Abstract
Autophagy is an essential component of host innate and adaptive immunity. Viruses have developed diverse strategies for evading or utilizing autophagy for survival. The response of the autophagy pathways to virus invasion is poorly documented. Here, we report on the induction of autophagy initiated by the pathogen receptor HSP90AA1 (heat shock protein 90 kDa α [cytosolic], class A member 1) via the AKT-MTOR (mechanistic target of rapamycin)-dependent pathway. Transmission electron microscopy and confocal microscopy revealed that intracellular autolysosomes packaged avibirnavirus particles. Autophagy detection showed that early avibirnavirus infection not only increased the amount of light chain 3 (LC3)-II, but also upregulated AKT-MTOR dephosphorylation. HSP90AA1-AKT-MTOR knockdown by RNA interference resulted in inhibition of autophagy during avibirnavirus infection. Virus titer assays further verified that autophagy inhibition, but not induction, enhanced avibirnavirus replication. Subsequently, we found that HSP90AA1 binding to the viral protein VP2 resulted in induction of autophagy and AKT-MTOR pathway inactivation. Collectively, our findings suggest that the cell surface protein HSP90AA1, an avibirnavirus-binding receptor, induces autophagy through the HSP90AA1-AKT-MTOR pathway in early infection. We reveal that upon viral recognition, a direct connection between HSP90AA1 and the AKT-MTOR pathway trigger autophagy, a critical step for controlling infection.
Keywords: AKT-MTOR pathway, autophagy, avibirnavirus, HSP90AA1, viral protein VP2
Abbreviations: ANOVA, analysis of variance; ATG5, autophagy-related 5; BCA, bicinchoninic acid; BECN1, Beclin 1, autophagy-related; cDNA, complementary DNA; CoIP, coimmunoprecipitation; DMEM, Dulbecco's modified Eagle's medium; dsRNA, double-stranded RNA; EBSS, Earle's balanced salt solution; EIF2AK2, eukaryotic translation initiation factor 2-alpha kinase 2; EIF2S1, eukaryotic translation initiation factor 2, subunit 1 alpha; eGFP, enhanced green fluorescent protein; ER, endoplasmic reticulum; Gg, Gallus gallus (chicken); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GOPC, golgi-associated PDZ and coiled-coil motif containing; GST, glutathione S-transferase; HE-IBDV, heat-inactivated IBDV; hpi, hours post-infection; Hs, Homo sapiens (human); HSP90AA1, heat shock protein 90 kDa alpha (cytosolic), class A member 1; HSV-1, herpes simplex virus 1; IBDV, infectious bursal disease virus; IgG, immunoglobulin G; LPS, lipopolysaccharide; mAb, monoclonal antibody; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; MOI, multiplicity of infection; MTOR, mechanistic target of rapamycin (serine/threonine kinase); Ni-NTA, nickel-nitrilotriacetic acid; PAMP, pathogen-associated molecular patterns; PBS, phosphate-buffered saline; PI3K, phosphoinositide 3-kinase; PRR, pattern recognition receptors; RNAi, RNA interference; SDS, sodium dodecyl sulfate; siRNA, small interfering RNA; shRNA, short hairpin RNA; SQSTM1, sequestosome 1; TCID50, 50% tissue culture infectious doses; TLR, toll-like receptors; TSC, tuberous sclerosis complex; VP, viral protein; SVP, subviral particle
Introduction
Accumulated evidence for the antiviral role of autophagy suggests a strong connection between autophagy and innate or adaptive immunity.1 Autophagy activates various cellular defense responses, including direct digestion of intracytoplasmic virions,2-4 recognition of pathogen-associated molecular patterns (PAMP) through the delivery of viral genomes to endosomal toll-like receptors (TLRs),5 activation of innate immune signaling,6 regulation of inflammatory response, and facilitation of the appropriate antiviral adaptive immune responses.7-12 Viruses subvert autophagy through various strategies for inhibiting autophagosome formation and blocking autophagosome maturation.2,13-15 However, some viruses exploit autophagy for their own replication.16,17 Thus, there is an extremely complex interaction between autophagy and invading viruses.
Autophagy is a highly conserved cytoplasmic pathway in eukaryotes; it is required for both normal development and survival of nutrient deprivation.18 Autophagy includes macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy, which we will refer to as autophagy in this article, is the most well-characterized type in eukaryotic cells.19 Autophagy is involved in the engulfment of damaged or unwanted cellular organelles and protein aggregates into an autophagosome, which are delivered to a lysosome to be degraded. The MTOR kinase-dependent signaling pathway controls autophagy.20 Activation of the phosphoinositide 3-kinase (PI3K)-AKT-MTOR and AKT-tuberous sclerosis complex (TSC)-MTOR pathways inhibits autophagy, whereas the loss of signaling through this cascade removes the negative repression of MTOR.20 Therefore, there is a direct link between autophagy and the MTOR signaling pathway.
Upon intracellular pathogen invasion, autophagy can be activated as an innate immune mechanism for controlling infection.21 To date, a growing number of studies use pathogen recognition to reveal the effects of pathogen infection on autophagy. Pattern recognition receptors (PRR) detect pathogens by recognizing PAMP. Extracellular pathogens are recognized via TLRs on the cell surface or in endolysosomal compartments; consequently, functional autophagy is induced.22-24 Cytoplasmic pattern recognition systems act as intracellular pathogen detectors and degrade viral double-stranded RNA (dsRNA) genomes or replication intermediates.25 CD46, present on the cell membrane, shares PRRs with several viruses and bacteria, which is sufficient for inducing functional autophagy through the (CD46-Cyt-1)-Golgi-associated PDZ and coiled-coil motif containing protein (GOPC) pathway.26 Heat shock protein 90 (HSP90AA1), a highly conserved molecular chaperone, is also a PRR component that binds to lipopolysaccharide (LPS),27 dengue virus, and avibirnavirus.28,29 However, whether HSP90AA1 involvement regulates autophagy remains unknown.
In the present study, we address whether HSP90AA1 can induce autophagy, and how, and investigate the physiological relevance of this process to pathogen binding. We found that HSP90AA1 PRR to the viral protein VP2 of the avibirnavirus infectious bursal disease virus (IBDV) activate MTOR-dependent autophagy. Our study highlights the role of HSP90AA1 in linking pathogen recognition to the MTOR signaling and autophagy pathways.
Results
Intracellular autophagosome-packaged avibirnavirus virions
Transmission electron microscopy is a valid and important method for observing autophagosomes in cells;30,31 in this study, it showed that avibirnavirus particle and organelle were captured within membrane vesicles in infected DF-1 cells (Fig. 1A), similar to the previously reported autophagosomes that contained avibirnavirus virions in lymphoid cells and macrophages of infected Gallus gallus,32 indicating that autophagy was involved in virion degradation. Correspondingly, after DF-1 cells, transfected with enhanced green fluorescent protein–light chain 3 (eGFP-LC3) for 24 h, were infected with IBDV or incubated with heat-inactivated IBDV (HE-IBDV), the puncta or ring-like structures of eGFP-LC3 and LysoTracker Red-labeled autophagosomes per cell were calculated using confocal microscopy. Fig. 1B and C show that in comparison with mock-infected cells, the ring-like and small puncta eGFP-LC3 structures were significantly increased in the cytoplasm of IBDV-infected or HE-IBDV-treated DF-1 cells at 2 h postinfection (hpi, P < 0.01), and subsequently reverted to normal condition in the cytoplasm of IBDV-infected cells but not HE-IBDV-treated cells at 4 hpi. To verify whether IBDV virions were the target of autophagy, IBDV-infected, eGFP-LC3-labeled cells were stained with monoclonal antibody (mAb) to viral protein VP2 of IBDV. Confocal microscopy revealed that VP2 staining was surrounded by the ring-like eGFP-LC3 (Fig. 1D). Collectively, these data strongly support the premise that autophagosome formation is induced in IBDV-infected and HE-IBDV-treated cells and that IBDV virions are captured in autophagosomes.
Figure 1.
IBDV infection induces autophagosome accumulation in DF-1 cells. (A) Autophagic vacuoles in infected cells observed by transmission electron microscopy. Autophagic vacuole engulfs IBDV particle (black arrows) and organelle (asterisk) in cytoplasm of DF-1 cell infected with IBDV (MOI = 10) at 2 hpi. (B) DF-1 cells transfected with peGFP-LC3 for 48 h, and then infected with IBDV (MOI = 10) or treated with HE-IBDV. At 2 hpi or 4 hpi, cells were incubated with LysoTracker Red (50 nM) for 30 min; intracellular autophagic vacuoles were observed under confocal microscopy. Statistical analysis of the number of cells with >3 autophagic vacuoles. At 2 hpi or 4 hpi, autopgagic vacuoles were counted in IBDV-infected cells. Error bars: Mean ± SD of 3 independent tests. Two-way analysis of variance (ANOVA); ***P < 0.001 compared to control. (C) DF-1 cells transfected with peGFP-LC3 for 48 h, and then infected with IBDV (MOI = 10) or treated with HE-IBDV. At 2 hpi or 4 hpi, cells were incubated with LysoTracker Red (50 nM) for 30 min; intracellular autophagic vacuoles were observed under confocal microscopy. Scale bars: 10 10 μm.mu;m. (D) DF-1 cells transfected with peGFP-LC3 and infected with IBDV at 9 h post-transfection. The cells were fixed, immunostained with anti-VP2 mAb, and autophagosomes were observed under confocal microscopy. Scale bar: 10 μ.
IBDV virions adhering to the cell membrane triggered autophagy at the early stage of infection
The first generation of IBDV progeny virions is assembled at about 6 hpi.32 Therefore, to determine whether autophagy is triggered at the early stage of infection, we examined cells infected with IBDV at a multiplicity of infection (MOI) of 10 from 0 to 8 hpi to investigate the changes to endogenous LC3-II, an autophagosome marker.30 Fig. 2A to E shows that in comparison to mock-infected cells, the amount of intracellular LC3-II in IBDV-infected cells was strongly increased at 1 hpi, decreased remarkably at 4 hpi and recovered significantly at 8 hpi (P < 0.01). However, intracellular LC3-II expression in HE-IBDV-treated cells was upregulated persistently from 1 hpi to 4 hpi (P < 0.001) and began to decrease from 4 hpi. Similar LC3-II expression profiles were observed in IBDV-infected or HE-IBDV-treated 293T cells (Fig. S1). In addition to LC3, we examined the autophagic flux by detecting the accumulation of SQSTM1 (sequestosome 1), an autophagy substrate.30 SQSTM1 in IBDV-infected cells decreased persistently from 1 hpi to 6 hpi (P < 0. 001) and began to increase at 8 hpi, indicating that the autophagic flux in IBDV-infected cells was induced after 1 hpi and began to be inhibited at 8 hpi (Figs. 2A to C, F, and S1). Differently, SQSTM1 in HE-IBDV-treated cells decreased persistently from 1 hpi to 8 hpi (P < 0.001), but did not change obviously in mock-infected cells. Interestingly, the upregulation of LC3-II conversion and decrease of SQSTM1 accumulation were also detected in the VP2-transfected 293T cells at 48 h after transfection (P < 0.001, Fig. 2H). To exclude the possibility that contaminations induce autophagy, IBDV virions purified with density gradient centrifugation were used to induce cellular autophagy. The similar autophagic induction was also observed in DF-1 cells infected with the purified infectious IBDV virions and in DF-1 cells treated with the purified heat-inactivated IBDV virions (Fig. 2D and G). Moreover, the viral structural proteins VP2 in IBDV-infected cells decreased greatly at 2 hpi and began to increase at 4 hpi (Fig. 2D), indicating that the viral structural proteins were degraded at the early stage of infection and synthesized gradually following production of IBDV infectious progeny. Taken together, these data indicated that cellular autophagy was induced by IBDV virions at the early stage of virus infection.
Figure 2.
Characterization of IBDV-triggered autophagosome accumulation. (A) IBDV promotes increase of LC3-II and decrease of SQSTM1 within 1 hpi but inhibits it from 4 hpi. DF-1 cells were infected with IBDV (MOI = 10). (B) Increase of LC3-II and decrease of SQSTM1 was constantly promoted in HE-IBDV-treated cells. (C) LC3 amount and SQSTM1 accumulation in mock-infected cells were unaffected. Cells were harvested and analyzed by immunoblotting using anti-LC3, anti-SQSTM1, anti-VP2, or anti-GAPDH antibody. (D) DF-1 cells were infected with the purified infectious IBDV virions or treated with the purified heat-inactivated IBDV virions (MOI = 10), respectively. The purified infectious IBDV increases the amount of LC3-II within 1 hpi but inhibits it from 4 hpi. However, levels of LC3-II increase constantly in the purified HE-IBDV-treated cells. M, mock infected. HI, purified heat-inactivated IBDV virions. PI, purified infectious IBDV virions. (E, F) The ratio of LC3-II or SQSTM1 to GAPDH was normalized to control conditions in (A to C). (G) The ratio of LC3-II to GAPDH was normalized to control conditions in (D). (H) LC3-II increases and SQSTM1 decreases in VP2-transfected cells. 293T cells were transfected with the vector pFlag-VP2 for 48 h, harvested and analyzed by immunoblotting using anti-LC3, anti-SQSTM1, anti-VP2, or anti-GAPDH antibody. Error bars: Mean ± SD of 3 independent tests. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001 compared to control.
Cellular autophagy affected IBDV replication
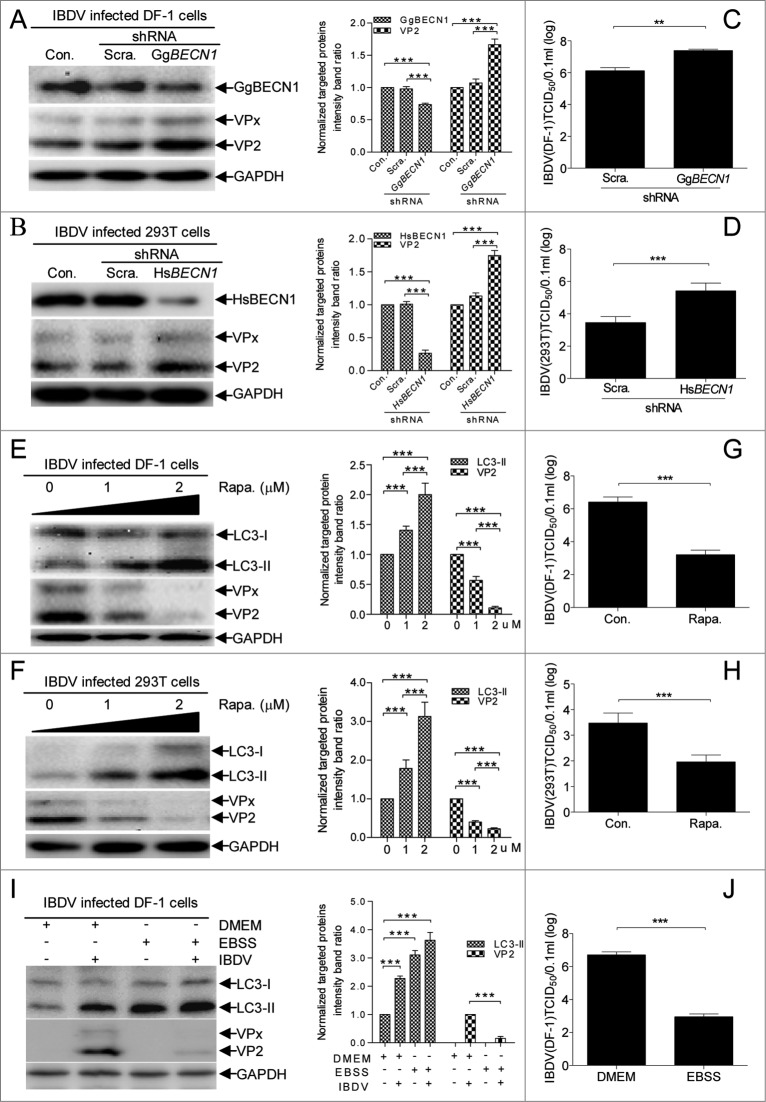
To investigate whether cellular autophagy regulates IBDV replication, we examined the effect of inhibiting autophagy on viral protein accumulation and viral progeny yields using a median tissue culture infectious dose (TCID50) detection assay. Target-specific RNA interference (RNAi) was used to knock down BECN1, a gene central to autophagy that encodes a protein necessary for autophagy.33 Fig. 3A and B show that compared to cells transfected with scrambled and ineffective small interfering RNAs (siRNAs), there was significantly decreased BECN1 expression (P < 0.001) and increased VP2 expression (P < 0.001) in DF-1 cells or 293T cells transfected with BECN1 siRNAs. Likewise, virus titer assays showed that BECN1 downregulation increased virus yields significantly at 24 hpi in DF-1 cells or 293T cells (Fig. 3C and D, P < 0.01), indicating that inhibition of autophagy promotes IBDV replication. In contrast, following rapamycin- or starvation-induced autophagy, amount of LC3-II increased significantly in infected DF-1 cells (P < 0.001, Fig. 3E and I) or 293T cells (P < 0.001, Fig. 3F) and the IBDV progeny titer decreased dramatically (Fig. 3G and H, and J, P < 0.001), revealing that induction of autophagy greatly inhibits IBDV replication. Collectively, these findings confirm that cellular autophagy is involved in regulating IBDV replication.
Figure 3.
Effect of autophagy on IBDV replication. (A, B) BECN1 knockdown increases the accumulation of IBDV-encoded VP2 in 293T or DF-1 cells. DF-1 cell and 293T cell monolayers were transfected with negative control (Con.), Scrambled shRNA (Scra.) or Gallus gallus (chicken) BECN1 shRNA (GgBECN1 shRNA) or Homo sapiens (human) BECN1 (HsBECN1 shRNA) for RNA knockdown, respectively, and were infected with IBDV (MOI = 10). At 8 hpi, cells were harvested and analyzed by western blotting with anti-LC3, anti-VP2, and anti-GAPDH antibodies. The ratio of VP2 or BECN1 to GAPDH was normalized to control conditions. (C, D) DF-1 or 293T cells transfected with Scra. or HsBECN1 shRNA, or GgBECN1 shRNA were infected with IBDV (MOI = 0.01) for 24 h. Progeny virus yields in DF-1 cells were determined by TCID50 assay. (E, F) Induction of autophagy with rapamycin (Rapa.) reduced IBDV replication in DF-1 and 293T cells. DF-1 and 293T cells were pretreated in DMEM containing 1 1 μM mu;M or 2 2 μM mu;M rapamycin for 4 h and then infected with IBDV (MOI = 10) for 1 h. After 1 hpi, cells were incubated in DMEM containing 1 1 μM mu;M or 2 2 μM mu;M rapamycin for 7 h, then harvested, lysed, and processed for western blotting with anti-LC3, anti-VP2, and anti-GAPDH antibodies. The ratio of LC3-II or VP2 to GAPDH was normalized to control conditions. (G, H) DF-1 or 293T cells were infected with IBDV (MOI = 0.01) and cultured in DMEM containing 2 2 μM mu;M rapamycin for 24 h to yield progeny virus; virus titers in DF-1 cells were determined by TCID50 assay. (I) Induction of autophagy with starvation reduced IBDV replication in DF-1cells. DF-1 cells were pretreated in EBSS for 2 h. Then the cells were infected with IBDV at MOI of 10 and incubated in EBSS. After cultured for 8 h, cells were harvested and detected by western blotting with anti-LC3, anti-VP2, and anti-GAPDH antibodies. The ratio of LC3-II or VP2 to GAPDH was normalized to control conditions. (J) DF-1 cells pretreated with EBSS for 2 h were infected with IBDV (MOI = 0.01) for 1 h and cultured in EBSS for 24 h to yield progeny virus; virus titers in DF-1 cells were determined by TCID50 assay. Error bars: Mean ± SD of 3 independent tests. Two-way ANOVA; ***P < 0.001 compared to control.
IBDV-induced autophagy by inactivating the AKT-MTOR pathway
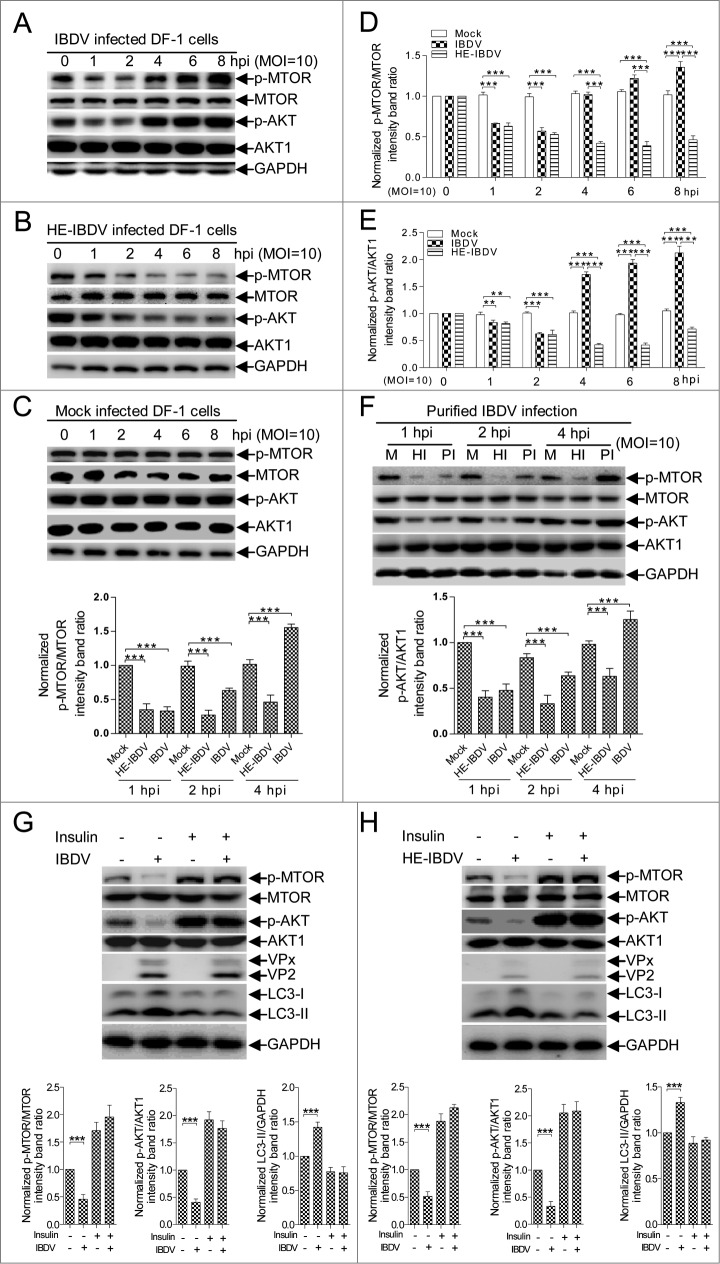
We examined AKT and MTOR activity to investigate whether autophagy induced in the early stage of IBDV infection is involved in the MTOR-dependent signaling pathway. Compared to mock-infected DF-1 cells or 293T cells, AKT and MTOR phosphorylation in IBDV-infected cells was downregulated (P < 0.001) from 1 hpi to 2 hpi, but upregulated (P < 0.001) at 4 hpi (Fig. 4A to E and S2A to E). However, phosphorylated (p) AKT and MTOR were persistently downregulated (P < 0.001) in HE-IBDV-treated cells. The similar profiles were confirmed in cells infected with purified IBDV or treated with purified HE-IBDV (Fig. 4F). These data indicate that IBDV-induced autophagy upon inactivating the AKT-MTOR pathway in the early stage of infection, although AKT and MTOR phosphorylation decreased and then recovered.
Figure 4.
MTOR and AKT were inactivated at the early stage of IBDV infection. (A) DF-1 cells infected with IBDV (MOI = 10). Cells were harvested at 1, 2, 4, 6, and 8 h, followed by immunoblotting with anti-p-MTOR, anti-MTOR (total protein), anti-p-AKT, anti-AKT1, or anti-GAPDH antibody. (B) Cells incubated with HE-IBDV for 1, 2, 4, 6, and 8 h followed by immunoblotting as in (A). (C) Cells mock-infected and analyzed by immunoblotting as in (A). (D, E) The ratio of p-MTOR to MTOR or p-AKT to AKT was normalized to mock infection and set at 1.0. (F) Cells were infected with purified IBDV or treated with purified HE-IBDV for 1, 2, and 4 h, and analyzed by immunoblotting using the antibody in (A). (G) DF-1 cells pretreated with insulin (500 nM) for 1 h, infected with IBDV and cultured for 2 h, and processed by immunoblotting using the corresponding antibody in (A). (H) DF-1 cells pretreated with insulin (500 nM) for 1 h, incubated with HE-IBDV for 2 h, and processed for immunoblotting using the antibody in (A). The ratio of LC3 to GAPDH, p-MTOR to MTOR, or p-AKT to AKT was normalized to control conditions and set at 1.0. Error bars: Mean ± SD of 3 independent tests. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001 compared to control.
To validate the role of AKT and MTOR in IBDV-dependent induction of autophagy, we activated the AKT-MTOR pathway with insulin before infecting cells with IBDV or treating them with HE-IBDV. Fig. 4G and H showed that following insulin treatment, there was no obvious decrease in AKT and MTOR phosphorylation (P > 0.05) in IBDV-infected or HE-IBDV-treated cells compared to mock-infected cells and IBDV infection had no significant changes in cells treated with or without insulin. Correspondingly, the level of LC3-II in IBDV-infected and HE-IBDV-treated cells did not increase remarkably (P > 0.05) compared to mock-infected cells (Fig. 4G and H), indicating that AKT-MTOR signaling pathway activation suppressed IBDV-induced autophagy. Collectively, our observations confirm that inactivating the AKT-MTOR pathway in the early stage of IBDV infection stimulates autophagy.
VP2 was critical for induction of autophagy via the AKT-MTOR pathway
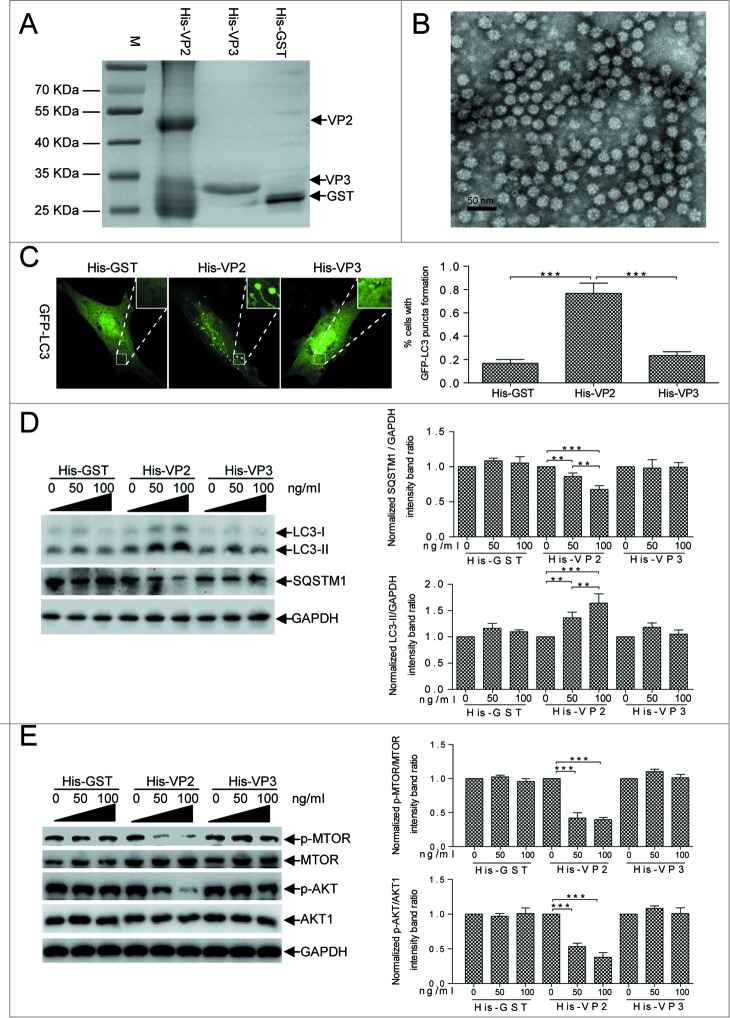
In host cells, cellular autophagy might be involved in the recognition of incoming IBDV virions during the invading stage. VP2 and VP3 are IBDV capsid proteins: VP2 localizes on the external surface of mature virions34 and VP3 is a major inner capsid protein.35 Consequently, we examined whether VP2 activated autophagy effectively via the AKT-MTOR pathway. The observation of transmission electron microscope showed that the purified Escherichia coli-expressed fusion protein His-tagged VP2 formed subviral particles (SVPs) with an approximate 25 nm diameter (Fig. 5A and B). We detected a significant increase in autophagosomes by observing the ring-like eGFP-LC3 structures in His-VP2-treated DF-1 cells compared to His-VP3 or His-GST-treated cells (P < 0.001, Fig. 5C). We incubated cells with the purified fusion proteins His-VP2 SVPs, His-VP3, and His-glutathione S-transferase (GST) separately for 1 h and subsequently examined the endogenous LC3-II and SQSTM1 levels. Fig. 5D shows that LC3-II was upregulated and SQSTM1 was downregulated dose-dependently in His-VP2-treated cells (P < 0.001). In contrast, LC3-II and SQSTM1 expression was not disrupted in His-VP3 or His-GST-treated cells (P > 0.05). Moreover, we determined AKT and MTOR phosphorylation levels within 4 h after cells were treated with VP2 to clarify the signaling pathway involved in VP2-induced cellular autophagy. Fig. 5E shows that p-AKT and p-MTOR decreased significantly (P < 0.001) in a dose-dependent manner in VP2-treated cells and that there was no detectable change in His-GST or VP3-treated cells (P > 0.05), indicating that VP2 inhibited AKT-MTOR phosphorylation. Together, these results confirm that the external capsid protein VP2 is sufficient for activating autophagy effectively by decreasing AKT and MTOR activity.
Figure 5.
IBDV VP2 was sufficient for inducing autophagy via AKT and MTOR dephosphorylation. (A) His-tagged VP2 or VP3 expressed in E. coli BL21 and purified in Ni-NTA columns. The purified products were separated using SDS-PAGE and stained with Coomassie brilliant blue. (B) The subviral particles of the purified His-VP2 protein expressed in E.coli. Scale bar: 50 nm. (C) DF-1 cells transfected with eGFP-LC3 for 24 h and incubated with His-VP2 (100 ng/mL), His-VP3 (100 ng/mL), or His-GST (100 ng/mL) for 2 h and observed under confocal microscopy. The ratio of cells with >3 autophagic vacuoles was determined. Scale bars: 10 10 μm.mu;m. Error bars: Mean ± SD of 3 independent tests. (D, E) DF-1 cells incubated in DMEM containing His-GST, His-VP2, or His-VP3 for 4 h, were analyzed by western blotting with anti-LC3, anti-SQSTM1, anti-GAPDH, anti-p-MTOR, anti-MTOR, anti-p-AKT, and anti-AKT1 antibodies. The ratio of LC3 or SQSTM1 to GAPDH, p-MTOR to MTOR, and p-AKT to AKT were normalized to control conditions. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001 compared to the control.
HSP90AA1 contributed to IBDV-induced autophagy via the AKT-MTOR pathway
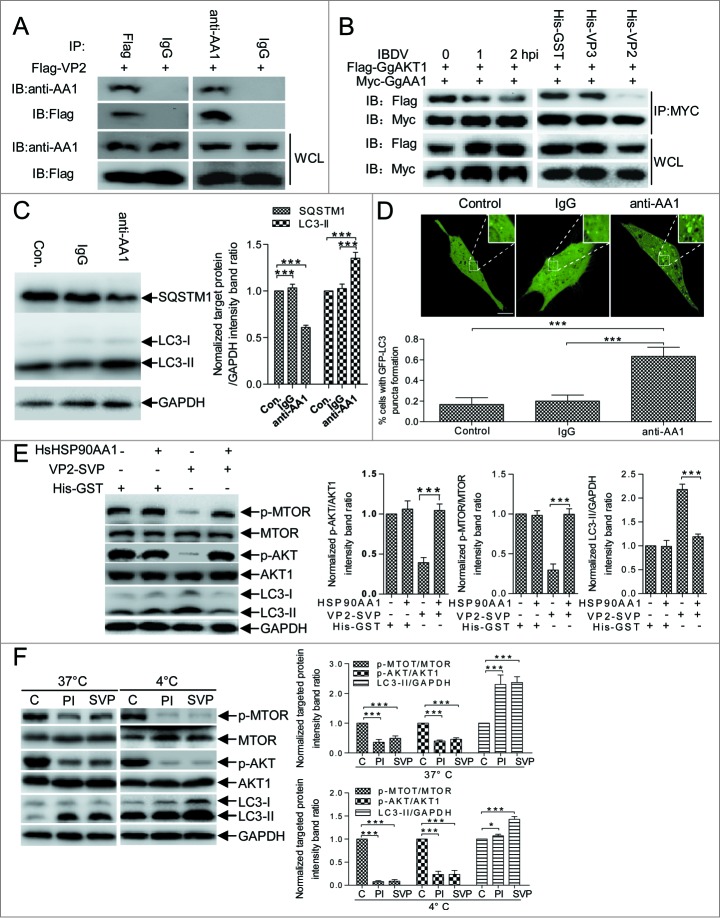
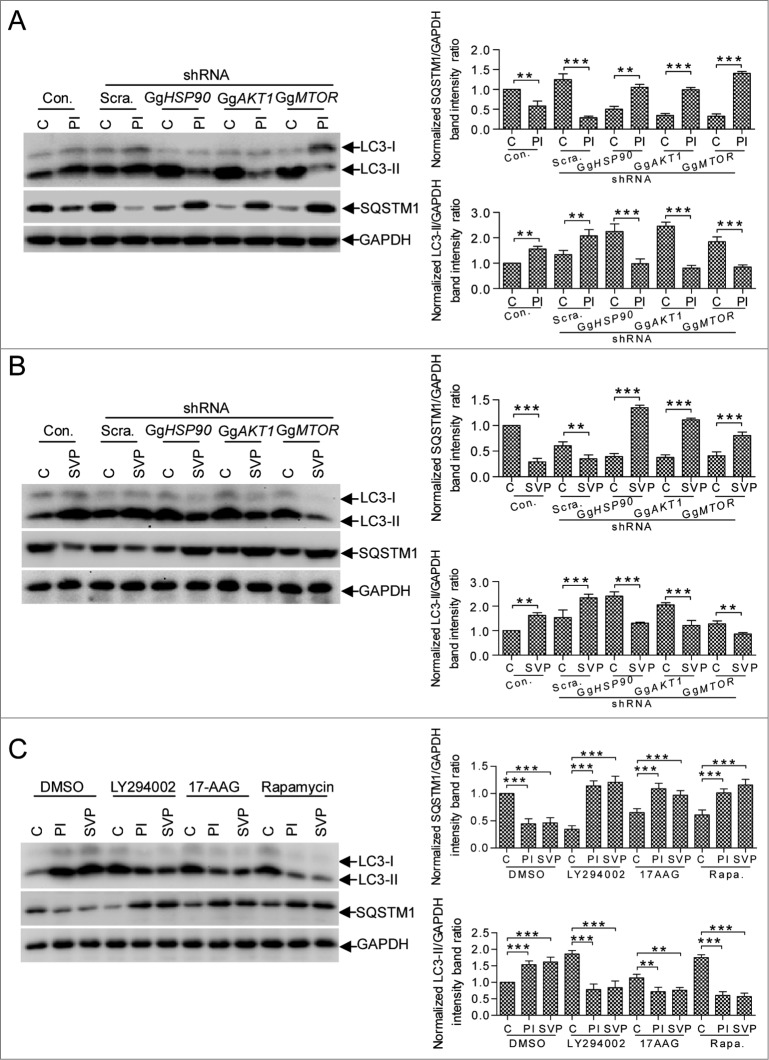
Cell membrane surface-distributed GgHSP90AA1 is a pathogen receptor that recognizes IBDV;29 cytoplasmic HSP90 is important for maintaining AKT kinase activity through its interaction with AKT.36 We examined whether cellular GgHSP90AA1 mediated VP2-induced autophagy via the AKT-MTOR pathway. We detected VP2 and GgHSP90AA1 interaction, and AKT-MTOR pathway and autophagy cross-linking using anti-HSP90AA1 antibody (anti-AA1) to mimic VP2. Coimmunoprecipitation (CoIP) showed that VP2 interacted with host HSP90AA1 (Fig. 6A), interestingly, GgAKT and GgHSP90AA1 has a weaker interaction in IBDV-infected and His-VP2 treated cells in comparison with that in normal cells (Fig. 6B). LC3-II was upregulated and SQSTM1 was downregulated in anti-HSP90AA1-treated DF-1 cells, but there were no obvious changes in irrelevant immunoglobulin G (IgG)-treated and control cells (Fig. 6C). Likewise, immunofluorescence assay revealed significantly increased autophagosomal vacuoles in anti-HSP90AA1-treated cells transfected with eGFP-LC3 (P < 0.01, Fig. 6D), but not in irrelevant IgG-treated cells. Correspondingly, the VP2 SVP-treated DF-1 cells had a significant decrease of p-MTOR and p-AKT expression (P < 0.001) and a remarkable increase of LC3-II (P < 0.001), however, the similar change did not showed in mock, recombinant HsHSP90AA1 protein (HsHsp90AA1), and SVP-HsHSP90AA1 complex-treated cells (Fig. 6E). Moreover, after DF-1 cells were treated with SVP or infected with IBDV virion at 4°C and 37°C, the concentration of p-MTOR and p-AKT had a remarkable decrease (P < 0.001) in comparison with the mock-treated DF-1 cells (Fig. 6F). These data indicated that the membrane-distributed HSP90AA1 binding to the SVP impaired MTOR signaling activity was a temperature-independent. To validate HSP90AA1-AKT-MTOR pathway mediation of IBDV-induced autophagy, we examined autophagy following IBDV infection and VP2 treatment by constructing DF-1 cells that did not express GgHSP90AA1, AKT, or MTOR using the short hairpin RNA (shRNA) approach (Fig. S3). Fig. 7A and B shows that in the control or scrambled shRNA transfectant, both IBDV infection and VP2-SVP treatment decreased SQSTM1 significantly and increased LC3-II remarkably (P < 0.001). Conversely, in IBDV-infected and VP2-SVP-treated DF-1 cells with knockdown of GgHSP90AA1, GgAKT, or GgMTOR, both IBDV infection and VP2-SVP treatment resulted in increase of SQSTM1 and decrease of LC3-II (P < 0.001). Similar results were revealed in DF-1 cells treated with the specific inhibitors of HSP90AA1 (17-AAG), AKT (LY294002) or MTOR (rapamycin) (Fig. 7C). These data demonstrate that induction of autophagy was regulated via the HSP90AA1-AKT-MTOR signaling pathway by VP2 binding to HSP90AA1 in the early stage of IBDV infection (Fig. 8).
Figure 6.
HSP90AA1 binding to VP2 triggers autophagy via AKT-MTOR dephosphorylation. (A) DF-1 cells transfected with pFlag-VP2 for 24 h. Whole cell lysates (WCL) were used for CoIP and western blotting with anti-Flag or anti-HSP90AA1 antibody (anti-AA1) and irrelevant IgG (Control). (B) DF-1cells cotransfected with Myc-GgHSP90AA1 (Myc-GgAA1) and Flag-GgAKT1 for 48 h. Transfected cells were infected with IBDV for 1 or 2 h, or were incubated in DMEM containing His-VP2(100 ng/ml), His-VP3(100 ng/ml) or His-GST(100 ng/ml). Whole cell lysates of each sample were used for CoIP with anti-MYC antibody and western blotting with anti-Flag or anti-MYC antibody. (C) Western blotting performed using anti-LC3 antibody and anti-SQSTM1 mAb on lysates from DF-1 cells cultured in uncoated plates or in coated plates with anti-HSP90AA1 or irrelevant isotype control IgG for 4 h. The ratio of SQSTM1 or LC3-II to GAPDH was normalized to control conditions. (D) DF-1 cells transfected with peGFP-LC3 for 24 h and cultured in plates coated with negative control, IgG, or anti-HSP90AA1 for 4 h. Autophagic vacuoles were analyzed under confocal microscopy. The ratio of cells containing >3 ring-like GFP structures was determined. Scale bars: 10 10 μm.mu;m. Error bars: Mean ± SD of 3 independent tests. (E) DF-1 cells were incubated respectively with the His-GST, mixture of His-GST and HSP90AA1 (His-GST:HSP90AA1 = 1:2.5), mixture of SVP and HSP90AA1 (SVP:HSP90AA1 = 1:2.5) or SVP for 2 h, and analyzed by immunoblotting with anti-LC3, anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT1, or anti-GAPDH antibody. (F) DF-1 cells incubated with His-GST (100 ng/ml), SVP (100 ng/ml), or purified IBDV (MOI = 10 ) for 2 h at 37°C or 4°C. Cells were analyzed by immunoblotting using the antibody in (E). The ratio of p-MTOR to MTOR, p-AKT to AKT or LC3-II to GAPDH was normalized to mock infection and set at 1.0. Two-way ANOVA; ***P < 0.001 compared to control. C, His-GST; PtdIns, purified IBDV; SVP, His-VP2 subviral particle.
Figure 7.
HSP90AA1, AKT, and MTOR are critical for autophagic induction at the early stage of IBDV infection. DF-1 cells transfected with Con. (Control), Scra. (Scrambled shRNA), GgHSP90 (GgHSP90AA1 shRNA), GgAKT1 (GgAKT1 shRNA), and GgMTOR (GgMTOR shRNA) for RNA knockdown were infected with IBDV (A), and pretreated with purified His-VP2 subviral particles (B) for 2 h and analyzed by immunoblotting with anti-LC3, anti-SQSTM1 or anti-GAPDH antibodies. The purified His-GST was as a control. (C) DF-1 cells were pretreated by specific inhibitors 17-AAG (HSP90AA1), LY294002 (AKT) and rapamycin (MTOR) for 4 h, and incubated with His-GST (100 ng/ml), SVP (100 ng/ml) or purified IBDV (MOI = 10) for 2 h. The cells were harvested and analyzed by immunoblotting with anti-LC3, anti-SQSTM1 or anti-GAPDH antibody. The ratio of LC3 or SQSTM1 to GAPDH was normalized to control conditions. Two-way ANOVA; ***P < 0.001 compared to control. C, His-GST; PI, purified IBDV; SVP, His-VP2 subviral particle.
Figure 8.
Proposed model of IBDV-induced autophagy via the HSP90AA1-AKT-MTOR pathway. IBDV-encoded VP2 binding to cell surface HSP90AA1 and leads to disassociation of phosphorylated AKT from HSP90AA1. The disassociated AKT then loses phosphorylation and results in dephosphorylation of MTOR. The dephosphorylated MTOR then activates autophagosome formation. The autophagosome engulfs IBDV virions, delivering them to lysosomes for final degradation.
Discussion
IBDV and autophagy
Several reports have demonstrated that viruses have evolved strategies for regulating autophagy by encoding certain proteins, such as ICP34.5 of herpes simplex virus 1 (HSV-1), Nef of HIV, and M2 of influenza A virus.2,13,37 Orvedahl and colleagues report that Sindbis virus infection induces autophagy via the autophagy-related 5 gene (ATG5).38 In our study, transmission electron microscopy revealed IBDV particles inside autophagosomes and autolysosomes, which were engulfed into ring-like eGFP-LC3 structures as observed under confocal microscopy. Induction of autophagy by starvation or rapamycin decreased VP2 accumulation and IBDV titers; in contrast, inhibition of autophagy through BECN1 knockdown (Fig. 3) increased VP2 accumulation and viral titers. These results suggest that activation of host cell autophagy is a defense response to IBDV invasion.
PAMP recognition and HSP90AA1-mediated autophagy
To cause infection, viruses must deliver their nucleic acids across the cell membrane into the target cell cytoplasm. TLR3, TLR4, and TLR7 in mammals and PGLYRP1 (peptidoglycan recognition protein 1)-LE in Drosophila induce autophagy upon ligand binding.5,23,24,39 The molecular chaperone HSP90AA1 facilitates the maturation of a wide range of proteins and plays a crucial role in various cellular functions. Of interest is the fact that HSP90AA1 is also a pathogen receptor that binds to LPS27 and dengue virus.28 Importantly, GgHSP90AA1 is also a cell receptor complex component of IBDV particles.29 However, the molecular connections between such receptors and autophagy remain poorly understood. In our study, autophagy was induced immediately within 2 h following avibirnavirus infection (Figs. 1 and 2); HSP90AA1 strengthened the formation of early and mature autophagosomes that efficiently sequestered and degraded IBDV particles immediately (Fig. 6). These autophagic vacuoles, containing IBDV virions, were previously observed in IBDV-infected macrophages and lymphocytes.32 This demonstrates that GgHSP90AA1 controls early avibirnavirus infection by xenophagy.
Notably, of the avibirnavirus-encoded proteins, only VP2 interacted with GgHSP90AA1 and triggered autophagy (Figs. 5 and 6). Mature VP2 is an outer capsid protein and the primary host-protective immunogen of the IBDV virion.34 In addition, the hypervariable region of VP2 (amino acids 206-350) is presumably responsible for interaction with cellular receptors and restriction of infectivity.40 Therefore, our results suggest that GgHSP90AA1 recognizes VP2 as a PAMP and triggers autophagy in the early stage of IBDV infection, and that HSP90AA1 may initiate the innate cellular defense against invading pathogens. This also suggests an attractive hypothesis: viral genomic dsRNA is released from the IBDV virion by autophagic degradation and is ready for subsequent viral replication in the early stage of infection upon inhibition of autophagy initiated from 2 hpi (Figs. 1 and 2).
Detection of MTOR and PAMP
In all eukaryotes, the MTOR signaling pathway couples energy and nutrient abundance to execute cell growth and division.41 As a highly conserved kinase in the phosphatidylinositol 3-kinase family, MTOR leads to a swift response to various environmental cues, regulating cell metabolism and immune responses by regulating the kinase AKT.42,43 Endoplasmic reticulum (ER) stress suppresses autophagy via negative regulation of the AKT-TSC-MTOR pathway.44 Activation of the PI3K-AKT-MTOR signaling pathway inhibits the autophagy required for both normal development and survival of nutrient deprivation.45 Extracellular and intracellular environmental factors and autophagy form a direct link via the MTOR-dependent pathway.
Recent data also demonstrate that hepatitis C virus induces autophagy by inactivating the AKT-TSC-MTOR pathway based on ER stress;17 Sindbis virus induces autophagy by suppressing MTOR signaling,46 avian influenza viruses result in autophagic cell death by inhibiting MTOR,47 and vesicular stomatitis virus induces autophagy by regulating the PI3K-AKT signaling pathway.48 Despite that ICP34.5 and Us11 of HSV-1 antagonize autophagy based on the antiviral EIF2AK2/PKR (eukaryotic translation initiation factor 2-α kinase 2)-EIF2S1 (eukaryotic translation initiation factor 2, subunit 1 α, 35kDa) kinase signaling pathway,2,14 it is worthwhile to note MTOR also plays an important role in regulating autophagy in response to virus infection. Although the central role of autophagy in innate immune control of viruses,19 the connection between pathogen recognition and MTOR-dependent autophagy in these reports were poorly understood. In the present report, we discovered that not only HSP90AA1 knockdown, but also AKT1 or MTOR knockdown inactivated the induction of autophagy in IBDV-infected and VP2-treated cells (Fig. 7), revealing that HSP90AA1 regulates a unique AKT-MTOR molecular cascade pathway in the early stage of avibirnavirus infection to activate autophagic machinery. Our research chemically demonstrates for the first time the connection between HSP90AA1 and the AKT-MTOR-dependent signaling pathway in virus invasion (Fig. 8). In other words, IBDV invasion initiated the HSP90AA1-AKT-MTOR signaling pathway to induce autophagy.
In summary, both avibirnavirus VP2 and anti-HSP90AA1 antibody are sufficient to induce autophagy solely through the AKT-MTOR pathway, but VP2 is insufficient to initiate induction of autophagy after HSP90AA1, AKT1, and MTOR knockdown. Our study has direct proof that functional autophagy was induced in response to avibirnavirus invasion through AKT-MTOR pathway inactivation mediated by HSP90AA1-VP2 complex. However, we note that avibirnavirus inhibited autophagy at the late stage of infection successfully. Revealing the mechanism of this process requires further detailed study. Our study raises the possibility that controlling the AKT-MTOR pathway might serve as a strategy for triggering the process of antiviral autophagy. This work also highlights the role of HSP90AA1 in linking pathogen recognition signaling to MTOR-dependent autophagy, which is crucial for controlling infection.
Materials and Methods
Cells and virus stock
DF-1 and 293T cells from ATCC were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, 11995) supplemented with 10% fetal bovine serum (Gibco-BRL Life Technologies, 10099-141). IBDV strain NB (1.0 × 107 TCID50/0.1 mL) was stored in our lab. 49 IBDV virions were produced on DF-1 cell monolayer and purified by cesium chloride density gradient centrifugation.50 IBDV heat inactivation was performed at 90°C for 10 min in a water bath and inoculated on DF-1 monolayer to detect the infectivity of inactivated virus.
Antibodies and reagents
Mouse anti-VP2 polyclonal antibodies and mouse anti-VP3 mAb were maintained in our lab. Rabbit anti-LC3B (2775), anti-p-AKT (Ser473) (4060), and anti-p-MTOR (Ser2448) (5536) mAbs were purchased from Cell Signaling Technology. Rabbit Anti-AKT1 (ab32505) and anti-MTOR (ab2732) antibodies were purchased from Abcam. Rabbit anti-HSP90AA1 mAb (3670-1), rabbit anti-SQSTM1 (3340-1), anti-MYC (R1208-1) and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 2251-1) antibody were purchased from Epitomics. Donkey anti-mouse Alexa Fluor 647 (A-31571) was obtained from Invitrogen. HSP90AA1 protein (1145-hnce) was purchased from Sino Biological Inc. Rapamycin (9904) and LY294002 (S1737) were purchased from Cell Signaling Technology and Beyotime Institute of Biotechnology, respectively. Insulin (91077C) and 17-AAG (A8476) was purchased from Sigma-Aldrich. Nickel–nitrilotriacetic acid (Ni-NTA) agarose beads (30210) were obtained from Qiagen. Na2HPO4 (10039-32-4), KH2PO4(7778-77-0), NaCl (7647-14-5 ) and KCl (7447-40-7) were purchased from Sinopharm. Tris base (0497-1Kg) and sodium dodecyl sulfate (SDS;0227-500g) were purchased from Amresco.
Virus infection
DF-1 or 293T cells were infected with IBDV or purified IBDV virions at a multiplicity of infection (MOI) of 10 or 0.01 according to experiment requirements. The cells were then incubated in complete DMEM at 37°C for different durations according to experiment requirements. HE-IBDV was added to the cells as required. For induction of autophagy or MTOR activation experiments, cells were pretreated with rapamycin, starvation, or insulin for 4 h prior to viral infection. Virus titer was determined on DF-1 cell monolayers as TCID50/0.1 mL by observing the cytopathic effect as previously described.51
Expression plasmids and shRNA constructs
The chicken LC3B gene (GenBank accession number: NM_001031461) was PCR-amplified from total cellular RNA of chicken DF-1 cell with gene-specific primers and cloned into the pEGFP-C3 vector (Clontech, 6082-1). IBDV genes VP2 was amplified from IBDV genomic cDNA and cloned into pCI-Neo (PN; Promega, E1841) and pET28a vectors (PE; Novagen, 69864-3) using specific primers. GST was amplified from the pGEX4T-1 plasmid and cloned into pET28a vector as a control. Table S1 lists the primers used in the PCR amplification.
BECN1, HSP90AA1, AKT1, and MTOR knockdown in DF-1 cells or BECN1 knockdown in 293T cells was performed using the vector-based shRNA approach.52 shRNA targeting sequences against BECN1, HSP90AA1, AKT1, and MTOR were designed using online design tools (http://rnaidesigner.lifetechnologies.com/rnaiexpress/design.do, Block-iT RNAi Designer; Invitrogen); we selected 3 short target sequences with the best scores for each gene, while human BECN1 was selected from the Sigma-Aldrich RNAi sequence database. Table S2 lists the oligonucleotides used. The shRNAs were cloned into the RNAi-Ready pSIREN-RetroQ vector (Clontech, 632455) containing EcoRI and BamHI sites .
Cell transfection with silencing shRNA
DF-1 or 293T cells grown to 80% to 90% confluence were transfected with shRNA constructs using Lipofectamine 2000 (Invitrogen, 11668-019). Two days later, GFP-positive cells were evaluated using repeated fluorescence-activated cell sorting until more than 90% of cells were GFP-positive. GFP-positive cells were then expanded in DMEM with 200 ng/μL G418 and stored in complete DMEM without G418 until used.
Transmission electron microscopy
Ultrathin samples from IBDV- or mock-infected DF-1 cells, and negatively staining samples from purified IBDV particles or VP2 subviral particles were processed as previously described.53,54 Then these samples were observed using a Hitachi H-9500 transmission electron microscope (Hitachi High-Technologies Corporation) at 80 kV.
Confocal microscopy
DF-1 cells were seeded in 35-mm glass-bottom dishes (Shengyou Biotechnology) and transfected with eGFP-LC3 or control plasmids. At 24 h post-transfection, cells were incubated with Earle balanced salt solution medium (EBSS; Gibco, 24010-043) and complete DMEM supplemented with 10% fetal bovine serum, respectively, then mock-infected or infected with IBDV for the indicated duration. Cells were then incubated with 50 nM LysoTracker Red DND-99 (Invitrogen, L-7528) for 30 min, rinsed 3 times with phosphate-buffered saline (PBS; 8.4 mM Na2HPO4, 1.5 mM KH2PO4, 136.9 mM NaCl, 2.7 mM KCl) and immediately analyzed under a Zeiss LSM 510 laser confocal microscope (Zeiss). Only cells with at least 5 GFP dots or ring-like structures were scored as positive.55 The ratios of LysoTracker Red-positive ring-like structures were calculated from 20 cells infected with IBDV, HE-IBDV or mock-infection for 2 or 4 h.
Western blotting
Cells were harvested and lysed immediately in lysis buffer (2% SDS, 1% Triton X-100 [Sigma, 9002-93-1], 50 mM Tris-HCl and 150 mM NaCl, pH 7.5). Lysate protein concentration was measured by bicinchoninic acid (BCA) assay (Pierce, #23225). Equal amounts of total protein from different samples were separated on SDS–polyacrylamide gel electrophoresis and the protein bands were transferred onto polyvinylidene fluoride membranes (Milipore, ISEQ00010). After blocking with 5% nonfat dry milk containing 0.1% Tween 20 (Sigma, 9005-64-5) for 1 h at 37°C, the membranes were incubated with primary antibody for 2 h at 37°C, followed by horseradish peroxidase–conjugated anti-mouse/rabbit IgG (Kirkegaard & Perry Laboratories, 074-1806/074-1506) and visualized using a SuperSignal West Femto Substrate Trial Kit (Thermo Scientific/Pierce, 34096).
His-VP2 and His-VP3 expression and purification
GST and IBDV VP2 and VP3 genes were cloned into the PET-28a vector. His-GST, His-VP2, and His-VP3 were expressed in E. coli BL-21 and purified using Ni-NTA resin. The purified proteins were eluted with elution buffer containing 8 M urea, which was removed using a dialysis bag against buffer (50 mM NaH2PO4, 300 mM NaCl) with gradually reduced concentrations of urea (6 M, 3 M, buffer alone). The subviral particle of purified His-VP2 protein was detected by transmission electron microscope as previously stated.53,54 The concentrations of the purified proteins were measured with the BCA assay. The 3 purified proteins were then detected using western blotting with the corresponding antibody.
HSP90AA1 receptor stimulation
The experiment was performed as previously stated with some modifications.26 Briefly, cell culture plates were precoated with 20 μg/mL anti-HSP90AA1 mAb (Anti-HSP90) or rabbit IgG in PBS for 3 h at 37°C. Following 2 washes with PBS, plates were seeded with DF-1 cells, which were cultured in DMEM with 0.5 nM/mL insulin for 4 h. Cells cultured without insulin were used as the control. The cells were harvested and lysed with lysis buffer (2% SDS, 1% Triton X-100, 50 mM Tris-HCl, 150 mM NaCl, pH 7.5), and western blotting was performed using anti-SQSTM1, anti-LC3, anti-AKT1, and anti-MTOR mAbs to monitor autophagy.
Statistical analysis
The statistical significance of differences between groups were analyzed by 2-way ANOVA using the SPSS software (version 17.0). A P value of < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from China Agriculture Research System (Grant No. CARS-41-K11), National Key Technology R & D Program of China (Grant No. 2015BAD12B01) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplemental Material
Supplemental data for this article can be accessed on the pub-lisher's website.
References
- 1. Puleston DJ, Simon AK. Autophagy in the immune system. Immunology 2014; 141:1-8; PMID:23991647; http://dx.doi.org/ 10.1111/imm.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007; 1:23-35; PMID:18005679; http://dx.doi.org/ 10.1016/j.chom.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 3. Talloczy Z, Virgin HWT, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2006; 2:24-9; PMID:16874088; http://dx.doi.org/ 10.4161/auto.2176 [DOI] [PubMed] [Google Scholar]
- 4. Smith JD, de Harven E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. III. Cytochemical localization of lysosomal enzymes in infected cells. J Virol 1978; 26:102-9; PMID:206717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27:1110-21; PMID:18337753; http://dx.doi.org/ 10.1038/emboj.2008.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem 2008; 283:33175-82; PMID:18772134; http://dx.doi.org/ 10.1074/jbc.M804478200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. . Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol 2013; 14:480-8; PMID:23525089; http://dx.doi.org/ 10.1038/ni.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang M, Covar J, Zhang NY, Chen W, Marshall B, Mo J, Atherton SS. Virus spread and immune response following anterior chamber inoculation of HSV-1 lacking the Beclin-binding domain (BBD). J Neuroimmunol 2013; 260:82-91; PMID:23611643; http://dx.doi.org/ 10.1016/j.jneuroim.2013.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. . Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264-8; PMID:18849965; http://dx.doi.org/ 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- 10. Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005; 307:593-6; PMID:15591165; http://dx.doi.org/ 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- 11. Shin JN, Eissa NT. Autophagy gets the 'NOD' to enhance bacterial handling and antigen presentation. Immunol Cell Biol 2010; 88:343-5; PMID:20157329; http://dx.doi.org/ 10.1038/icb.2010.19 [DOI] [PubMed] [Google Scholar]
- 12. English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, et al. . Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480-7; PMID:19305394; http://dx.doi.org/ 10.1038/ni.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, et al. . Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009; 6:367-80; PMID:19837376; http://dx.doi.org/ 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol 2013; 87:859-71; PMID:23115300; http://dx.doi.org/ 10.1128/JVI.01158-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al. . Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 2013; 10; PMID:24401568; http://dx.doi.org/ 10.4161/auto.27286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alonso C, Galindo I, Cuesta-Geijo MA, Cabezas M, Hernaez B, Munoz-Moreno R. African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res 2013; 173:42-57; PMID:23262167; http://dx.doi.org/ 10.1016/j.virusres.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H, Kang R, Wang J, Luo G, Yang W, Zhao Z. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy 2013; 9:175-95; PMID:23169238; [http://dx.doi.org/ 10.4161/auto.22791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463-77; PMID:15068787; http://dx.doi.org/ 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 19. Richetta C, Faure M. Autophagy in antiviral innate immunity. Cell Microbiol 2013; 15:368-76; PMID:23051682; http://dx.doi.org/ 10.1111/cmi.12043 [DOI] [PubMed] [Google Scholar]
- 20. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol 2009; 19:359-78; PMID:19750559; http://dx.doi.org/ 10.1002/rmv.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin DM, Yuk JM, Lee HM, Lee SH, Son JW, Harding CV, Kim JM, Modlin RL, Jo EK. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol 2010; 12:1648-65; PMID:20560977; http://dx.doi.org/ 10.1111/j.1462-5822.2010.01497.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007; 27:135-44; PMID:17658277; http://dx.doi.org/ 10.1016/j.immuni.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 2012; 36:658-67; PMID:22464169; http://dx.doi.org/ 10.1016/j.immuni.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oh JE, Lee HK. Modulation of pathogen recognition by autophagy. Front Immunol 2012; 3:44; PMID:22566926; http://dx.doi.org/ 10.3389/fimmu.2012.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al. . Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 2009; 6:354-66; PMID:19837375; http://dx.doi.org/ 10.1016/j.chom.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 27. Triantafilou K, Triantafilou M, Dedrick RL. A CD14-independent LPS receptor cluster. Nat Immunol 2001; 2:338-45; PMID:11276205; http://dx.doi.org/ 10.1038/86342 [DOI] [PubMed] [Google Scholar]
- 28. Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol 2005; 79:4557-67; PMID:15795242; http://dx.doi.org/ 10.1128/JVI.79.8.4557-4567.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin TW, Lo CW, Lai SY, Fan RJ, Lo CJ, Chou YM, Thiruvengadam R, Wang AH, Wang MY. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J Virol 2007; 81:8730-41; PMID:17522206; http://dx.doi.org/ 10.1128/JVI.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eskelinen EL, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 2011; 7:935-56; PMID:21566462; http://dx.doi.org/ 10.4161/auto.7.9.15760 [DOI] [PubMed] [Google Scholar]
- 32. Kaufer I, Weiss E. Electron-microscope studies on the pathogenesis of infectious bursal disease after intrabursal application of the causal virus. Avian Dis 1976; 20:483-95; PMID:183649; http://dx.doi.org/ 10.2307/1589381 [DOI] [PubMed] [Google Scholar]
- 33. Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy 2009; 5:713-6; PMID:19372752; http://dx.doi.org/ 10.4161/auto.5.5.8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chevalier C, Lepault J, Erk I, Da Costa B, Delmas B. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J Virol 2002; 76:2384-92; PMID:11836416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maraver A, Ona A, Abaitua F, Gonzalez D, Clemente R, Ruiz-Diaz JA, Castón JR, Pazos F, Rodriguez JF. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J Virol 2003; 77:6438-49; PMID:12743301; http://dx.doi.org/ 10.1128/JVI.77.11.6438-6449.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A 2000; 97:10832-7; PMID:10995457; http://dx.doi.org/ 10.1073/pnas.170276797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. . Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 2009; 186:255-68; PMID:19635843; http://dx.doi.org/ 10.1083/jcb.200903070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orvedahl A, MacPherson S, Sumpter R, Jr., Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 2010; 7:115-27; PMID:20159618; http://dx.doi.org/ 10.1016/j.chom.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. Int Immunol 2010; 22:143-8; PMID:20089584; http://dx.doi.org/ 10.1093/intimm/dxp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Letzel T, Coulibaly F, Rey FA, Delmas B, Jagt E, van Loon AA, Mundt E. Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J Virol 2007; 81:12827-35; PMID:17881448; http://dx.doi.org/ 10.1128/JVI.01501-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/ 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009; 122:3589-94; PMID:19812304; http://dx.doi.org/ 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cobbold SP. The mTOR pathway and integrating immune regulation. Immunology 2013; 140:391-8; PMID:23952610; http://dx.doi.org/ 10.1111/imm.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. RQin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 2010; 6:239-47; PMID:20104019; http://dx.doi.org/ 10.4161/auto.6.2.11062 [DOI] [PubMed] [Google Scholar]
- 45. Stephan JS, Herman PK. The regulation of autophagy in eukaryotic cells: do all roads pass through Atg1? Autophagy 2006; 2:146-8; PMID:16874100; http://dx.doi.org/ 10.4161/auto.2.2.2485 [DOI] [PubMed] [Google Scholar]
- 46. Mohankumar V, Dhanushkodi NR, Raju R. Sindbis virus replication, is insensitive to rapamycin and torin1, and suppresses Akt/mTOR pathway late during infection in HEK cells. Biochem Biophys Res Commun 2011; 406:262-7; PMID:21316343; http://dx.doi.org/ 10.1016/j.bbrc.2011.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma J, Sun Q, Mi R, Zhang H. Avian influenza A virus H5N1 causes autophagy-mediated cell death through suppression of mTOR signaling. J Genet Genomics 2011; 38:533-7; PMID:22133684; http://dx.doi.org/ 10.1016/j.jgg.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 48. Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009; 30:588-98; PMID:19362021; http://dx.doi.org/ 10.1016/j.immuni.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng X, Hong L, Li Y, Guo J, Zhang G, Zhou J. In vitro expression and monoclonal antibody of RNA-dependent RNA polymerase for infectious bursal disease virus. DNA Cell Biol 2006; 25:646-53; PMID:17132096; http://dx.doi.org/ 10.1089/dna.2006.25.646 [DOI] [PubMed] [Google Scholar]
- 50. Muller H, Becht H. Biosynthesis of virus-specific proteins in cells infected with infectious bursal disease virus and their significance as structural elements for infectious virus and incomplete particles. Journal of virology 1982; 44:384-92; PMID:6292499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei L, Hou L, Zhu S, Wang J, Zhou J, Liu J. Infectious bursal disease virus activates the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway by interaction of VP5 protein with the p85alpha subunit of PI3K. Virology 2011; 417:211-20; PMID:21723579; http://dx.doi.org/ 10.1016/j.virol.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 52. Wang XJ, Li Y, Huang H, Zhang XJ, Xie PW, Hu W, Li DD, Wang SQ. A simple and robust vector-based shRNA expression system used for RNA interference. PLoS One 2013; 8:e56110; PMID:23405258; http://dx.doi.org/ 10.1371/journal.pone.0056110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee CC, Ko TP, Lee MS, Chou CC, Lai SY, Wang AH, Wang MY. Purification, crystallization and preliminary X-ray analysis of immunogenic virus-like particles formed by infectious bursal disease virus (IBDV) structural protein VP2. Acta Crystallogr Section D, Biol Crystallogr 2003; 59:1234-7; PMID:12832770; http://dx.doi.org/ 10.1107/S090744490300859X [DOI] [PubMed] [Google Scholar]
- 54. Chen CS, Suen SY, Lai SY, Chang GR, Lu TC, Lee MS, Wang MY. Purification of capsid-like particles of infectious bursal disease virus (IBDV) VP2 expressed in E. coli with a metal-ion affinity membrane system. J Virol Methods 2005; 130:51-8; PMID:16040134; http://dx.doi.org/ 10.1016/j.jviromet.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 55. Zhu B, Xu F, Li J, Shuai J, Li X, Fang W. Porcine circovirus type 2 explores the autophagic machinery for replication in PK-15 cells. Virus Res 2012; 163:476-85; PMID:22134092; http://dx.doi.org/ 10.1016/j.virusres.2011.11.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.