Abstract
Dysregulation of autophagy contributes to neuronal cell death in several neurodegenerative and lysosomal storage diseases. Markers of autophagy are also increased after traumatic brain injury (TBI), but its mechanisms and function are not known. Following controlled cortical impact (CCI) brain injury in GFP-Lc3 (green fluorescent protein-LC3) transgenic mice, we observed accumulation of autophagosomes in ipsilateral cortex and hippocampus between 1 and 7 d. This accumulation was not due to increased initiation of autophagy but rather to a decrease in clearance of autophagosomes, as reflected by accumulation of the autophagic substrate SQSTM1/p62 (sequestosome 1). This was confirmed by ex vivo studies, which demonstrated impaired autophagic flux in brain slices from injured as compared to control animals. Increased SQSTM1 peaked at d 1–3 but resolved by d 7, suggesting that the defect in autophagy flux is temporary. The early impairment of autophagy is at least in part caused by lysosomal dysfunction, as evidenced by lower protein levels and enzymatic activity of CTSD (cathepsin D). Furthermore, immediately after injury both autophagosomes and SQSTM1 accumulated predominantly in neurons. This was accompanied by appearance of SQSTM1 and ubiquitin-positive puncta in the affected cells, suggesting that, similar to the situation observed in neurodegenerative diseases, impaired autophagy may contribute to neuronal injury. Consistently, GFP-LC3 and SQSTM1 colocalized with markers of both caspase-dependent and caspase-independent cell death in neuronal cells proximal to the injury site. Taken together, our data indicated for the first time that autophagic clearance is impaired early after TBI due to lysosomal dysfunction, and correlates with neuronal cell death.
Keywords: autophagy, autophagy flux, lysosome, neuronal cell death, traumatic brain injury
Abbreviations: ACTB, actin; β; AIF1/IBA1, allograft inflammatory factor 1; AIFM1, apoptosis-inducing factor, mitochondrion-associated, 1; APC, adenomatous polyposis coli; ATG12, autophagy-related 12; ATG5, autophagy-related 5; ATG7, autophagy-related 7; CAPS12, caspase 12; CASP3, caspase 3; CCI, controlled cortical impact; CD68, CD68 molecule; CSPG4, chondroitin sulfate proteoglycan 4; CTSD, cathepsin D; GFP, green fluorescent protein; LAMP1, lysosomal-associated membrane protein 1; LAMP2, lysosomal-associated membrane protein 2; LC3, microtubule associated protein 1 light chain 3; RBFOX3, RNA binding protein, fox-1 homolog (C. elegans) 3; SPTAN1, spectrin, α, non-erythrocytic 1; SQSTM1, sequestosome 1; TBI, traumatic brain injury; ULK1, unc-51 like autophagy activating kinase 1
Introduction
Macroautophagy (hereafter called autophagy) is a highly conserved cellular degradative process by which cells remove damaged organelles and toxic macromolecules. Autophagy is initiated by the formation of a crescent-shaped membrane structure called the phagophore, which gradually elongates and sequesters parts of the cytoplasm including damaged macromolecules and organelles. Elongating ends of the phagophore subsequently fuse to form a double-membrane vesicle called the autophagosome, which then fuses with the lysosome leading to degradation of its contents by lysosomal hydrolases.1-4
Although under certain circumstances pathologically increased autophagy has been implicated in cell death,5-7 under most circumstances autophagy is considered to be a cytoprotective mechanism. Basal levels of autophagy are important for maintaining cellular homeostasis and appear to be essential for normal cellular function and survival of terminally differentiated cells such as neurons. Mice with neural tissue specific knockout of the essential autophagy genes Atg5 (autophagy-related 5) or Atg7 (autophagy-related 7) develop severe neurodegeneration, leading to abnormal motor function and reflexes.8,9 Impaired autophagy has been implicated in neurodegenerative disorders such as Parkinson, Alzheimer, and Huntington diseases and in lysosomal storage disorders.10-17 The pathophysiology of these diseases is associated with autophagy defects contributing to accumulation of ubiquitin-positive protein aggregates and to neuronal cell dysfunction and death. In lysosomal storage diseases, defects in autophagy are secondary to deficiencies in specific lysosomal hydrolases and consequent impairment of the lysosomal function.16,17
Traumatic brain injury is one of the most common causes of death and long-term impairment among young adults.18 Brain trauma initiates delayed progressive tissue damage through a cascade of molecular and cellular events leading to neuronal cell death.18-20 The role of autophagy in this secondary neurodegeneration is uncertain. Increased markers of autophagy have been reported in the brain following TBI;21-24 however, its cell-type specificity and the mechanism of induction remain unclear. Moreover, the function of autophagy following TBI is controversial, with both beneficial and detrimental roles suggested.25-28
Here we examined levels of autophagy and autophagic flux following TBI induced by controlled cortical impact in wild-type and transgenic GFP-Lc3 autophagy reporter mice. Our data demonstrate that LC3 and autophagosomes accumulate in ipsilateral cortex and hippocampus within hours after injury, and remain elevated for at least 1 wk. Accumulation of autophagosomes after TBI is not due to increased initiation of autophagy, but rather to a temporary impairment of autophagic clearance associated with decreased lysosomal function after TBI. Markers of autophagy remain elevated at later time points, but eventually autophagic flux is restored. Additionally, our analysis demonstrates that initially autophagosomes accumulate specifically in neurons and colocalize with markers of apoptotic cell death. This suggests that early after TBI impaired autophagy may play a detrimental role. Therefore, treatments that either decrease pathological accumulation of autophagosomes or increase their degradation may be neuroprotective after TBI.
Results
Autophagosomes accumulate in the brain after TBI
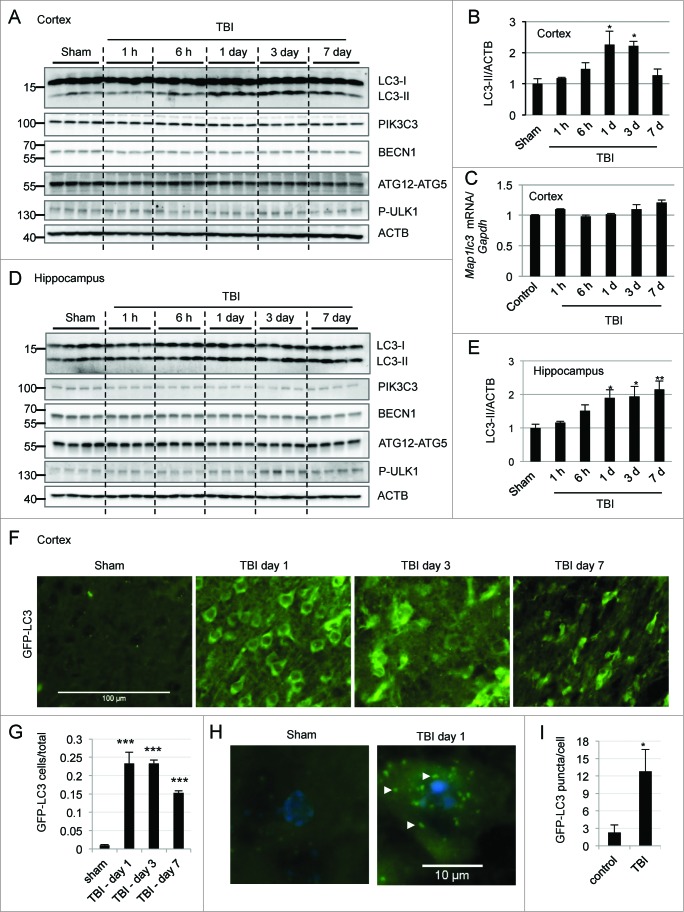
To examine induction of autophagy after TBI, we determined levels of the autophagy marker protein MAP1LC3B/LC3 (microtubule-associated protein 1 light chain 3) in the ipsilateral cortex by western blot. Conversion of LC3-I to LC3-II by the addition of phosphatidylethanolamine is essential for the formation of autophagosomes,4,29,30 and can serve as a marker of autophagy. We found a time-dependent increase in the levels of LC3-II, which peaked between 1 and 3 d after injury and then gradually decreased by d 7 (Fig. 1A,upper panel and Fig. 1B). Confirming that lipidated LC3 associates with membranes after TBI, we observed accumulation of LC3-II in the crude lysosomal/membrane fraction but not in the cytosolic fraction prepared from the cortex of injured mice as compared to sham (Fig. S1). No substantial changes in Map1lc3 mRNA were apparent in the injured cortex as compared to uninjured controls (Fig. 1C). A time-dependent increase in LC3-II was also observed in the ipsilateral hippocampus of injured mice (Fig. 1D and E), suggesting that a direct mechanical injury was not necessary for the induction of autophagy markers.\raster(96%)="rgFigKAUP_A_981787_F0001_B"
Figure 1.
For figure legend, see page 2211.
In order to investigate the potential mechanism of autophagy after TBI we examined levels of proteins involved in autophagosome formation in the injured cortex and hippocampus. Two protein complexes—the PIK3C3/VPS34 (phosphatidylinositol 3-kinase, catalytic subunit type 3)-BECN1/Beclin 1 complex and the ULK1 (unc-51 like autophagy activating kinase 1) complex are involved in regulation and initiation of the autophagic process. Additionally, ATG12 (autophagy-related 12)–ATG5 conjugation is necessary for phagophore elongation.4 No significant increases in the levels of PIK3C3, BECN1, ATG12–ATG5 conjugate, or phospho-ULK1 were observed in the injured cortex as compared to sham-controls (Fig. 1A and Fig. S2A-D). Rather, we noticed a gradual decrease in ATG12–ATG5 conjugate. mRNA levels of Becn1 and Atg12 remained unaltered in the injured cortex as compared to uninjured controls (Fig. S2E and F). Together, these data indicate that autophagy initiation is not increased after TBI.
Similarly, we did not observe any increases in PIK3C3, BECN1, or ATG12–ATG5 conjugate levels in the hippocampus after injury (Fig. 1D and Supplementary Figure S2G-I). Rather, we noticed a small decrease in PIK3C3 at d 1 and 3, and BECN1 at d 7, in the injured hippocampus. ATG12–ATG5 conjugate also decreased slightly. We did observe a slight increase in phospho-ULK1 level in the hippocampus at d 3 and 7 after injury (Fig. 1D and Supplementary Fig. S2J). However, given a decrease in downstream mediators, this is unlikely to result in increased autophagosome formation. Therefore, as in the cortex, increased initiation of autophagy cannot account for the observed accumulation of LC3-II.
In order to confirm upregulation of autophagy markers following TBI, we performed image analysis of transgenic C57BL6 mice ubiquitously expressing GFP-tagged LC3. To ensure that we could detect induction of autophagy in this model, we treated naïve mice with rapamycin, an MTOR inhibitor and inducer of autophagy, and performed direct image analysis of GFP-LC3 fluorescence. As expected, rapamycin treatment led to significantly (P < 0.05) increased numbers of GFP-LC3-positive cortical cells as compared to vehicle-treated controls (Fig. S3A and B). At higher magnification we also observed increased numbers of intracellular GFP-LC3-positive puncta corresponding to phagophores and autophagosomes (Fig. S3C and D). This was further confirmed by the immunofluorescence staining of cortical sections prepared from vehicle and rapamycin-treated GFP-Lc3 mice using GFP antibody. We observed a significant increase in GFP staining, which colocalized with the endogenous GFP-LC3 signal in the cortices of rapamycin-treated mice as compared to cortices of naïve and vehicle-treated controls (Fig. S3E and F).
Following CCI we observed significantly (P < 0.001) higher numbers of GFP-positive cells in the injured cortex (Fig. 1F and G) and hippocampus (Fig. S4A) as compared to sham. The numbers of GFP-LC3-positive cells peaked at d 1 and 3 after injury and decreased but still remained substantially higher at d 7. At higher magnification we observed significant (P < 0.05) accumulation of punctate GFP-LC3-positive autophagic structures in the injured cortex (Fig. 1H and I). We further confirmed our findings using LC3 antibody in injured and sham wild-type animals (Fig. S4B and C) and GFP antibody in injured and sham GFP-Lc3 transgenic mice (Fig. S4D and E). Taken together, these data demonstrate that LC3 and phagophores or autophagosomes accumulate within the ipsilateral hemisphere after TBI in areas both proximal and distal from the injury site.
Autophagosomes accumulate at different times in neurons, activated microglia, and oligodendrocytes
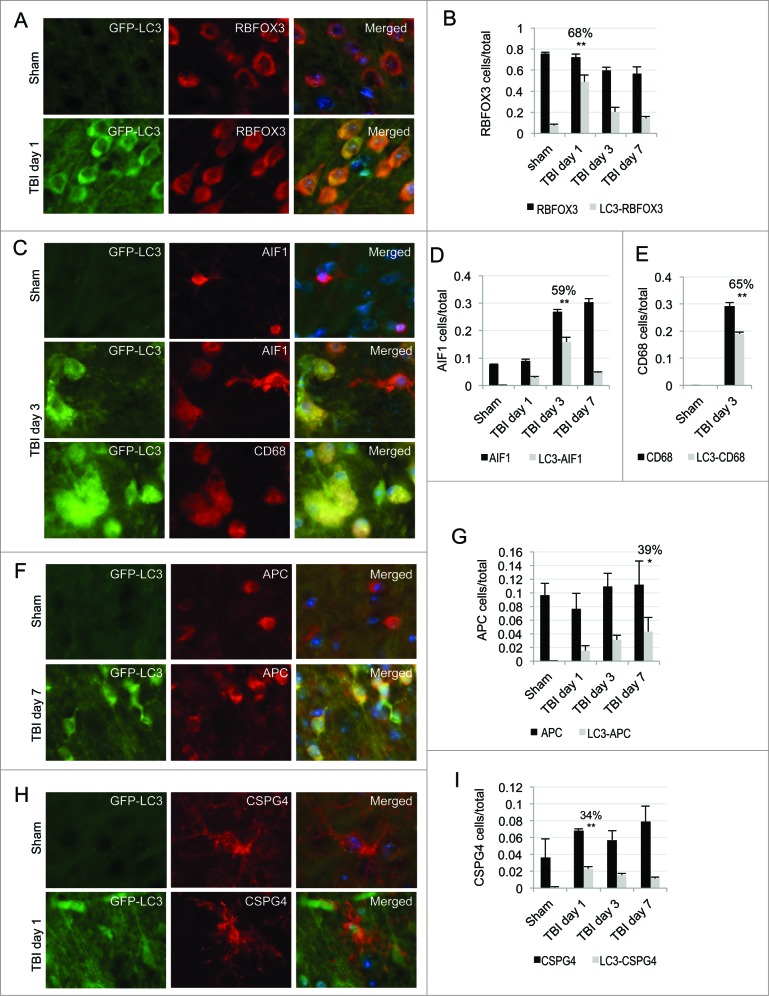
Next we determined the cell-type specificity for autophagosome accumulation in the cortex. We performed immunofluorescence analysis using different cell type markers in GFP-Lc3 transgenic mice at d 1, 3 and 7 after TBI. We observed significant colocalization of the GFP-LC3 signal with the neuronal marker RBFOX3/NeuN (RNA binding protein, fox-1 homolog [C. elegans] 3) in the cortex at d 1 following injury (68% of RBFOX3-positive neurons were GFP-LC3 positive, P < 0.001; Fig. 2A and B). These data suggest that early after CCI injury phagophores or autophagosomes accumulate specifically in neurons. This accumulation was transient; by d 3 and 7 progressively fewer neurons were GFP-LC3 positive (Fig. 2B, Fig. S5A). Conversely, at d 3 significantly higher numbers of autophagosomes accumulated in microglia, with 59% of AIF1/IBA1 (allograft inflammatory factor 1)-expressing cells positive for GFP-LC3 (P < 0.001; Fig. 2C and D). AIF1 is expressed by both ramified and activated microglia. The GFP-LC3 signal predominantly colocalized in microglia with amoeboid morphology in the injury area, suggesting phagophores and/or autophagosomes accumulate in activated microglia. To confirm this, we stained GFP-Lc3 mouse brain sections with antibody against the CD68 (CD68 molecule) antigen expressed in activated microglia/macrophages. We observed high colocalization of the GFP-LC3 signal with CD68 (65%, P < 0.001), indicating that phagophores and/or autophagosomes specifically accumulated within activated microglia in the injury area (Fig. 2C and E) at d 3. Despite an increase in total numbers of AIF1-positive cells at d 7 after injury, colocalization with GFP-LC3 decreased at this time point, suggesting that accumulation of autophagosomes in microglia was transient after TBI (Fig. 2D, Fig. S5B). We also observed a gradual increase in colocalization of the GFP-LC3 signal with the oligodendrocyte marker APC/CC1 (adenomatous polyposis coli), reaching 39% at 7 d after injury (p = 0.002; Fig. 2F and G and Fig. S5C). The GFP-LC3 signal also colocalized with oligodendrocyte precursor marker CSPG4/NG2 (chondroitin sulfate proteoglycan 4) at d 1 (35%, P < 0.001), then gradually decreased at d 3 and 7 following injury (Fig. 2H and I and Fig. S5D). We hypothesize that this could be due to the differentiation of GFP-LC3-positive oligodendrocyte precursor cells into the mature (APC positive) form. Conversely, we observed poor colocalization of the GFP-LC3 signal with the astrocyte marker GFAP (under 20%; Fig. S6) at all time points examined, suggesting that autophagy was not affected by TBI in astrocytes.\raster(98%)="rgFigKAUP_A_981787_F0002_B"
Figure 2.
Accumulation of autophagosomes is cell-type specific in the cortex after TBI. Images (20×) of GFP-LC3 mouse cortical brain sections stained with antibodies against neuronal marker RBFOX3/NeuN (A), microglial and activated microglial markers AIF1/IBA1 and CD68, respectively (C), oligodendrocyte marker APC/CC1 (F) or oligodendrocyte precursor marker CSPG4/NG2 (H). Corresponding quantification of numbers of cells single positive for each of the cell type markers (black bars) and cells double-positive (gray bars) for GFP-LC3 and RBFOX3, P < 0.001 (B), AIF1, P < 0.001 (D), CD68, P < 0.001 (E), APC, P < 0.01 (G) and CSPG4, P < 0.001 (H) normalized to total cell numbers. The percentages of double-positive versus single-positive cells are indicated at the time points with highest significance. Data are presented as mean ± SE; n = 3; at least 1,000 cells were quantified per mouse per experiment.
Autophagosome accumulation is due to impaired autophagy flux after TBI
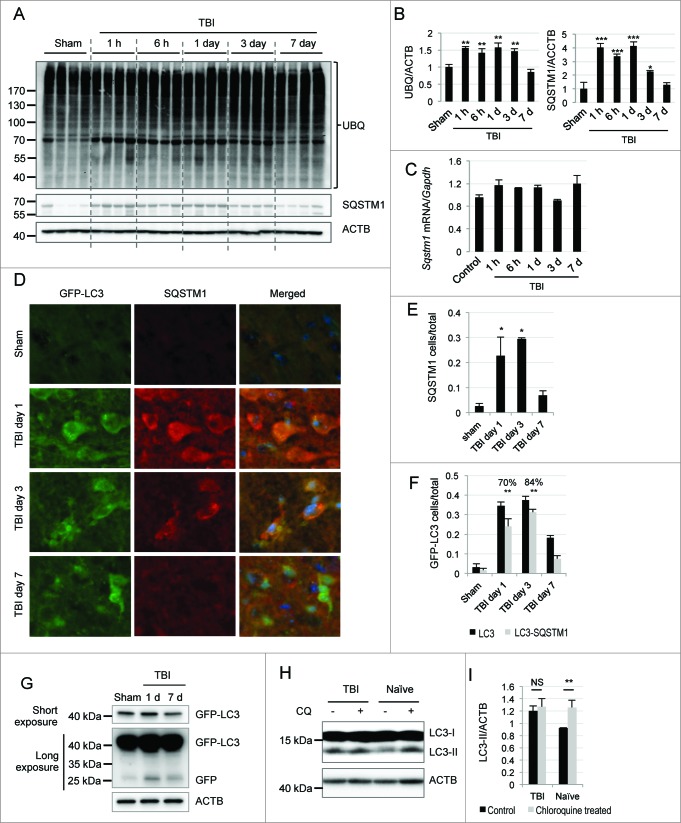
Our data suggested that initiation of autophagy is not increased and may, in fact, be slightly suppressed following TBI. Therefore, we investigated whether impairment of autophagy flux may contribute to the observed accumulation of LC3-II and autophagosomes. Ubiquitinated cargo including injured organelles and potentially toxic protein aggregates are delivered to autophagosomes by the receptor protein SQSTM1/p62.31,32 On the one hand, stimulation of autophagy flux causes depletion of SQSTM1 along with other autophagic substrates. On the other hand, when autophagic clearance is impaired SQSTM1 accumulates within cells.33 To determine whether autophagosomes may accumulate after TBI due to impaired autophagic turnover, we examined levels of SQSTM1 by protein gel blot. There was a marked increase in SQSTM1 protein levels in both ipsilateral cortex and hippocampus within 1 h after injury. SQSTM1 remained elevated through d 3 after injury but declined to baseline by d 7 (Fig. 3A and B and Fig. S7A and B). No significant changes in Sqstm1 mRNA levels were apparent (Fig. 3C). This is consistent with autophagic protein degradation being impaired immediately after TBI but restored at later time points. Consistent with a defect in protein degradation, we observed general accumulation of ubiquitinated proteins (Fig. 3A and B). Similarly to SQSTM1, levels of ubiquitinated proteins gradually decreased. However, unlike SQSTM1, they remained above sham levels at 7 d after injury. Since ubiquitinated proteins are also degraded by the proteasome, their persistence could be due to the previously described impairment in proteasomal degradation after TBI.34,35 Another possibility is that there has not been sufficient time after restoration of flux to clear all accumulated potential autophagy cargo.\raster="rgFigKAUP_A_981787_F0003_B"
Figure 3.
For figure legend, see page 2215.
To confirm our findings, we performed immunohistochemical analysis of SQSTM1 levels in GFP-Lc3 mice. Consistent with the western blot data, we observed markedly higher SQSTM1 signal in injured mouse cortex as compared to shams (Fig. 3D and E). The SQSTM1 signal strongly colocalized with GFP-LC3 in the injured cortex at d 1 and 3 (with, respectively, 70% [P < 0.001] and 84% [P < 0.001] of all GFP-LC3-positive cells also positive for SQSTM1; Fig. 3D and F). Similar data were obtained when wild-type control and TBI brain sections were costained with antibodies against LC3 and SQSTM1 (Fig. S7C). To make sure that transient accumulation of SQSTM1 is not a normal consequence of induction of autophagy in the brain, we examined levels of SQSTM1 in the cortex of GFP-Lc3 mice treated with rapamycin. Unlike TBI, rapamycin led to a decrease in the number of SQSTM1-positive cells (Fig. S7D). There was also no increase in colocalization of GFP-LC3 and SQSTM1 (Fig. S7E and F).
Furthermore, we assessed the extent of GFP processing in GFP-Lc3 mice after injury. We observed accumulation of both free GFP and GFP-LC3 in the cortex at d 1 after injury, which decreased by d 7 (Fig. 3G and Fig. S8A). Unlike TBI, treatment with rapamycin did not increase free GFP levels in the cortex of GPF-Lc3 mice (Fig. S8B and C). Accumulation of free GFP after TBI is similar to the previously reported observation that a partial block of lysosomal function can lead to accumulation of both GFP and GFP-LC3.36,37 Taken together, these results suggest that autophagosome clearance is partially impaired but not fully blocked in the cortex at early time points after TBI.
To directly confirm impairment of autophagy flux after TBI, we determined autophagy flux in brain slices from control and injured mice ex vivo. We incubated ipsilateral brain sections cut from injured (1 d after TBI) or control mice for 2 h in the presence or absence of chloroquine, which increases lysosomal pH and thereby inhibits lysosomal degradation. Our data demonstrated a significant increase in LC3-II in brain sections from naȉve animals treated with chloroquine as compared to untreated sections (P < 0.01). Levels of LC3-II were elevated in untreated sections from TBI animals and did not further increase upon addition of chloroquine (Fig. 3H and I). These data confirm that autophagy flux and autophagosome clearance are impaired in the brain after TBI.
In neurodegenerative diseases, impairment in autophagic turnover leads to accumulation of ubiquitinated proteins and protein aggregates, which can contribute to neuronal cell death.9,38 Similarly, we observed an increased ubiquitin signal at d 1 after TBI. 66% of ubiquitin-positive cells were also positive for SQSTM1 (P < 0.01, Fig. S9A and B), suggesting that impaired autophagy flux contributed to accumulation of ubiquitinated proteins. We also observed numerous ubiquitin and SQSTM1 double-positive puncta in those cells, possibly indicating the presence of protein aggregates (Fig. S9C). Therefore, defects in autophagic turnover after TBI may contribute to accumulation of potentially toxic ubiquitinated proteins and protein aggregates.
Lysosomal malfunction contributes to disruption of autophagy after TBI
Under normal circumstances autophagosomes and their cargo are degraded within lysosomes by lysosomal hydrolases.1,2,14 Impaired autophagosomal clearance after TBI led us to hypothesize that lysosomal function may be affected in the injured brain. First we examined if injury to the brain may cause disruption of lysosomal integrity and leakage of lysosomal enzymes into the cytosol. We isolated crude lysosomal and cytosolic fractions from the cortices of injured and sham mice and determined the level of the soluble lysosomal enzyme CTSD in those fractions by protein gel blot. Both precursor and mature CTSD were detected in the lysosomal fractions of sham and injured animals. In the cytosolic fraction only a faint band of CTSD was detected in either sham or injured cortex (Fig. S10A). This observation indicates that lysosomal integrity is likely intact following TBI.
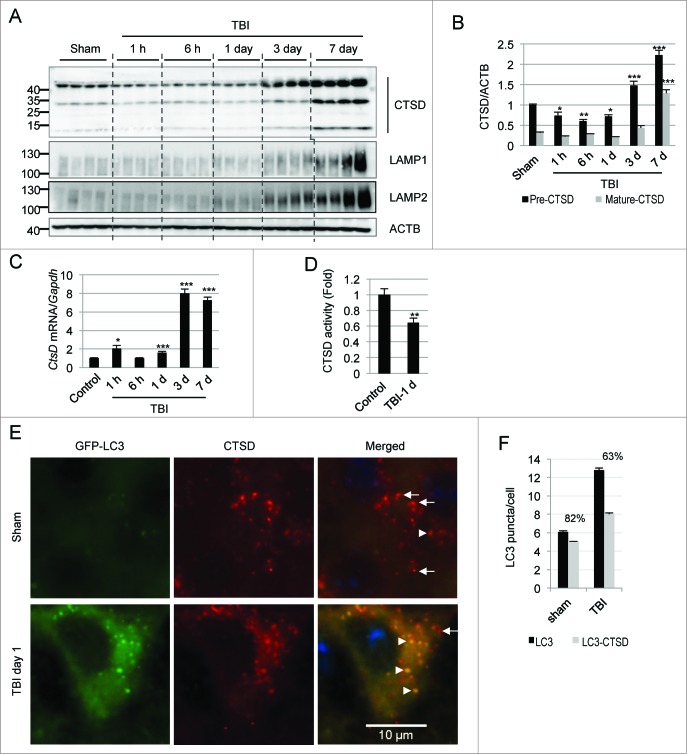
Next we determined levels of lysosomal proteins in total protein lysates from injured and control cortex by western blot. We noticed slightly lower levels of CTSD in TBI cortex as compared to sham from 1 to 24 h after injury (Fig. 4A and B). CTSD markedly increased at d 3 and 7 following injury. A similar expression pattern was observed in injured hippocampus (Fig. S10B and C). Ctsd mRNA levels remained either unaltered or slightly elevated in the cortex from 1 to 24 h following injury, but similarly to the protein they were strongly upregulated at d 3 and 7 (Fig. 4C). Unlike CTSD, levels of lysosomal membrane proteins, LAMP1 and LAMP2 (lysosomal-associated membrane protein 1 and 2), did not change at d 1 after TBI (Fig. 4A and Fig. S10D and E) as compared to sham, suggesting that the size of the lysosomal compartment was not altered at that time. On the other hand, levels of LAMP1 and LAMP2 also increased starting at d 3 and 7 after injury. The increase in lysosomal proteins correlated with proliferation of microglia after TBI and could be due to the expanded lysosomal compartment characteristic of these cells.39 This was confirmed by immunofluorescence analysis where we found elevated levels of CTSD and its strong colocalization with AIF1-positive microglial cells in the injured cortex at d 3 and 7 as compared to sham (82% and 80%, respectively, P < 0.001; Fig. S11A and B).
Figure 4.
TBI leads to lysosomal dysfunction. (A) Western blot analysis of CTSD in cortical tissue lysates from sham and TBI animals. (B) Densitometric analysis of precursor (black bars) and mature (gray bars) forms of CTSD with respect to the loading control ACTB. n = 4, *P < 0.05, **P < 0.01, ***P < 0.001. (C) Relative mRNA level (qPCR) of CtsD in the cortex of uninjured control and injured mice normalized to loading control Gapdh; n = 3, *P < 0.05, ***P < 0.001 vs. sham. (D) CTSD enzyme activity determined by in vitro fluorometric assay in the crude lysosomal fraction prepared from sham and injured mouse cortices. n = 5, **P < 0.01. (E) High magnification (60×) images of cells in the cortex of GFP-Lc3 mice stained with antibody against CTSD. Accumulation of GFP-LC3 and CTSD double-positive structures (arrowheads) and depletion of single CTSD-positive lysosomes (arrows) is apparent after TBI. (F) Quantification of GFP-LC3 puncta and double-positive GFP-LC3/CTSD puncta in sham and TBI mouse cortex. Percentage of overlap is indicated. n = 3; data are presented as mean ± SE.
In order to determine if decreased levels of CTSD early after injury may lead to decreased lysosomal function and thus potentially contribute to the observed impairment of autophagic degradation, we compared enzymatic activity of CTSD in the cortex from injured and control animals. We found significantly lower CTSD enzyme activity in cortical extracts (Fig. S11C) as well as in isolated crude lysosomal fractions from injured animals at d 1 after TBI as compared to controls (P < 0.01; Fig. 4D). This lower enzymatic function indicates decreased lysosomal activity after TBI, which could at least in part account for the observed impairment in autophagy flux.
Since lysosomal function can affect fusion of autophagosomes with lysosomes, we also examined colocalization of lysosomes and autophagosomes after injury. High-resolution analysis of brain sections from injured mice revealed decreased colocalization of GFP-LC3-positive autophagosomes with CTSD-positive lysosomes as compared to sham animals (Fig. 4E and F). There were also very few unfused lysosomes remaining in the injured cortices as compared to sham. Therefore, decreased CTSD protein levels and activity after TBI may contribute to a backlog of unfused or partially fused autophagosomes that cannot be efficiently processed by lysosomes.
Impaired autophagy contributes to neuronal cell death after TBI
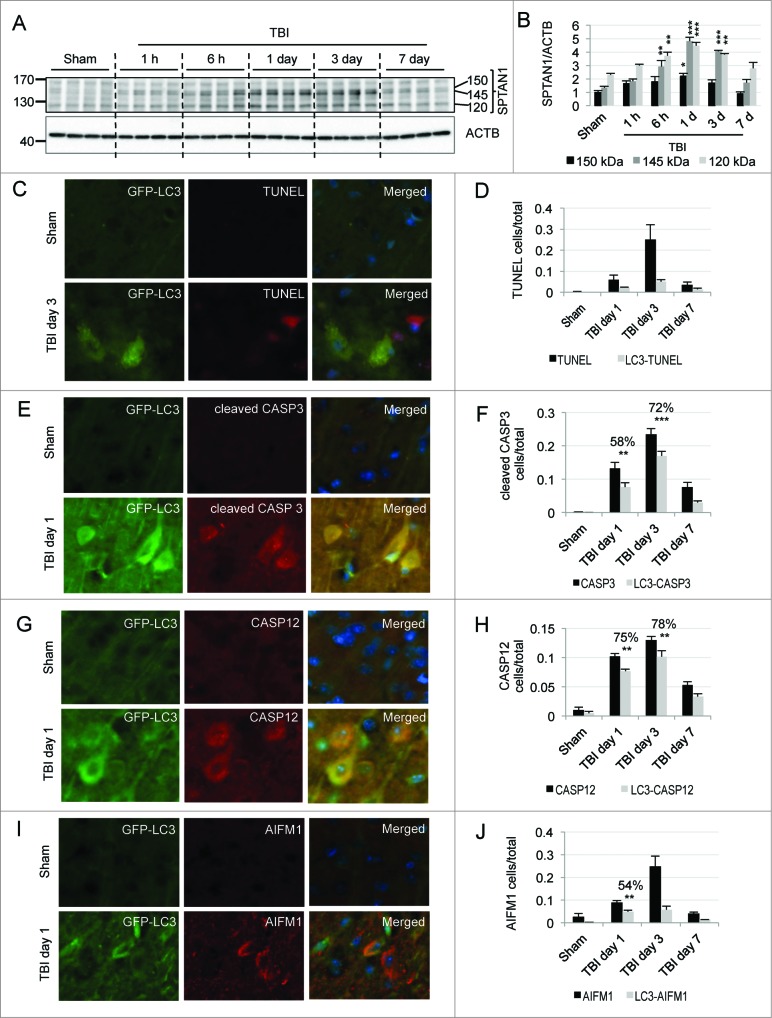
TBI is associated with severe neurodegeneration. Increased levels of SPTAN1 (spectrin, α, non-erythrocytic 1) breakdown products were detected in the cortex, with a peak at d 1 to 3 following TBI (Fig. 5A and B). This indicates the potential presence of both apoptotic and nonapoptotic cell death in the injured cortex at time points correlating with maximal impairment of autophagy. To examine if inhibition of autophagy flux may contribute to cell death after TBI we assessed levels of cell death markers in GFP-Lc3 mice. At d 1 to 3 after injury, we observed many TUNEL-positive cells in the brain areas with high GFP-LC3 accumulation. However, we were able to detect only a few GFP-LC3 and TUNEL double-positive cells. This may be due to the fact that TUNEL is a very late cell death marker, detected at a time when other cellular proteins including LC3 may no longer be present (Fig. 5C and D and Fig. S12A).
Figure 5.
Impairment of autophagy after TBI contributes to neuronal cell death. (A) Western blot analysis of SPTAN1 breakdown products of 150 kDa, 145 kDa, and 120 kDa in cortical tissue from sham and TBI mouse brain. (B) Corresponding densitometric analysis of SPTAN1 bands with respect to ACTB. n = 4, *P < 0.05, **P < 0.01, ***P < 0.001. (C, E, G) Images (20×) of GFP-Lc3 mouse cortical brain sections from sham and TBI mice stained for cell death markers: TUNEL (C), cleaved CASP3 (E), CASP12 (G) and AIFM1 (I). Corresponding quantification of cells single positive for indicated cell death markers (black bars) and double positive for GFP-LC3 (gray bars) and TUNEL (D), Cleaved CASP3, **P < 0.01, ***P < 0.001 (F), CASP12, P < 0.001 at both d 1 and 3 (H), and AIFM1, P < 0.001 (J). The percentages of double-positive versus single-positive cells are indicated at the most significant time points. n = 3; data are represented as mean ± SE.
To overcome this obstacle, we examined colocalization of GFP-LC3 with earlier markers of different types of cell death. CASP3/caspase 3, the major executioner caspase, is cleaved to an active form during apoptosis. We observed significant accumulation of cells expressing cleaved CASP3 in the cortex at d 1 and 3 after injury (Fig. 5E and F and Fig. S12B). Cleaved CASP3 strongly colocalized with GFP-LC3 (with 58% and 72% cleaved CASP3-positive cells also positive for GFP-LC3 at d 1 and 3 after TBI, P < 0.001 and p = 0.003, respectively). At d 1 after TBI the majority of GFP-LC3-positive cells were neurons (Fig. 2A and B) and at this time point cleaved CASP3 was mostly present in GFP-LC3-positive cells with neuronal morphology. Therefore impaired autophagic flux correlates with induction of neuronal apoptosis in the same cells after injury.
Next we examined expression of the ER stress associated CASP12/caspase 12. ER stress is influenced by autophagy,40 and has been reported to be involved in apoptotic cell death after TBI.41 We observed an increase in CASP12 levels in the injured brain as well as significant colocalization of CASP12 with GFP-LC3-positive cells in the cortex at d 1 and d 3 after TBI (75% and 78%, respectively, P < 0.001 for both time points; Fig. 5G and H and Fig. S13A). Confirming that CASP12-positive cells were defective in autophagy flux, the CASP12 signal also colocalized with SQSTM1 at d 1 and 3 after injury (Fig. S13B and C). Consistent with the hypothesis that impairment of autophagy may contribute to ER stress and to neuronal cell death, at d 1 after TBI the majority of CASP12 and SQSTM1 double-positive cells had neuronal morphology.
Finally we examined if autophagy may also be impaired in cells undergoing caspase-independent cell death. We stained brain sections from GFP-Lc3 mice with antibody against AIFM1/AIF (apoptosis-inducing factor, mitochondrion-associated, 1). We noticed a gradual increase in AIFM1-positive cells, 54% of which colocalized with GFP-LC3 at d 1 after TBI (P < 0.001; Fig. 5I and J and Fig. S14A). Despite total greater numbers of AIFM1-positive cells, the degree of colocalization between AIFM1 and GFP-LC3 decreased at later time points. Since neurons are the predominant cell type showing accumulation of autophagosomes 1 d after TBI, this indicates that a block of autophagic clearance may specifically correlate with caspase-independent neuronal cell death in this system. This was further confirmed by immunofluorescence analysis, which demonstrated marked colocalization of SQSTM1 with AIFM1 (Fig. S14B). Taken together these data suggest that autophagic impairment correlates with and may contribute to the induction of both caspase-dependent and -independent neuronal apoptosis after TBI.
Discussion
Previous studies demonstrated increased autophagic markers in the brain after traumatic injury in both human and animal models. However, the mechanism leading to this phenotype remained unknown. Our data demonstrate that since upstream regulators and mediators of autophagy are unchanged or slightly decreased after TBI, increased initiation of autophagy cannot account for the observed accumulation of autophagosomes. On the other hand, accumulation of autophagy substrates and impaired autophagy flux ex vivo suggest that autophagy flux is inhibited after TBI. This is at least in part due to lysosomal impairment observed after TBI. Thus our data provide the first insight into the cellular mechanisms leading to accumulation of autophagosomes after TBI.
Dysfunction of autophagy has been implicated in neuronal cell loss in neurodegenerative diseases and in lysosomal storage diseases.5,7,11,13-17,42-45 In lysosomal storage diseases, defects in specific lysosomal hydrolases lead to lysosomal dysfunction and, as a consequence, inhibition of autophagy.16,17 Lysosomal function abnormalities have also been reported in some neurodegenerative diseases and proposed to contribute to pathological accumulation of autophagosomes and to neuronal dysfunction and death.46 In the present study we demonstrate for the first time that lysosomal function is impaired at the early time points after TBI. We propose that this contributes to defects in autophagic clearance, which in turn may have an impact on neuronal cell death. Thus, our data reveal a potential common mechanism contributing to neuronal cell death due to chronic (neurodegenerative and lysosomal storage diseases) and acute (TBI) insults.
In the present study we also identify the specific cell types in which phagophores and/or autophagosomes accumulate at different time points after TBI. We demonstrate that phagophores and/or autophagosomes predominantly accumulate within neurons at early time points (d 1) and within activated microglia and oligodendrocytes at later time points (d 3 and 7, respectively) after injury. It is well documented that autophagic degradation is essential for neuronal survival. Therefore, the initial accumulation of autophagosomes within neurons is likely contributing to neuronal cell death. This is supported by the strong colocalization of both GFP-LC3 and SQSTM1 with markers of neuronal cell death, including cleaved CASP3, CASP12, and AIFM1. These data also indicate that the early impairment of autophagic clearance in neurons may contribute to both caspase-dependent and caspase-independent cell death. Induction of endoplasmic reticulum stress (ER stress) and activation of CASP12 following TBI have been previously reported.41 Since autophagy is also activated by and can help relieve ER stress,40 we hypothesize that the observed block in autophagy may further contribute to ER stress after TBI. Conversely, since ER stress causes translational arrest,47 lower levels of lysosomal enzymes including CTSD may reflect an ER stress-mediated decrease in protein translation. This decline in translation could potentially lead to a deleterious positive feedback loop between a block in autophagy and ER stress after TBI. During caspase-independent cell death, AIFM1 translocates from the mitochondrial inner membrane to the cytosol.48 As damaged mitochondria are targeted by autophagy, colocalization of AIFM1 with GFP-LC3 and SQSTM1 suggests the possibility that impaired autophagic clearance may contribute to accumulation of damaged mitochondria after TBI.
At d 3 after TBI both GFP-LC3 and SQSTM1 accumulate predominantly in activated microglia. Defective autophagy has been recently suggested to contribute to inflammation by activating the NFKB pathway in cancer and other diseases.49 Specifically, SQSTM1 can directly stimulate the NFKB pathway through its interaction with TRAF6.50 Furthermore, in M2 macrophages autophagy can selectively degrade NFKB RELA/p65, thereby reducing production of pro-inflammatory cytokines.51 Thus, a block of autophagosome clearance and the resulting accumulation of SQSTM1 within activated microglia may contribute to the induction of deleterious neuroinflammatory responses after TBI. Recently, induction of autophagy by GSK3B inhibitors has been shown to decrease neuroinflammation following ischemic brain injury.52 We hypothesize that restoration of autophagy flux may also attenuate inflammatory responses after TBI.
Although the number of autophagosomes remains elevated at later time points (7 d) after TBI, accumulation of SQSTM1 and to some extent ubiquitin seem to resolve. This suggests that at this time point autophagic flux may be restored. This could in part reflect death of the affected neuronal cells, while increased lysosomal activity in the proliferating microglia could allow restoration of autophagic flux in this cell type. Additionally activation of other autophagic pathways like chaperone-mediated autophagy could also contribute to eventual clearance of SQSTM1. This possibility is consistent with the increase in the level of LAMP2, which is involved in chaperone-mediated autophagy. The remaining elevation in number of autophagosomes at the 7-d time point could indicate that following restoration of flux there has not been sufficient time to clear all accumulated autophagosomes. Alternatively, it is possible that autophagosome synthesis may be increased at d 7 and potentially beyond. At least in the hippocampus, this could be mediated by increased phospho-ULK1 levels.
Although beneficial effects of autophagy after TBI have been suggested by some previous studies, in others, pharmacological inhibition of autophagy after TBI has neuroprotective effects.21,23,26-28 Our data indicate a transient block of autophagic clearance at the early time points after TBI, which is than relieved later after injury. Therefore, the contradictory observations of both beneficial and detrimental effects of autophagy after TBI described by previous studies may reflect this time-dependent alteration in autophagic flux in the injured brain. We expect that early after TBI accumulation of autophagosomes reflects a deleterious effect: impaired lysosomal function and autophagosome clearance lead to accumulation of ubiquitinated proteins and protein aggregates, thus contributing to neuronal cell death. At later time points when autophagic flux is restored, autophagy may be neuroprotective. Based on our data, we propose that early interventions aimed at reduction in autophagosome overload, by either restoring lysosomal function or by decreasing autophagosome synthesis and activating other degradative pathways, may be beneficial after TBI. We hypothesize that such interventions could both directly decrease the extent of neuronal cell death as well as attenuate neuroinflammation. Conversely, later after TBI when autophagic flux is restored, induction of autophagy could be neuroprotective.
Materials and Methods
Controlled cortical impact
All surgical procedures and animal experiments were performed according to the protocols approved by the Animal Care and Use Committee of the University of Maryland. CCI TBI was performed under surgical anesthesia (2–3% isoflurane evaporated in a gas mixture containing 70% N2O and 30% O2) in male C57BL6/J mice (20–25 g) or transgenic C57BL6/J mice expressing GFP-Lc3 as previously described.53 The injury device consists of a microprocessor-controlled pneumatic impactor, driven by compressed air, with a 3.5-mm diameter tip. A 10-mm midline incision was made over the skull, the skin and fascia were refracted and a 4-mm craniotomy was made on the central aspect of the left parietal bone. Moderate injury was induced by the impact velocity of 6 m/s and a deformation depth of 2 mm. Sham animals underwent the same procedure as injured mice except for the impact.
Rapamycin injection
Rapamycin (Sigma, 37094) was dissolved in DMSO and then diluted in vehicle containing 0.25% PEG400 (Sigma, 202398) and 0.25% Tween 80 (Sigma, P4780). The final concentration of DMSO was adjusted to 0.1%. Rapamycin was injected intraperitoneally in one group (n = 3) at a dose of 5 mg/Kg. Mice of the control group (n = 3) were injected with the vehicle only. Twenty-four h later animals were anesthetized and processed for immunohistochemistry and protein gel blot.
Immunohistochemistry
At 24 h, 3 d and 7 d after injury or 24 h after rapamycin injection mice were anesthetized with isofluorane and transcardially perfused with cold saline and then with 4% paraformaldehyde (pH 7.4). Brains were removed and post-fixed in paraformaldehyde for 24 h and then protected in 30% sucrose. Twenty-μm frozen sections were cut and mounted on glass slides. For immunofluorescence analysis sections were blocked with 5% goat serum (Millipore, S26-LITER) or donkey serum (Sigma, D9663) in 1(X) phosphate-buffered saline (Quality Biological, Inc., 119–069–101) containing 0.25% Triton X-100 (Sigma, X100), incubated overnight with primary antibodies and then with secondary antibodies for 2 h at room temperature. Nuclei were stained with DAPI.
Primary antibodies used in this study include LC3 (1:200; Novus, NB100–2220), SQSTM1 (1:200; Progen, GP62-C), RBFOX3/NeuN (1:500; Millipore, MAB377), AIF1/Iba-1 (1:1000; Wako, 019–19741), GFAP (1:1000; Dako, Z0334), APC/CC1 (1:1000; Abcam, ab16794), CSPG4/NG2 (1:500; Chemicon, AB5320), CTSD/cathepsin D (1:100; SantaCruz Biotechnology, sc-6486), ubiquitin (1:200; Cell Signaling Technology, 3936), AIFM1/AIF (1:250; 5318), CASP12/caspase 12 (1:200; Cell Signaling Technology, 2202), cleaved CASP3/caspase 3 (1: 200; Cell Signaling Technology, 9661), GFP (1:250; Clontech, 632592), CD68 (1:250; AbD Serotec, MCA1957GA). Secondary antibodies used are from Invitrogen: Alexa Fluor 488 goat anti-rabbit (A11034), Alexa Fluor 546 goat anti-mouse (A11030), Alexa Fluor 568 goat anti-guinea pig (A11075), Alexa Fluor 633 goat anti-mouse (A21052) and Alexa Fluor 546 donkey anti-goat (A11056).
TUNEL assay was performed on frozen brain sections using ApopTag In Situ Apoptosis Detection Kit (Millipore, S7165) as per the manufacturer's protocol.
Image acquisition and quantification
Images were acquired using a fluorescence Nikon Ti-E inverted microscope, at 20× (CFI Plan APO VC 20× NA 0.75 WD 1 mm) or 60× (CFI Plan APO VC 60× NA 1.4 Oil) magnification; at emission wavelengths of 460 nm (DAPI), 535 nm (GFP-LC3 and alexa fluor 488), 620 nm (alexa fluor 546) and 670 nm (alexa fluor 633). Exposure times were kept constant for all sections in each experiment. All 60× images were acquired as z-stacks and focused using the Extended Depth of Focus module of Elements software (Nikon). Background for all images was subtracted using Elements. All images were quantified using Elements: nuclei were identified using Spot Detection algorithm; cells expressing GFP-LC3 or positive for any of the immunofluorescence markers were identified using Detect Regional Maxima algorithm, followed by global thresholding. The number of positive cells was normalized to the total number of cells imaged. Intracellular puncta were detected using Spot Detection and normalized to the number of cells imaged. Numbers of GFP-LC3 cells were quantified in both the cortex and hippocampus. All other quantifications were performed in the cortex. At least 1,000–2,000 cells were quantified per mouse per experiment. For additional details on image quantification please see the Supplementary Methods section.
Western blot analysis
For western blot analysis, mice were anesthetized, perfused with ice-cold saline, and decapitated. Hippocampus and ∼5 mm of the cortical area surrounding the ipsilateral injury site were collected and homogenized in RIPA buffer (Teknova, R3792) containing protease inhibitor (Roche, 11836170001) and phosphatase inhibitor (Sigma, P5726). Homogenates were centrifuged at 20,000 g for 20 min at 4°C to collect the tissue lysate. Protein concentration was measured using BCA reagent (Pierce, 23225). Twenty μg of protein was resolved in either 18% or 4–15% SDS-PAGE gels (Bio-Rad, 345–0025 and 345–0029) and transferred onto PVDF membrane (Millipore, IPVH00010). Membranes were blocked with 5% nonfat milk, probed with primary antibodies overnight at 4°C and incubated with HRP-conjugated secondary antibodies (KPL, 474–1506, 474–1806, 14–16–06 and 14–13–06) at room temperature for 1 h. Protein bands were then detected using a chemiluminiscence kit (Pierce, 34076) and visualized using a Chemi-doc system (Bio-Rad). Bands were analyzed by Image Lab software (Bio-Rad).
Primary antibodies used in this study are: LC3 (1:1000; Novus, NB100–2220), PIK3C3/VPS34 (1:1000; Invitrogen, 382100), BECN1/Beclin 1 (1:1000; Santa Cruz Biotechnology, sc-11427), CTSD/cathepsin D (1:1000; Santa Cruz Biotechnology, sc-6486), SQSTM1 (1:1000; BD Bioscience, 610832), ubiquitin (1:1000; Cell Signaling Technology, 3936), phospho-ULK1 (1:1000; Cell Signaling Technology, 5869), SPTAN1/spectrin (1:5000; Enzo Life Science International, BML-FG6090), ATG5 (1:1000; Sigma, A0731), ACTB/β-actin (1:10,000; Sigma, A1978), GFP (1:1000; Clontech, 632592), and TUBB3/tubulin (1:5000; Covance, MMS-435P). Antibodies to LAMP1 (1:1000; 1D4B) and LAMP2 (1:1000; ABL-93) were developed by J Thomas August and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA USA.
Subcellular fractionation
For subcellular fractionation brains perfused with cold saline were removed at 24 h and 7 d after injury, homogenized in buffered ice-cold sucrose solution containing 0.32 M sucrose (Fisher Scientific, BP220–212), 10 mM HEPES (HyClone, SH30237.01) and protease and phosphatase inhibitors. Homogenates were centrifuged at 800 g for 10 min at 4°C to spin down nuclei. Supernatant fractions were then centrifuged at 20,000 g for 20 min at 4°C to spin down the crude lysosomal fractions. Supernatant fractions from this step were further centrifuged at 100,000 g for 1 h at 4°C to spin down the crude membrane fraction. Pellet fractions obtained at each step were resuspended in homogenization buffer. Both supernatant and suspended pellet fractions were recentrifuged to minimize cross contamination from the different subcellular fractions. At the end all pellet fractions were resuspended in homogenizing buffer and analyzed by protein gel blot.
LC3 processing assay in brain slices
LC3 flux assay in brain sections was adapted from a previously described method.37,54 24-h post-TBI mice were anesthetized, sacrificed using a guillotine and the brains were collected in oxygenated (95% O2–5% CO2) artificial cerebral spinal fluid containing 125 mM NaCl, 1.25 mM NaH2PO4, 2.5 mM KCl, 20 mM glucose, 25 mM NaHCO3, 1 mM CaCl2 and 1 mM MgSO4. 400-μm ipsilateral brain sections were cut using a Leica 1000 Plus vibratome (Leica Microsystems Inc., Buffalo Grove, IL, USA) and incubated in oxygenated artificial cerebral spinal fluid with or without chloroquine (100 μM; Sigma, C6628) for 2 h at room temperature. After incubation, sections were homogenized in RIPA buffer containing protease and phosphatase inhibitors and centrifuged at 20,000 g for 20 min at 4°C to collect the lysate. Protein concentration was measured by the BCA method and lysates were analyzed by western blot.
Real-time PCR
Total RNA isolated from ipsilateral cortex using Trizol reagent (Invitrogen, 15596–018) was converted into cDNA using the Verso™ cDNA Kit (Thermo Scientific, AB1453B) as per the manufacturer's instruction. cDNA TaqMan® Universal Master Mix II (Applied Biosystems, 4440040) was used to perform quantitative real-time PCR amplification. Briefly, reactions were performed in duplicate by mixing 2 × TaqMan® Universal Master Mix II, 1 μL of cDNA (corresponding to 50 ng RNA/reaction) and 20 × TaqMan® Gene Expression Assay, in a final volume of 20 μL. TaqMan® Gene Expression assays for the following genes were used for mouse: Gapdh (Mm99999915_g1), Map1lc3b (Mm00782868_sH), Atg12 (Mm00503201_m1), Becn1 (Mm01265461_m1), Sqstm1 (Mm00448091_m1) and Ctsd (Mm00515586_m1) (Applied Biosystems). Reactions were amplified and quantified by using a 7900HT Fast Real-Time PCR System and the corresponding software (Applied Biosystems). The PCR profile consisted of 1 cycle at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Gene expression was normalized to Gapdh, and the relative quantity of mRNAs was calculated based on the comparative Ct method.55
Cathepsin D assay
The CTSD/cathepsin D assay was performed using a fluorometric CTSD assay kit from Abcam (ab65302) as per the manufacturer's instruction. Briefly, mice were anesthetized, perfused with ice-cold saline, decapitated, and cortical tissue of approximately 5-mm diameter surrounding the site of injury was dissected and homogenized in ice-cold cell lysis buffer provided in the kit. Tissue homogenates were centrifuged at 15,000 g for 5 min at 4°C. Protein concentration was estimated by the BCA method. Fifty ng of protein were incubated with the CTSD substrate mixture at 37°C for 1 h. Fluorescence released from the synthetic substrate by tissue CTSD was estimated in a fluorescence plate reader (Synergy Hybrid, Biotek) at Ex/Em = 328/460 nm.
Statistical analysis
Data were statistically analyzed using the Sigma Plot software (Version 12) and GraphPad Prism (version 4). One-way ANOVA was performed followed by appropriate post-hoc test (Bonferroni, Tukey's or SNK t-test) for parametric (normality and equal variance passed) data. Kruskal-Wallis ANOVA based on ranks followed by Dunn's post-hoc test was used for nonparametric (normality and/or equal variance failed) data. For experiments with only 2 groups 2-tailed Mann-Whitney Rank Sum Test (nonparametric) or 2-tailed unpaired Student t-test was performed. A P value ≤ 0.05 was considered statistically significant.
Acknowledgments
We thank Dr. Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) and Dr. Beth Levine (UT Southwestern Medical center, Dallas TX) for the GFP-Lc3 mice; Drs. Junfang Wu, David Lane, and Bogdan Stoica for technical assistance and advice; Dr. Shruti Kabadi for help with statistical analysis; Katherine Cardiff and Titilola Akintola for help with animal husbandry and histological tissue preparation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by startup funds from the Department of Anesthesiology, University of Maryland School of Medicine, Baltimore MD to MML and R01 NS06183901A2 to AIF.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID: 11099404; http://dx.doi.org/ 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klionsky DJ. Cell biology: regulated self-cannibalism. Nature 2004; 431:31-2; PMID: 15343317; http://dx.doi.org/ 10.1038/431031a [DOI] [PubMed] [Google Scholar]
- 3. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728-41; PMID: 22078875; http://dx.doi.org/ 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- 4. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID: 21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 5. Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci 2006; 29:528-35; PMID: 16859759; http://dx.doi.org/ 10.1016/j.tins.2006.07.003 [DOI] [PubMed] [Google Scholar]
- 6. Higgins GC, Devenish RJ, Beart PM, Nagley P. Transitory phases of autophagic death and programmed necrosis during superoxide-induced neuronal cell death. Free Radic Biol Med 2012; 53:1960-7; PMID: 22982049; http://dx.doi.org/ 10.1016/j.freeradbiomed.2012.08.586 [DOI] [PubMed] [Google Scholar]
- 7. Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, et al. . Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci U S A 2010; 107:14164-9; PMID: 20660724; http://dx.doi.org/ 10.1073/pnas.1009485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. . Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; PMID: 16625204; http://dx.doi.org/ 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- 9. Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. . Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; PMID: 16625205; http://dx.doi.org/ 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- 10. Klionsky DJ. Neurodegeneration: good riddance to bad rubbish. Nature 2006; 441:819-20; PMID: 16778876; http://dx.doi.org/ 10.1038/441819a [DOI] [PubMed] [Google Scholar]
- 11. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004; 306:990-5; PMID: 15528435; http://dx.doi.org/ 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butler D, Nixon RA, Bahr BA. Potential compensatory responses through autophagiclysosomal pathways in neurodegenerative diseases. Autophagy 2006; 2:234-7; PMID: 16874061; http://dx.doi.org/ 10.4161/auto.2729 [DOI] [PubMed] [Google Scholar]
- 13. Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006; 443:780-6; PMID: 17051204; http://dx.doi.org/ 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- 14. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID: 18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19:983-97; PMID: 23921753; http://dx.doi.org/ 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- 16. Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet 2008; 17:119-29; PMID: 17913701; http://dx.doi.org/ 10.1093/hmg/ddm289 [DOI] [PubMed] [Google Scholar]
- 17. Settembre C, Fraldi A, Rubinsztein DC, Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy 2008; 4:113-4; PMID: 18000397; http://dx.doi.org/ 10.4161/auto.5227 [DOI] [PubMed] [Google Scholar]
- 18. Werner C, Engelhard K. Pathophysiology of traumatic brain injury. Br J Anaesth 2007; 99:4-9; PMID: 17573392; http://dx.doi.org/ 10.1093/bja/aem131 [DOI] [PubMed] [Google Scholar]
- 19. Loane DJ, Faden AI. Neuroprotection for traumatic brain injury: translational challenges and emerging therapeutic strategies. Trends Pharmacol Sci 2010; 31:596-604; PMID: 21035878; http://dx.doi.org/ 10.1016/j.tips.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cernak I, Noble-Haeusslein LJ. Traumatic brain injury: an overview of pathobiology with emphasis on military populations. J Cereb Blood Flow Metab 2010; 30:255-66; PMID: 19809467; http://dx.doi.org/ 10.1038/jcbfm.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clark RS, Bayir H, Chu CT, Alber SM, Kochanek PM, Watkins SC. Autophagy is increased in mice after traumatic brain injury and is detectable in human brain after trauma and critical illness. Autophagy 2008; 4:88-90; PMID: 17957135; http://dx.doi.org/ 10.4161/auto.5173 [DOI] [PubMed] [Google Scholar]
- 22. Diskin T, Tal-Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma 2005; 22:750-62; PMID: 16004578; http://dx.doi.org/ 10.1089/neu.2005.22.750 [DOI] [PubMed] [Google Scholar]
- 23. Liu CL, Chen S, Dietrich D, Hu BR. Changes in autophagy after traumatic brain injury. J Cereb Blood Flow Metab 2008; 28:674-83; PMID: 18059433; http://dx.doi.org/ 10.1038/sj.jcbfm.9600587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadasivan S, Dunn WA, Jr., Hayes RL, Wang KK. Changes in autophagy proteins in a rat model of controlled cortical impact induced brain injury. Biochem Biophys Res Commun 2008; 373:478-81; PMID: 18486600; http://dx.doi.org/ 10.1016/j.bbrc.2008.05.031 [DOI] [PubMed] [Google Scholar]
- 25. Erlich S, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol Dis 2007; 26:86-93; PMID: 17270455; http://dx.doi.org/ 10.1016/j.nbd.2006.12.003 [DOI] [PubMed] [Google Scholar]
- 26. Lai Y, Hickey RW, Chen Y, Bayir H, Sullivan ML, Chu CT, Kochanek PM, Dixon CE, Jenkins LW, Graham SH, et al. . Autophagy is increased after traumatic brain injury in mice and is partially inhibited by the antioxidant gamma-glutamylcysteinyl ethyl ester. J Cereb Blood Flow Metab 2008; 28:540-50; PMID: 17786151; http://dx.doi.org/ 10.1038/sj.jcbfm.9600551 [DOI] [PubMed] [Google Scholar]
- 27. Wang YQ, Wang L, Zhang MY, Wang T, Bao HJ, Liu WL, Dai DK, Zhang L, Chang P, Dong WW, et al. . Necrostatin-1 suppresses autophagy and apoptosis in mice traumatic brain injury model. Neurochem Res 2012; 37:1849-58; PMID: 22736198; http://dx.doi.org/ 10.1007/s11064-012-0791-4 [DOI] [PubMed] [Google Scholar]
- 28. Luo CL, Li BX, Li QQ, Chen XP, Sun YX, Bao HJ, Dai DK, Shen YW, Xu HF, Ni H, et al. . Autophagy is involved in traumatic brain injury-induced cell death and contributes to functional outcome deficits in mice. Neuroscience 2011; 184:54-63; PMID: 21463664; http://dx.doi.org/ 10.1016/j.neuroscience.2011.03.021 [DOI] [PubMed] [Google Scholar]
- 29. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID: 11060023; http://dx.doi.org/ 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. . A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488-92; PMID: 11100732; http://dx.doi.org/ 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 31. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62SQSTM1 binds directly to Atg8LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID: 17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 32. Bjorkoy G, Lamark T, Johansen T. p62SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2006; 2:138-9; PMID: 16874037; http://dx.doi.org/ 10.4161/auto.2.2.2405 [DOI] [PubMed] [Google Scholar]
- 33. Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62A170SQSTM1 by autophagy. Autophagy 2008; 4:1063-6; PMID: 18776737; http://dx.doi.org/ 10.4161/auto.6826 [DOI] [PubMed] [Google Scholar]
- 34. Yao X, Liu J, McCabe JT. Alterations of cerebral cortex and hippocampal proteasome subunit expression and function in a traumatic brain injury rat model. J Neurochem 2008; 104:353-63; PMID: 17944870 [DOI] [PubMed] [Google Scholar]
- 35. Yao X, Liu J, McCabe JT. Ubiquitin and ubiquitin-conjugated protein expression in the rat cerebral cortex and hippocampus following traumatic brain injury (TBI). Brain Res 2007; 1182:116-22; PMID:17936732; http://dx.doi.org/ 10.1016/j.brainres.2007.08.076 [DOI] [PubMed] [Google Scholar]
- 36. Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 2011; 7:188-204; PMID: 21107021; http://dx.doi.org/ 10.4161/auto.7.2.14181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID: 22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, et al. . Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol 2010; 191:537-52; PMID: 21041446; http://dx.doi.org/ 10.1083/jcb.201005012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakanishi H. Neuronal and microglial cathepsins in aging and age-related diseases. Ageing Res Rev 2003; 2:367-81; PMID: 14522241; http://dx.doi.org/ 10.1016/S1568-1637(03)00027-8 [DOI] [PubMed] [Google Scholar]
- 40. Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. . Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26:9220-31; PMID: 17030611; http://dx.doi.org/ 10.1128/MCB.01453-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larner SF, Hayes RL, McKinsey DM, Pike BR, Wang KK. Increased expression and processing of caspase-12 after traumatic brain injury in rats. J Neurochem 2004; 88:78-90; PMID: 14675152; http://dx.doi.org/ 10.1046/j.1471-4159.2003.02141.x [DOI] [PubMed] [Google Scholar]
- 42. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; PMID: 18191218; http://dx.doi.org/ 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Q, Mao Z. Dysregulation of autophagy and Parkinson's disease: the MEF2D link. Apoptosis 2010; 15:1410-4; PMID: 20165919; http://dx.doi.org/ 10.1007/s10495-010-0475-y [DOI] [PubMed] [Google Scholar]
- 44. Janda E, Isidoro C, Carresi C, Mollace V. Defective autophagy in Parkinson's disease: role of oxidative stress. Mol Neurobiol 2012; 46:639-61; PMID: 22899187; http://dx.doi.org/ 10.1007/s12035-012-8318-1 [DOI] [PubMed] [Google Scholar]
- 45. Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy 2012; 8:719-30; PMID: 22647656; http://dx.doi.org/ 10.4161/auto.19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dehay B, Martinez-Vicente M, Caldwell GA, Caldwell KA, Yue Z, Cookson MR, Klein C, Vila M, Bezard E. Lysosomal impairment in Parkinson's disease. Mov disord 2013; 28:725-32; PMID: 23580333; http://dx.doi.org/ 10.1002/mds.25462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999; 397:271-4; PMID: 9930704; http://dx.doi.org/ 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- 48. Norberg E, Orrenius S, Zhivotovsky B. Mitochondrial regulation of cell death: processing of apoptosis-inducing factor (AIF). Biochem Biophys Res Commun 2010; 396:95-100; PMID: 20494118; http://dx.doi.org/ 10.1016/j.bbrc.2010.02.163 [DOI] [PubMed] [Google Scholar]
- 49. White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 2010; 22:212-7; PMID: 20056400; http://dx.doi.org/ 10.1016/j.ceb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J 2000; 19:1576-86; PMID: 10747026; http://dx.doi.org/ 10.1093/emboj/19.7.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang CP, Su YC, Hu CW, Lei HY. TLR2-dependent selective autophagy regulates NF-kappaB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell Death Differ 2013; 20:515-23; PMID: 23175187; http://dx.doi.org/ 10.1038/cdd.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou X, Zhou J, Li X, Guo C, Fang T, Chen Z. GSK-3beta inhibitors suppressed neuroinflammation in rat cortex by activating autophagy in ischemic brain injury. Biochem Biophys Res Commun 2011; 411:271-5; PMID: 21723251; http://dx.doi.org/ 10.1016/j.bbrc.2011.06.117 [DOI] [PubMed] [Google Scholar]
- 53. Zhao Z, Loane DJ, Murray MG, 2nd, Stoica BA, Faden AI. Comparing the predictive value of multiple cognitive, affective, and motor tasks after rodent traumatic brain injury. J Neurotrauma 2012; 29:2475-89; PMID: 22924665; http://dx.doi.org/ 10.1089/neu.2012.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McMahon J, Huang X, Yang J, Komatsu M, Yue Z, Qian J, Zhu X, Huang Y. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J Neurosci 2012; 32:15704-14; PMID: 23136410; http://dx.doi.org/ 10.1523/JNEUROSCI.2392-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.