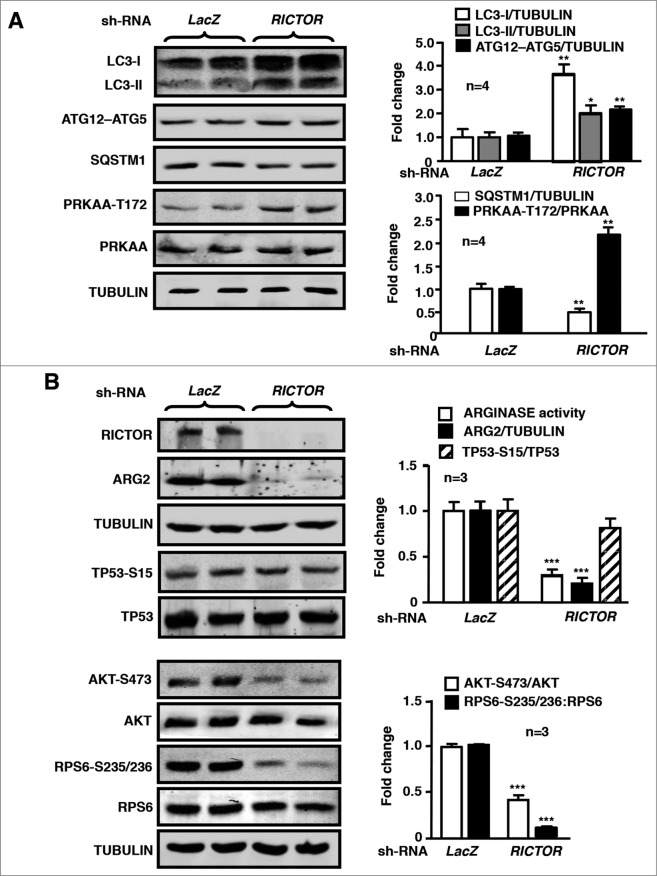
Interestingly, in senescent endothelial cells, silencing RICTOR enhanced the autophagy marker LC3-II and ATG12–ATG5 levels, while it decreased SQSTM1 levels, along with increased activation of PRKAA (Fig. 8A) and decreased activation of AKT and RPS6KB1 as expected (Fig. 8B). In contrast, TP53-S15 levels reflecting TP53 activation were not affected by silencing RICTOR in the senescent cells (Fig. 8B). ARG2 expression was also strongly inhibited upon RICTOR silencing (Fig. 8B). The results from Fig. 7 and 8 demonstrate a feed-forward regulation between MTOR signaling and ARG2 in senescent endothelial cells.
Figure 8.
Silencing RICTOR in senescent HUVECs enhances autophagy and PRKAA signaling while reducing ARG2 protein level and signaling of TP53, AKT, and RPS6KB1. The senescent HUVECs were transduced either with rAd/U6-LacZshRNA as control or rAd/U6- RICTORshRNA. Eighty-8 h (serum-free during the last 16 h) post transduction, cells were subjected to immunoblotting analysis of RICTOR, LC3-I/-II, ATG12–ATG5, SQSTM1, and tubulin, AKT-S473 and total AKT, RPS6-S235/236 and total RPS6, PRKAA-T172 and total PRKAA, TP53-S15 and total TP53, ARG2 and tubulin. Quantification of the immunoblotting signals is shown in the bar graphs in the right panels (n = 3 to 4 as indicated in the figures). *P < 0.05, **P < 0.01, ***P < 0.001 vs. LacZshRNA control.
Arg2 deficiency in mouse enhances endothelial autophagy in atherosclerosis
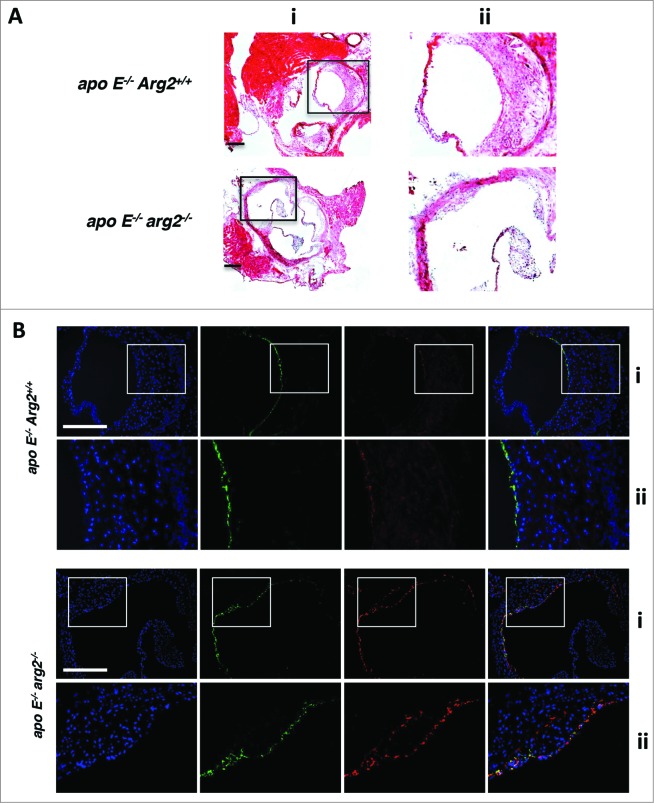
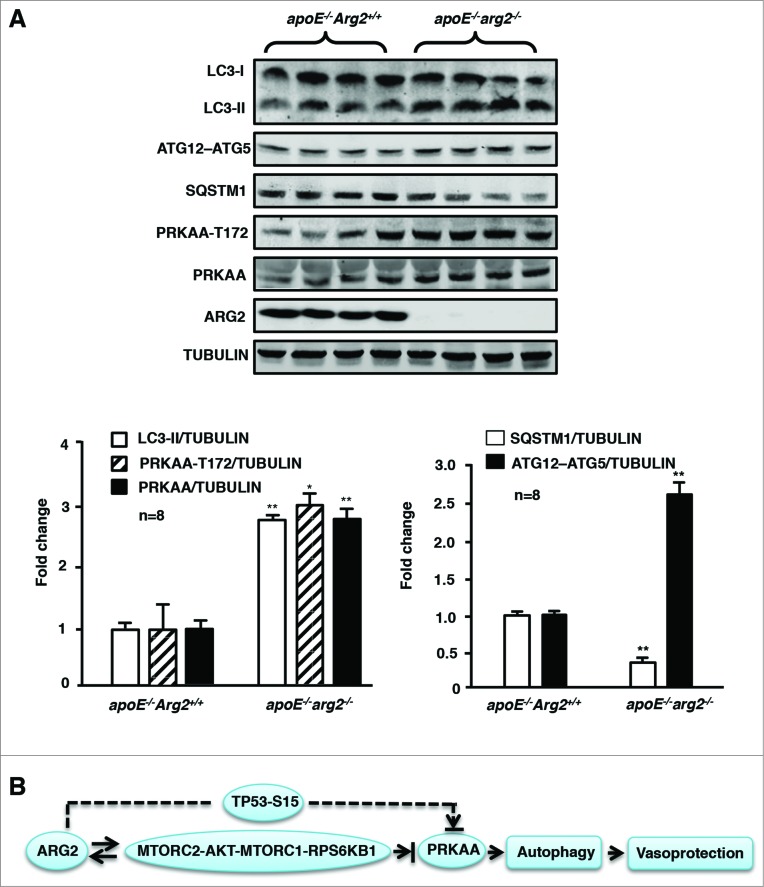
Deficiency in the Arg2 gene in the atherosclerosis-prone apoe−/− mice (apoe−/−arg2−/−) resulted in decreased atherosclerotic lesion formation (Fig. 9A) and reveals more stable plaque features as previously shown by our study.24 In the current study, immunofluorescence confocal microscopy revealed an increase in LC3-II staining in the aortic roots, which colocalized with VWF-expressing endothelial cells in the aortas from the apoe−/−arg2−/− mouse (Fig. 9B). This result demonstrates that endothelial cells have increased levels of autophagosomes in the apoe−/−arg2−/− mice as compared to the control apoe−/−Arg2+/+ animals. Consistent with the immunostaining data, immunoblotting displayed enhanced levels of the autophagosome marker LC3-II and ATG12–ATG5 levels while there were decreased SQSTM1 levels, along with increased levels of the activated T172-PRKAA as well as total PRKAA (Fig. 10A).
Figure 9.
Ablation of Arg2 in atherosclerosis-prone apoe−/− mice enhances autophagy in endothelial cells in the aortas. apoe−/−Arg2+/+ and apoe−/−arg2−/− mice were fed a high-fat diet for 10 wk. AR cryosections (7 μm) in adjacent samples were subjected to (A) hematoxylin and eosin staining. (ii) Is the enlargement of the insert in (i). Scale bars = 0.2 mm. (B) immunofluorescence costaining of the LC3-I/-II (red) and endothelial cell marker VWF (green) followed by counterstaining with DAPI for nuclei (blue). Representative images from 6 individual animals of each genotype are shown. The merged images are also shown. Images in panels (i) correspond to the images in (Aii) of the corresponding genotype. Panels (ii) are the enlargement of the inserts in the corresponding (Bi). Scale bars = 0.2 mm.
Figure 10.
Ablation of Arg2 in atherosclerosis-prone apoe−/− mice enhances PRKAA signaling and autophagy in the aortas. apoe−/−Arg2+/+ and apoe−/−arg2−/− mice were fed a high-fat diet for 10 weeks. (A) Aortas from mice of both genotypes were cleaned of perivascular tissues and subjected to immunoblotting analysis of LC3-I/-II, ATG12–ATG5, SQSTM1, PRKAA-T172 and total PRKAA, ARG2, and TUBULIN. Quantification of the signals is shown in the corresponding lower panels (n = 8 each group). *P < 0.05, **P < 0.001 vs. apoe−/−Arg2+/+. (B) Schematic summary of the major findings of this study. A feed-forward crosstalk between elevated ARG2 expression and RICTOR activates the MTORC2-AKT-MTORC1-RPS6KB1 signaling cascade, which leads to inhibition of PRKAA and subsequently inhibition of endothelial autophagy in senescent endothelial cells. ARG2 can also activate the TP53 pathway in parallel with MTOR signaling, resulting in inhibition of PRKAA and suppression of autophagy. ARG2 exerts these effects independently of its L-arginine ureahydrolase activity. The reduced PRKAA activation by ARG2 results in impairment of endothelial autophagy and acceleration of endothelial senescence and atherosclerosis.
Discussion
Emerging evidence suggests that functional autophagy protects against damaging signals imposed by various stressors in aging and age-associated cardiovascular diseases.30 Pharmacological or genetic inhibition of autophagy increases cardiomyocyte death, whereas upregulation of autophagy enhances cell survival after ischemia/reperfusion injury.31,32 Adequate autophagy induction reduces atherosclerosis, which is attributable to protection of endothelial and smooth muscle cell damage and foam cell formation.9,10 Our recent studies provide evidence that the enzyme ARG2 accelerates endothelial senescence and adhesion molecule expression in vascular aging through NOS3-uncoupling22 and promotes macrophage pro-inflammatory responses in atherosclerosis and obesity.24 Most recently, a pleiotropic effect of ARG2 in smooth muscle cell apoptosis and senescence in atherosclerosis, which is notably independent of its L-arginine ureahydrolase activity, has been described by our group.23 In line with these results, our current study further explored the potential regulatory function of ARG2 in endothelial autophagy formation in advanced atherosclerosis. The results show that ARG2 suppresses autophagy in endothelial cells under both serum-growth and serum-starvation conditions, which is also independent of the L-arginine ureahydrolase activity, since overexpression of the inactive ARG2 in young endothelial cells suppresses autophagy as the WT ARG2 does. Moreover, pharmacological inhibition of arginase activity in senescent cells does not affect autophagy, whereas silencing ARG2 enhances autophagy in senescent cells. Given that the inactive mutant H160F does not cause NOS3-uncoupling in endothelial cells as demonstrated by our previous study,22 these results demonstrate that the effects of ARG2 on autophagy as shown in the current study are not due to reduced NO production. Moreover, we showed in the current study that knocking out the Arg2 gene in atherosclerosis-prone Apoe-deficient mice enhances autophagy in the aortas, which is associated with reduced atherosclerotic lesions and a more stable plaque characteristic as previously shown.24
It is noteworthy that the role of autophagy in atherosclerosis is still unclear and currently under intensive investigation. Both beneficial and detrimental roles of autophagy in atherosclerosis have been reported, which are associated with the extent of autophagy activity.9 Evidence is accumulating that adequate activated autophagy in vascular cells including endothelial and smooth muscle cells (SMC) protects against endothelial and SMC injury/death by risk factors, contributing to plaque stability, while inhibition of autophagy accelerates apoptosis, necrosis, and senescence conferring plaque vulnerability.9 In contrast, excessive activation of autophagy by overwhelming or continuous cellular stress causes an authophagic type of cell death, which is obviously detrimental.9 Our in vitro data using an autophagy inhibitor or genetic silencing of components that are essential for autophagosome formation reveals that suppression of autophagy promotes endothelial senescence. Moreover, the data from an animal model in our current study also reveal a positive correlation between augmentation of autophagy and the more stable plaque phenotype in apoe−/−arg2−/− mice, which supports the aforementioned notion about the role of autophagy in atherosclerosis and indicates that the augmentation of autophagy by Arg2-deficiency is not excessive and thus protective. However, a definite role of endothelial autophagy in atherosclerosis awaits studies by generating atherosclerosis-prone mice genetically deficient in endothelial autophagy.
The expected alterations resulting from increased arginase activity upon ectopic WT ARG2 expression, including decrease in L-arginine, increased polyamine as well as decreased NO production, have been reported to enhance autophagy by numerous other studies.33-36 However, our current study demonstrates that the net effect of ARG2 overexpression is inhibition of autophagy, which is attributable to the activation of MTORC1-RPS6KB1 and inhibition of PRKAA signaling independently of its L-arginine ureahydrolase activity. Taking into account the other studies, our current study suggests that the enzymatic activity-independent inhibitory effect of ARG2 on autophagy is dominant over the stimulatory effects on autophagy resulting from its activity-dependent decrease in L-arginine, increase in polyamine, and/or regulation of NO production.
The mechanisms underlying the pleiotropic and enzymatic activity-independent suppressing effects on autophagy in endothelial cells are mediated through inhibition of PRKAA signaling, since ectopic expression of ARG2 or the inactive mutant H160F in young endothelial cells reduced PRKAA activation, and expression of a constitutive active PRKAA mutant rescued the autophagy in the endothelial cells, demonstrating that ARG2 decreases autophagy in endothelial cells through inhibition of PRKAA. This conclusion was further confirmed by the fact that in the senescent endothelial cells in which the ARG2 expression was increased autophagy levels and PRKAA activation were decreased, while silencing ARG2 enhanced PRKAA activation and also autophagy as assessed by combinatory detection of the autophagosome formation markers LC3-II and the ATG12–ATG5 conjugate, along with SQSTM1. In contrast to silencing ARG2, treatment of senescent cells with the arginase inhibitor BEC or nor-NOHA did not affect autophagy markers or signaling. These results suggest that ARG2, independently of its L-arginine ureahydrolase activity, decreases endothelial autophagy through inhibition of PRKAA signaling. Importantly, knockout of the Arg2 gene in apoe−/− mice increased PRKAA activation as well as endothelial autophagy in the aortas, demonstrating a critical role of ARG2-induced suppression of PRKAA and autophagy in atherogenesis.
PRKAA has been shown to enhance autophagy formation at least by 2 mechanisms: a direct activation of the key autophagy kinase ULK1 through phosphorylation at S317, which is involved in nucleation of the phagophore membrane11 and an indirect activation of autophagy through inhibition of MTORC1 signaling, which is known as a suppressor of autophagy.11 Our previous study and also our current work showed that ARG2 activates the MTORC1-RPS6KB1 pathway, resulting in accelerated endothelial aging.22 The negative relationship between MTORC1-RPS6KB1 activity and PRKAA activation regulated by ARG2 as shown in this study raises the question of whether PRKAA is an upstream target of MTORC1 in mammals as shown in the literature.16,37 To our surprise, silencing RPS6KB1 prevented ARG2s inhibitory effect on PRKAA activation. This result demonstrates for the first time that in human endothelial cells MTORC1-RPS6KB1 acts upstream of PRKAA and inhibits its activity. Similar findings are also reported in yeast, which show that Snf1 (yeast homolog of PRKAA) can be inhibited by TORC1.38 The exact mechanisms of how MTORC1 inhibits PRKAA in the endothelial cells require further investigation.
A further important finding of our study is that senescent endothelial cells as compared to the young cells express higher levels of RICTOR, a key component of the MTORC2 complex. Silencing ARG2 in the cells is able to inhibit its downstream signaling, including AKT, MTORC1, and RPS6KB1 and to restore PRKAA activation and autophagy. Interestingly, silencing RICTOR also inhibits ARG2 expression and enhances PRKAA activation as well as autophagy. These results demonstrate that a positive crosstalk between ARG2 and RICTOR leads to activation of MTORC2, and subsequently AKT, MTORC1, and RPS6KB1, resulting in inhibition of PRKAA and autophagy in endothelial cells, contributing to atherosclerosis. This finding explains our previous observation of the positive regulation of ARG2 and RPS6KB1 in endothelial aging.22 Future experiments shall investigate the mechanisms of ARG2-induced activation of MTORC2, i.e., upregulation of RICTOR expression.
TP53, the tumor suppressor, is a well-characterized stress-responsive transcription factor.39 The function of TP53 in regulation of autophagic flux is complex and to some degree is context dependent. Both stimulation and suppression of autophagy have been demonstrated.39 In nematodes, mice and human cells, pharmacological or genetic inhibition of TP53 triggers a canonical autophagic pathway.40 We have previously shown that TP53 is activated by ARG2 in human smooth muscle cells and is involved in the regulation of cell senescence and apoptosis.23 Similar to the smooth muscle cells, we show here that in endothelial cells, ARG2 also activates TP53 independently of its ureahydrolase activity, resulting in inhibition of PRKAA signaling and subsequently inhibition of autophagy. This effect of TP53 is independent of MTOR-RPS6KB1 signaling, since silencing TP53 or RPS6KB1 did not influence each other in young cells overexpressing ARG2. In the senescent endothelial cells, in which ARG2 and RPS6KB1 signaling are upregulated, silencing RICTOR was able to decrease its downstream signaling, i.e., activation of AKT, MTORC1, and RPS6KB1, as well as ARG2. However, it did not affect TP53 activation, suggesting that activation of the TP53 pathway in the senescent cells is mainly maintained by other pathways, which are independent of ARG2-MTOR2 signaling. The results also illustrate that the MTORC2-AKT-MTORC1-RPS6KB1 signaling cascade is the major pathway in inhibition of PRKAA and autophagy in senescent cells.
In conclusion, our current study provides evidence that ARG2 impairs endothelial autophagy independently of the L-arginine ureahydrolase activity through a feed-forward interaction with MTORC2 signaling, leading to reduced PRKAA activation and, subsequently, inhibition of autophagy in atherosclerosis (Fig. 10B). Modulation of autophagy function by targeting ARG2 gene expression may have strong implications for treatment of atherosclerotic coronary artery disease.
Materials and Methods
Materials and chemicals
All chemicals, if not specifically indicated, including those used for immunoblotting (mouse anti-tubulin monoclonal antibody) were obtained from Sigma. Bafilomycin A1 (19–148) was obtained from Millipore; BEC (cay-10170–5) was from Cayman Adipogen; Nor-NOHA (399275) was from Calbiochem. Rabbit antibodies against phospho-RPS6-S235/236 (2211s), RPS6 (2217s), phospho-AKT-S473 (9271s), AKT (9272s), phospho-TP53-S15 (9284s), phospho-PRKAA-T172 (2535s), PRKAA (2793s), LC3A/B (4108s), mouse anti-TP53 (2524s) and anti-ATG12 (4180s) were from Cell Signaling Technology; mouse anti-RPS6KB1 (611260) was from BD Transduction Laboratories; rabbit anti-ARG2 (sc-20151), rabbit anti-TP53 (sc-6243), mouse anti-RICTOR (sc-271081), rabbit anti-ULK1 (sc-33182), mouse anti-BECN1 (sc-48381) and goat anti-LC3A/B (sc-16756) were from Santa Cruz Biotechnology; rabbit anti-VWF/von Willebrand factor (ab6994) and anti-SQSTM1 (ab109012) were from Abcam. Alexa Fluor 680-conjugated anti-mouse IgG (A21057), Alexa Fluor 488-conjugated goat anti-rabbit IgG (A-11008) and Alexa Fluor 594-conjugated donkey anti-goat IgG (H+L) (A-11058) were from Molecular Probes/Invitrogen; IRDye800-conjugated anti-rabbit IgG (926–32211) was from LI-COR Biosciences; all cell culture media and materials were purchased from Gibco BRL. The plasmid pBudCE4.1-hARG1 (human) was kindly provided by Prof. Nan-Hong Tang (Union Hospital, Fujian Medical University, China).29
Recombinant adenovirus (rAd)
Generation of rAd-expressing shRNA targeting human RICTOR driven by the U6 promoter (rAd/U6-RICTORshRNA), and a HA-tagged constitutively active PRKAA1 driven by the CMV promoter (rAd/CMV-HA-PRKAA1) were carried out with Gateway Technology (Invitrogen Life Technologies) according to the manufacturer's instructions.41,42 The targeting sequences for RICTOR-shRNA, LC3-shRNA, ULK1-shRNA, and BECN1-shRNA are indicated in boldface below (only the sense strand is shown): ACTTGTGAAGAATCGTATCTT, GCTTACAGCTCAATGCTAATC, GCAGAACTACCAGCGCATTGA and GCTCAGTATCAGAGAGAATAC, respectively. The gene encoding constitutively active HA-PRKAA1 was amplified by polymerase chain reaction from the expression plasmid pEBG-PRKAA1 (1–312)43 which was purchased from Addgene, using the HA-tagged forward primer:
5′-GGGGTACCCCATGTACCCATAC GATGTTCCAGA TTACGCTGCCGAGAAGCAG
AAGCACGAC-3′ and the reverse primer:
5′-AAGGAAAAAAGCGGCCGCTTACTGTGCAAGAATTTTAATTAG-3′.
rAd/U6-LacZshRNA, rAd/U6-ARG2shRNA, rAd/U6-RPS6KB1shRNA, rAd/U6-TP53shRNA, rAd/CMV empty vector, rAd/CMV-Arg2 and rAd/CMV-Arg2-H160F (a L-arginine ureahydrolase inactive mutant of Arg2) were generated and characterized as previously described.22,23
Cell culture
Preparation of young and senescent HUVECs, and transduction of HUVECs by recombinant adenovirus were carried out as previously described.22,44 Briefly, cells of passage 1 to 3 (P1 to P3) were used as young cells. For preparation of senescent cells, the cells were further split in a ratio of 1:3 continuously over a period of several weeks till replicative senescence was reached as assessed by senescence-associated (SA)-ß-gal staining (P9 to P12). In the manuscript, nonsenescent endothelial cells are referred to as “young” cells. HAEC were purchased from PromoCell (C-12271) and cultured the same as HUVEC.
Animals
The apoe−/−Arg2+/+ mice from Jackson Laboratory and the generation of the apoe−/−arg2−/− mice, both in the C57BL/6J background, as well as the animal experimental procedures were as previously described.24 Briefly, to accelerate the atherosclerotic lesion formation, 10-wk-old male apoe−/−Arg2+/+ and apoe−/−arg2−/− mice were fed a high-fat diet (HF; energy content: 55% fat, 21% protein, and 24% carbohydrate; Harlan Teklad, TD.93075) for 10 wk. At 20 wk of age, animals were anesthetized with ketamine (100 mg/kg i.p.) and xylazine (10 mg/kg i.p.), the entire aorta from the heart to the iliac bifurcation were removed and dissected free from fat and adhering perivascular tissue for immunoblotting analysis. For immunostaining, the aortic roots (AR) were snap frozen in Optimal Cutting Temperature compound (Sakura Finetek, 4583) and the 7-μm thick cryosections of the AR were prepared. Housing and animal experimentation were approved by the Service de la sécurité alimentaire et des affaires vétérinaires, Etat de Fribourg.
Immunofluorescence staining of AR and endothelial cells
AR-cryosections (7 μm) were fixed with 4% paraformaldehyde and blocked with 2% bovine serum albumin (BSA; Fluka, 05479) in phosphate-buffered saline (137 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4, 1.8 mmol/L KH2PO4,) for 30 min. To assess the changes of LC3-I/-II levels in endothelial cells, costaining of LC3-I/-II with VWF was performed. The sections were first incubated for 2 h with both goat anti-LC3-I/-II and rabbit anti-VWF. After washing, the sections were incubated with Alexa Fluor 594-conjugated donkey anti-goat IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) at room temperature for 1 h. The sections were finally counterstained with 300 nmol/L DAPI (Invitrogen, D-1306) for 2 min. The immunofluorescence signals were visualized using a Leica DIM6000 confocal microscope. The same procedure was applied for the immunostaining of the HUVECs that were cultured on a coverslip, except that the cells were incubated with anti-LC3-I/-II antibody.
Immunoblotting
Cell or tissue lysate preparation, SDS-PAGE, and transfer of SDS gels to Immobilon-P membranes (Millipore, IPFL00010) were performed as previously described.44 The resultant membrane was first incubated with the corresponding primary antibody at room temperature for 2 h with gentle agitation after blocking with 5% skim milk. The blot was then further incubated with a corresponding anti-mouse (Alexa Fluor 680 conjugated) or anti-rabbit (IRDye 800 conjugated) secondary antibody. Signals were visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences). Quantification of the signals was performed using NIH Image 1.62 software (U. S. National Institutes of Health).
Statistical analysis
Data are given as mean ± SEM. In all experiments, n represents the number of experiments or animals. Statistical analysis was performed with the Student unpaired t-test or analysis of variance (ANOVA) with Bonferroni post test. Differences in mean values were considered significant at 2-tailed P ≤ 0.05. The number of treatment groups in each set of experiments and the n number (the number of repeated set of in vitro experiments conducted with cells or the number of animals of each genotype used) are indicated in each figure.
Acknowledgments
We are grateful to Mr. Jean Ruffieux for technical assistance with endothelial cell isolation and culture and Ms. Srividya Velagapudi for construction of shRNA-RICTOR.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Swiss National Science Foundation (310030_141070/1), Swiss Heart Foundation, and National Center of Competence in Research Program (NCCR-Kidney.CH). Yuyan Xiong and Yi Yu are supported by the Chinese Scholarship Council.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368:1845-6; PMID:23656658; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 3. Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S. et al. . Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 4. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol 2013; 20:59-66; PMID:23202584; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 2010; 584:1374-8; PMID: 20153326; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 7. Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I. Endothelial cell senescence in human atherosclerosis: role of telomere in endothelial dysfunction. Circulation 2002; 105:1541-4; PMID: 11927518; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 8. LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol 2012; 590:3305-16; PMID:22570377; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schrijvers DM, De Meyer GR, Martinet W. Autophagy in atherosclerosis: a potential drug target for plaque stabilization. Arterioscler Thromb Vasc Biol 2011; 31:2787-91; PMID:22096098; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 10. Ouimet M. Autophagy in obesity and atherosclerosis: interrelationships between cholesterol homeostasis, lipoprotein metabolism and autophagy in macrophages and other systems. Biochim Biophys Acta 2013; 1831:1124-33; PMID:23545567; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 11. Kim J, Kundu M, Viollet B, Guan KL. AMPK and MTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Z, Ming XF. MTOR signalling: the molecular interface connecting metabolic stress, aging and cardiovascular diseases. Obes Rev 2012; 13 Suppl 2:58-68; PMID:23107260; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 13. Laplante M, Sabatini DM. MTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID: 22500797; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang RC, Wei YJ, An ZY, Zou ZJ, Xiao GH, Bhagat G, White M, Reichelt J, Levine B. AKT-mediated regulation of autophagy and tumorigenesis through eclin 1 phosphorylation. Science 2012; 338:956-9; PMID: 23112296; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of MTORC1. Proc Natl Acad Sci U S A 2013; 110:E2950-E7; PMID:23878245; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esclatine A, Chaumorcel M, Codogno P. Macroautophagy signaling and regulation. Curr Top Microbiol Immunol 2009; 335:33-70; PMID:19802559; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 18. Armour SM, Baur JA, Hsieh SN, Land-Bracha A, Thomas SM, Sinclair DA. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany NY) 2009; 1:515-28; PMID:20157535s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu X, Chen K, Kobayashi S, Timm D, Liang Q. Resveratrol attenuates doxorubicin-induced cardiomyocyte death via inhibition of p70 S6 kinase 1-mediated autophagy. J Pharmacol Exp Ther 2012; 341:183-95; PMID:22209892; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:4180-9; PMID:17699586; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Z, Ming XF. Arginase: the emerging therapeutic target for vascular oxidative stress and inflammation. Front Immunol 2013; 4:149; PMID:23781221; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yepuri G, Velagapudi S, Xiong YY, Rajapakse AG, Montani JP, Ming XF, Yang Z. Positive crosstalk between arginase-II and S6K1 in vascular endothelial inflammation and aging. Aging Cell 2012; 11:1005-16; PMID:22928666; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 23. Xiong YY, Yu Y, Montani JP, Yang ZH, Ming XF. Arginase-II induces vascular smooth muscle cell senescence and apoptosis through p66Shc and p53 independently of its L-arginine ureahydrolase activity: implications for atherosclerotic plaque vulnerability. J Am Heart Assoc 2013; 2:e000096; PMID:23832324; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ming XF, Rajapakse AG, Yepuri G, Xiong YY, Carvas JM, Ruffieux J, Scerri I, Wu Z, Popp K, Li J, et al. . Arginase II promotes macrophage inflammatory responses through mitochondrial reactive oxygen species, contributing to insulin resistance and atherogenesis. J Am Heart Assoc 2012; 1:e000992; PMID: 23130157; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 1991; 266:17707-12; PMID:1832676 [PubMed] [Google Scholar]
- 26. Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E Cells. Cell Struct Funct 1998; 23:33-42; PMID:9639028; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 27. Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997; 243:240-6; PMID:9030745; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 28. Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagiclysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 1982; 79:1889-92; PMID:6952238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang N, Wang Y, Wang X, Zhou L, Zhang F, Li X, Chen Y. Stable overexpression of arginase I and ornithine transcarbamylase in HepG2 cells improves its ammonia detoxification. J Cell Biochem 2012; 113:518-27; PMID:21938740; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 30. Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol 2013; 305:H459-76; PMID:23748424; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemiareperfusion injury in cardiac myocytes. J Biol Chem 2006; 281:29776-87;PMID: 16882669; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 32. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007; 100:914-22; PMID:17332429; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 33. Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia-Arencibia M, Rose C, Luo S, Underwood BR, et al. . Complex inhibitory effects of nitric oxide on autophagy. Mol Cell 2011; 43:19-32; PMID:21726807; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia-Navas R, Munder M, Mollinedo F. Depletion of L-arginine induces autophagy as a cytoprotective response to endoplasmic reticulum stress in human T lymphocytes. Autophagy 2012; 8:1557-76;PMID: 22874569; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. . Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305-14; PMID:19801973; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 36. Vanrell MC, Cueto JA, Barclay JJ, Carrillo C, Colombo MI, Gottlieb RA, Romano PS. Polyamine depletion inhibits the autophagic response modulating Trypanosoma cruzi infectivity. Autophagy 2013; 9:1080-93; PMID:23697944; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of MTOR. Nat Rev Mol Cell Biol 2013; 14:133-9; PMID:23361334; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Klionsky DJ. The regulation of autophagy – unanswered questions. J Cell Sci 2011; 124:161-70; PMID:21187343; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 2013; 23:310-22; PMID: 23726895; http:dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 40. Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. . Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10:676-87; PMID:18454141; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ming XF, Viswambharan H, Barandier C, Ruffieux J, Kaibuchi K, Rusconi S, Yang Z. Rho GTPaseRho kinase negatively regulates endothelial nitric oxide synthase phosphorylation through the inhibition of protein kinase BAKT in human endothelial cells. Mol Cell Biol 2002; 22:8467-77; PMID:12446767; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ming XF, Rajapakse AG, Carvas JM, Ruffieux J, Yang Z. Inhibition of S6K1 accounts partially for the anti-inflammatory effects of the arginase inhibitor L-norvaline. BMC Cardiovasc Disord 2009; 9:12; PMID: 19284655; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. . Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; PMID:21205641; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rajapakse AG, Yepuri G, Carvas JM, Stein S, Matter CM, Scerri I, Ruffieux J, Montani JP, Ming XF, Yang Z. Hyperactive S6K1 mediates oxidative stress and endothelial dysfunction in aging: inhibition by resveratrol. PLoS One 2011; 6:e19237; PMID:21544240; http:dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.