Abstract
Autophagy is a catabolic lysosomal degradation process essential for cellular homeostasis and cell survival. Dysfunctional autophagy has been associated with a wide range of human diseases, e.g., cancer and neurodegenerative diseases. A large number of small molecules that modulate autophagy have been widely used to dissect this process and some of them, e.g., chloroquine (CQ), might be ultimately applied to treat a variety of autophagy-associated human diseases. Here we found that vacuolin-1 potently and reversibly inhibited the fusion between autophagosomes and lysosomes in mammalian cells, thereby inducing the accumulation of autophagosomes. Interestingly, vacuolin-1 was less toxic but at least 10-fold more potent in inhibiting autophagy compared with CQ. Vacuolin-1 treatment also blocked the fusion between endosomes and lysosomes, resulting in a defect in general endosomal-lysosomal degradation. Treatment of cells with vacuolin-1 alkalinized lysosomal pH and decreased lysosomal Ca2+ content. Besides marginally inhibiting vacuolar ATPase activity, vacuolin-1 treatment markedly activated RAB5A GTPase activity. Expression of a dominant negative mutant of RAB5A or RAB5A knockdown significantly inhibited vacuolin-1-induced autophagosome-lysosome fusion blockage, whereas expression of a constitutive active form of RAB5A suppressed autophagosome-lysosome fusion. These data suggest that vacuolin-1 activates RAB5A to block autophagosome-lysosome fusion. Vacuolin-1 and its analogs present a novel class of drug that can potently and reversibly modulate autophagy.
Keywords: vacuolin-1, autophagosomes, lysosomes, RAB5A, pH, endosomes
Abbreviations: ATG, autophagy-related; BAF, bafilomycin A1; CQ, chloroquine; CTSB, cathepsin B; CTSL, cathepsin L; EGFR, epidermal growth factor receptor; GFP, green fluorescent protein; GPN, glycyl-l-phenylalanine 2-naphthylamide; LAMP1, lysosomal-associated membrane protein 1; Leup, leupeptin; MAP1LC3, microtubule-associated protein 1 light chain 3; MTOR, mechanistic target of rapamycin; RFP, red fluorescent protein; tfLC3, tandem fluorescence-tagged LC3
Introduction
Among 3 types of autophagy, including microautophagy, chaperone-mediated autophagy, and macroautophagy, in mammals, macroautophagy (hereafter referred as autophagy) is the most common type. Autophagy is an evolutionarily conserved catabolic degradation cellular process in which misfolded proteins or damaged organelles are first sequestered by a double-membrane vesicle, called an autophagosome. Autophagosomes then fuse with lysosomes to form autolysosomes, inside which the sequestered contents are digested by lysosomal enzymes and recycled to maintain cellular homeostasis. Autophagy can also be markedly induced by a wide variety of stresses, e.g., nutrient starvation, infection, and aging, for cell survival. Dysfunctional autophagy has been associated with wide ranges of human diseases, e.g., cancer and neurodegenerative diseases.1-6
Basal autophagy activity is essential for cell homeostasis, and it is tightly controlled by a complicated interplay among several key machineries, including ULK1 or ULK2 complexes and the class III phosphatidylinositol-3 kinase complexes. The MTOR (mechanistic target of rapamycin) Ser/Thr kinase suppresses autophagy by inhibiting the ULK1/2 complex. Starvation, on the other hand, activates the AMPK (AMP-activated protein kinase) to inactivate MTOR, thereby inducing autophagy. The lipidation of the MAP1LC3 (microtubule-associated protein 1 light chain 3) (herein referred simply as LC3) is essential for autophagy induction as well. The conjugation of phosphatidylethanolamine (PE) to LC3 is sequentially controlled by the protease autophagy-related (ATG) 4, E1-like enzyme ATG7, and the E2-like enzyme ATG3, thereby converting the cytosolic LC3 (LC3-I) to the autophagic vesicle-associated form (LC3-II). Notably, lipidated LC3-II shows a punctate staining pattern and has faster electrophoretic mobility compared with diffused LC3-I.1,7 Although enormous progress has been made on the core molecular machineries of autophagy, mechanisms of autophagy induction, autophagosome maturation, and autophagosome-lysosome fusion still remain elusive in mammalian cells.8
RAB5, a small GTPase, is essential for endolysosomal system biogenesis, and RAB5 knockdown markedly reduced the number of early endosomes, late endosomes, and lysosomes, accompanied with a blockage in endocytosis of low-density lipoprotein. Moreover, RAB5-to-RAB7 conversion, aided by the class C VPS HOPS complex, is required for early and late endosomes progression.9-13 In addition, RAB5 was found to be involved in autophagy by regulating PIK3C3/VPS34 activity likely via PIK3CB/p110beta, a catalytic subunit of the class IA phosphoinositide 3-kinase, in response to growth factor limitation.14 Yet the precise role of RAB5 in autophagy regulation remains to be determined.15,16
Not surprisingly, a large number of chemicals either promote or inhibit autophagy. Some of these compounds have been widely used to dissect the mechanisms underlying autophagy.17 Popular autophagy inducers include MTOR kinase inhibitors, e.g., rapamycin and torin 1,18 and chemicals inhibiting inositol monophosphatase, e.g., lithium and carbamazepine.19 Notably, rapamycin is an immunosuppressant and has recently been used as an anticancer agent.20 Lithium has been used to treat Huntington disease and other related neurodegenerative disorders.21 Commonly used autophagy inhibitors include chloroquine (CQ), 3-methyladenine, wortmannin, and bafilomycin A1 (BAF).17,22-24 Higher basal autophagic activity detected in established tumor cells is found to promote the survival and growth of tumors by maintaining energy production under increased metabolic consumption and a hypoxic microenvironment, thereby enabling tumors escape chemotherapy and/or radiation.25 Therefore, numerous preclinical studies have found that inhibition of autophagy by CQ restores chemosensitivity and promotes tumor cell death by diverse anticancer therapies.26 Recently, many new autophagy chemical modulators have been identified by image-based screens with GFP-LC3 transfected cells. Although these are useful pharmacological tools to study autophagy and are potential therapeutic drugs for autophagy-related diseases, many of these compounds lack specificity, or potency, or both.27 Therefore, the search for specific and potent autophagy chemical modulators must continue in order to gain deep insight of autophagy and provide potential therapeutic drugs.
Here we found that vacuolin-1 potently and reversibly inhibited the fusion between autophagosomes and lysosomes, thereby inducing the accumulation of autophagosomes. This effect is likely due to vacuolin-1-induced RAB5A activation, vacuolar ATPase activity inhibition, and the subsequent lysosomal pH alkalinization. Amazingly, vacuolin-1 was much less toxic but at least 10-fold more potent in inhibiting autophagy compared with CQ.
Results
Vacuolin-1 inhibited autophagosome-lysosome fusion in HeLa cells
Prompted by the fact that many available autophagy chemical modulators lack either potency or specificity,26 we set up a fluorescence image-based assay to screen molecules affecting autophagy. To do so, we infected HeLa cells, an autophagy competent cell line, with lentiviruses carrying expression cassettes that encode tandem fluorescence-tagged LC3B (tfLC3B).28 Thus, the LC3-II positive autophagosomes are labeled with both GFP and RFP signals shown as yellow puncta, and after fusion with lysosomes, autolysosomes are shown as red only puncta because GFP loses its fluorescence in acidic pH. As shown in Figure S1, starvation greatly induced the increase of both yellow and red only puncta, yet treatment of cells with BAF, an inhibitor of the vacuolar proton pump that blocks the fusion of autophagosomes with lysosomes,29 or CQ markedly induced the accumulation of yellow puncta only, indicating that the autophagy is arrested at autophagosomes. These data indicate that tfLC3B-expressing HeLa cells can be used to monitor the progression of autophagy.
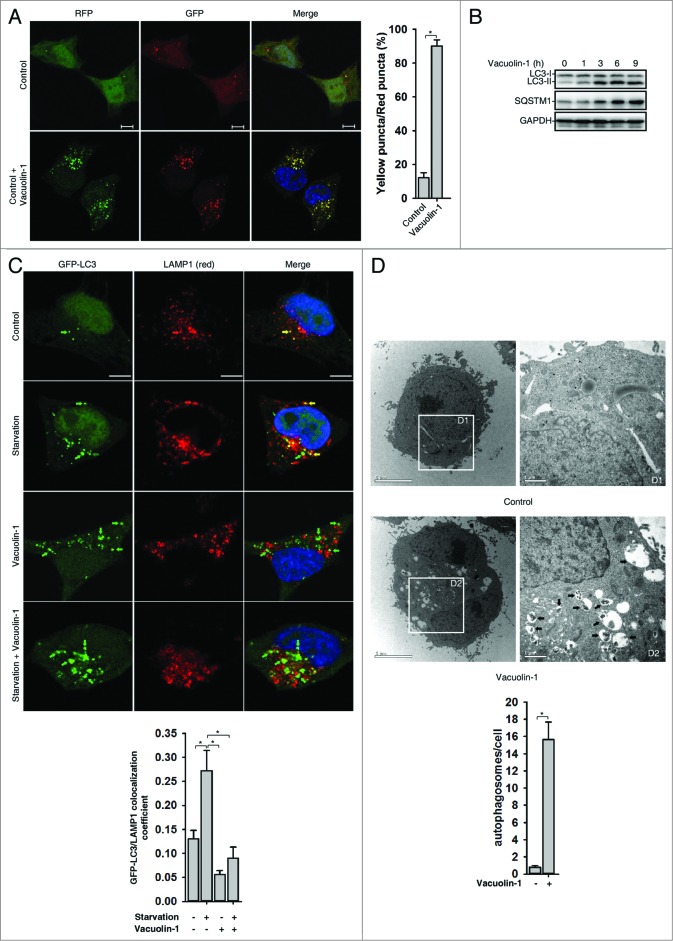
Next, we selected a panel of molecules that are commercially available and have been previously shown to affect vesicle trafficking or organelle morphology, and screened their effects on autophagy regulation in tfLC3B-expressing HeLa cells (data not shown). One molecule, vacuolin-1, potently induced LC3B yellow puncta, not red only puncta (Fig. 1A; Fig. S1). Western blot analyses further confirmed that lipidated LC3B-II was markedly increased in cells treated with vacuolin-1. SQSTM1/p62, an autophagic substrate,30 was also accumulated in cells treated with vacuolin-1, suggesting that vacuolin-1 inhibits the fusion between autophagosome and lysosomes (Fig. 1B). Indeed, GFP-LC3B puncta were greatly increased in vacuolin-1 treated cells and did not colocalize with lysosome-associated membrane protein 1 (LAMP1) (Fig. 1C), which was similar to the cells treated with BAF (Fig. S2). On the other hand, the majority of RFP-LC3B puncta induced by vacuolin-1 were colocalized with STX17-GFP, an autophagosome marker (Fig. S3).31,32 Similarly, vacuolin-1 induced the accumulation of both LC3B-II and SQSTM1 in the presence of autophagy inducer, starvation, or rapamycin (Fig. S4; Fig. 1C). To further exclude the possibility that accumulated LC3B-II or SQSTM1 is the result of inactivation of lysosomal proteases, we treated cells with leupeptin (leup), a protease inhibitor, in the presence or absence of vacuolin-1. As expected, vacuolin-1 treatment blocked the accumulation of autolysosomes induced by leup (Fig. S5), and cotreatment of cells with vacuolin-1 and leup only marginally increased the accumulation of LC3B-II and SQSTM1 compared with vacuolin-1 treatment alone (Fig. S6). Moreover, under the electron microscope, large numbers of autophagosomes were observed in vacuolin-1 treated HeLa cells maintained in normal culture conditions (Fig. 1D). Taken together, these data clearly demonstrated that vacuolin-1, similar to BAF or CQ, blocks the fusion between autophagosomes and autolysosomes, thereby leading to accumulation of LC3B-II positive autophagosomes.
Figure 1.
Vacuolin-1 inhibited the fusion between autophagosomes and lysosomes in HeLa cells. (A) Vacuolin-1 induced the accumulation of yellow LC3-II puncta in tfLC3B-expressing HeLa cells. Scale bar: 20 μM. Quantification of LC3 yellow puncta/red puncta (%) is expressed as mean ± S.E., n = ∼80 cells of 3 independent experiments. (B) Vacuolin-1 (1 μM) induced the accumulation of both LC3B-II and SQSTM1 in HeLa cells. (C) Vacuolin-1 (1 μM) markedly induced GFP-LC3B-II puncta in HeLa cells, which were not colocalized with RFP-LAMP1. Scale bar: 20 μM. The GFP-LC3B and LAMP1 colocalization coefficiency is expressed as mean ± S.E., n = ∼80 to 100 cells of 4 independent experiments. (D) Vacuolin-1 (1 μM) induced the accumulation of autophagic vacuoles as shown in the electron micrographs and highlighted in areas D1 and D2. Quantification of autophagosomes per cell are expressed as mean ± S.E., n = ∼20 to 40 cells. The *symbols indicate the results of the Student t test analysis, P < 0.05.
Vacuolin-1 potently and reversibly inhibited autophagy but showed little cell toxicity
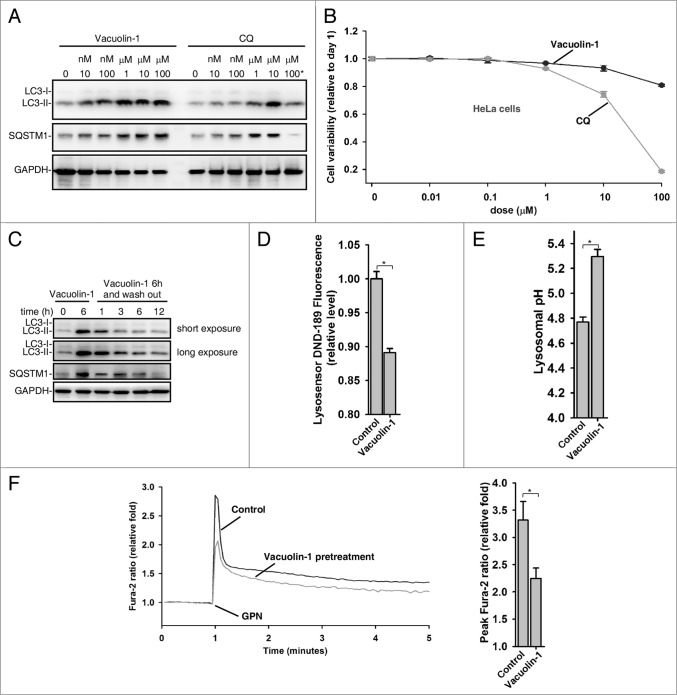
Vacuolin-1 is a cell-permeable and water-soluble triazine based compound. It has been previously reported that vacuolin-1 induced rapid homotypic fusion of endosomes and lysosomes to form large and swollen structures, yet it did not disturb cell cytoskeletal network.33-35 Amazingly, vacuolin-1 was at least 10 times more potent than CQ in suppressing autophagy (Fig. 2A; Fig. S7), yet it exhibited much less cell toxicity than CQ in a wide variety of cell types (Fig. 2B; Fig. S8). Interestingly, the fusion block between autophagosomes and lysosomes was completely relieved 3 h after removal of vacuolin-1 from the medium (Fig. 2C). Similarly, vacuolin-1 induced homotypic fusion between endosomes or lysosomes was recovered after removal of vacuolin-1 (data not shown). These data indicate that the effects of vacuolin-1 on autophagy inhibition or homortypic fusion are reversible.
Figure 2.
Vacuolin-1 reversibly inhibited autophagy by increasing lysosomal pH in HeLa cells. (A) Vacuolin-1 was a more potent autophagy inhibitor than CQ. Cells were treated with vacuolin-1 or CQ at the indicated dose for 6 h, and western blot analyses against LC3B and SQSTM1 were then performed. 100 μM CQ induced great cell loss is indicated by *. (B) Vacuolin-1 had much less cell toxicity than CQ in HeLa cells. Cells were treated with vacuolin-1 or CQ for the indicated doses for 48 h, and cell viability was then determined by MTT assay. (C) Vacuolin-1 (1 μM) reversibly inhibited autophagy. (D and E) Vacuolin-1 (1 μM) induced an increase of lysosomal pH in HeLa cells as determined by microplate reader measurement of Lysosensor DND-189 stained cells (D) or by a quantitative ratiometric LysoSensor Yellow/Blue DND-160 stained cells (E). (F) Vacuolin-1 (1 μM) pretreatment for 5 h markedly inhibited the ability of GPN (200 μM) to release lysosomal Ca2+ in Fura-2 loaded HeLa cells in the absence of extracellular Ca2+. The graphs in (B, D, E, or F) represent data from 3 independent experiments, and data are expressed as mean ± S.D., n = 3. The * symbols indicate the results of the Student t test analysis, P < 0.05.
Vacuolin-1 alkalinized lysosomal pH and decreased lysosomal Ca2+ content
We then explored the mechanisms underlying vacuolin-1-induced autophagy arrest. As lysosomal pH is essential for the fusion36 and lysosomes were enlarged by vacuolin-1 treatment,33-35 we examined whether vacuolin-1 affects lysosomal pH in these enlarged lysosomes. We first applied LysoSensor Green DND-189 (pKa = ∼5.2) to qualitatively measure lysosomal pH.37 LysoSensor Green DND-189 permeates cell membranes and accumulates in acidic intracellular organelles, and its fluorescence increases or decreases in acidic or alkaline environments, respectively. As shown in Figure 2D, vacuolin-1 treatment raised lysosomal pH in HeLa cells. We further quantified lysosomal pH using quantitative ratiometric LysoSensor Yellow/Blue DND-16038-stained cells, and found that lysosomal pH in vacuolin-1-treated HeLa cells was increased from pH 4.7 in control cells to pH 5.2 (Fig. 2E). Thus, it is clear that vacuolin-1 alkalinizes lysosomal pH.
Since lysosomes represent a major intracellular Ca2+ pool and Ca2+ and protons are tightly coupled in lysosomes,39 we also assessed whether vacuolin-1 treatment affects the lysosomal Ca2+ content. Treatment of cells with glycyl-l-phenylalanine 2-naphthylamide (GPN) selectively disrupts the lysosomal membrane,40 thereby releasing the lysosomal Ca2+.41 As shown in Figure 2F, pretreatment of cells with vacuolin-1 significantly lowered the ability of GPN to induce lysosomal Ca2+ release, suggesting that vacuolin-1 decreases lysosomal Ca2+ levels as well.
Vacuolin-1 inhibited general endosomal-lysosomal degradation in HeLa cells
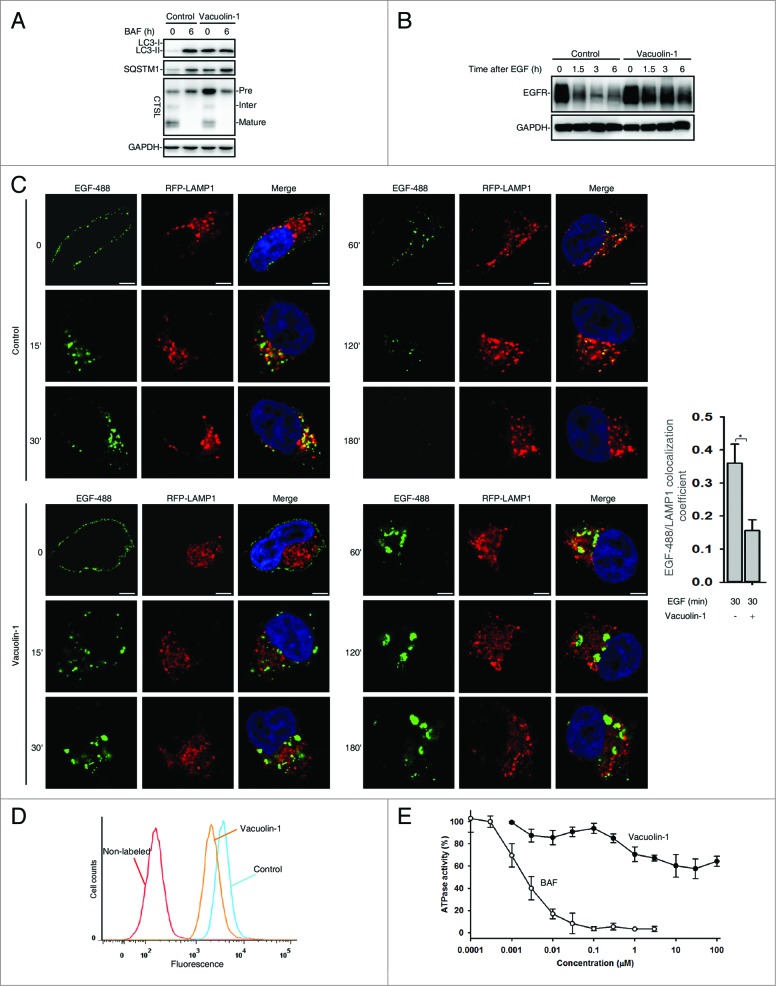
Since the increase of lysosomal pH normally compromises the lysosomal activity, we assessed whether vacuolin-1 affects the general lysosomal functions to inhibit autophagy maturation. The processing of CTSL (cathepsin L) from the precursor form to its mature form has been commonly used as a marker for lysosomal activity,42 and we found that the processing of CTSL into its mature form was not affected in vacuolin-1-treated HeLa cells, but vacuolin-1 did markedly increase the levels of immature form of CTSL (Fig. 3A). Similarly, treatment of cells with vacuolin-1 for 6h did not change the processing of CTSB (cathepsin B) (Fig. S9).42 These data indicate that the subtle increase of lysosomal pH induced by vacuolin-1 does not affect lysosomal protease activity.
Figure 3.
Vacuolin-1 inhibited general endosomal-lysosomal degradation in HeLa cells. (A) The processing of CTSL from the precursor form to its mature form in HeLa cells treated with or without vacuolin-1 (1 μM) or BAF (100 nM). (B) Vacuolin-1 (1 μM) inhibited EGF-induced EGFR degradation in HeLa cells. (C) Vacuolin-1 (1 μM) inhibited the fusion of EGFR endosomes with lysosomes in HeLa cells. Scale bar: 20 μM. The EGF-488/LAMP1 colocalization coefficiency is expressed as mean ± S.E., n = ∼40 cells of 2 independent experiments. (D) Vacuolin-1 (1 μM) inhibited the degradation of DQ-BSA-Green in HeLa cells. (E) Vacuolin-1 nonspecifically inhibited V-ATPase as assessed by the in vitro V-ATPase assay, whereas BAF markedly inhibited it. The activity of V-ATPase (1.05 ± 0.09 μmol/min/mg) at 0 μM of inhibitors is set to 100%. The data are expressed as mean ± S.D., n = 3. The * symbols indicate the results of the Student t test analysis, P < 0.05.
Next, an EGFR (epidermal growth factor receptor) degradation assay was performed to examine whether vacuolin-1 affects the general endosomal-lysosomal pathway. In this assay, HeLa cells were treated with EGF in the presence or absence of vacuolin-1. After EGF binds to its receptors, the receptor complex undergoes endocytosis and is targeted to lysosomes for degradation. As shown in Figure 3B, vacuolin-1 inhibited EGF-triggered EGFR degradation. Interestingly, EGFR was internalized normally but the endosomes containing EGFR failed to fuse with lysosomes; this explains the accumulation of EGFR in vacuolin-1 treated cells (Fig. 3C). Similarly, a DQ-BSA-green degradation assay was applied to measure the general endosomal-lysosomal degradation. DQ-BSA-green is a BSA labeled with a self-quenching fluorescent dye. After DQ-BSA-green is delivered to lysosomes via endocytosis, it is hydrolyzed into single dye-labeled peptides by lysosomal proteases, thereby relieving self-quenching and the fluorescence can subsequently be monitored by flow cytometry. As shown in Figure 3D, vacuolin-1 markedly inhibited the degradation of BSA by lysosomes, yet DQ-BSA-Green was present in endosomes but failed to be delivered to lysosomes (data not shown). Collectively, these results support that vacuolin-1 inhibits the general endosomal-lysosomal degradation, which is most likely because the alkalized lysosomal pH stops the fusion of endosomes with lysosomes.
We then assessed whether vacuolin-1 inhibits V-ATPase activity to increase lysosomal pH. Vacuolin-1 at concentration of 1 μM resulted in ∼30% inhibition of V-ATPase activity, yet higher concentration of vacuolin-1 failed to further inhibit it (Fig. 3E). Since hydrophobic substances at high concentrations sometimes have disturbing/inhibitory effects on the ATPase activity,43 the observed small inhibition of V-ATPase by high concentrations of vacuolin-1 is likely nonspecific. On the other hand, the ATPase activity dropped sharply from 100% to 0% within a concentration range of a factor of 100 for BAF, a potent V-ATPase inhibitor with IC50 = 3 nM (Fig. 3E). Taken together, these data suggest that vacuolin-1 is a weak V-ATPase inhibitor.
RAB5A was required for vacuolin-1-induced autophagy arrest and endosomal fusion
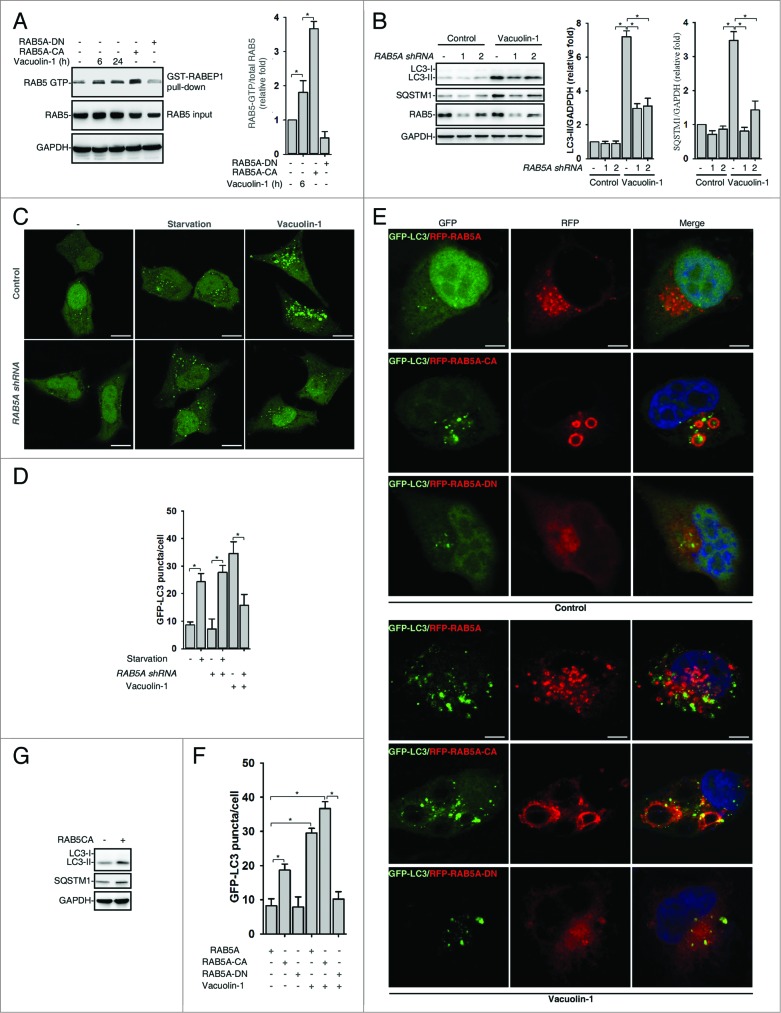
Vacuolin-1 also induced the homotypic fusion of endosomes or lysosomes, and RAB5, a small GTPase, is essential for endosome fusion.11 We thus examined whether vacuolin-1 activates RAB5 to regulate autophagy. Indeed, vacuolin-1 markedly activated RAB5, as shown by a GST-tagged RABEP1/RABaptin5 affinity isolation assay, which specifically interacts with RAB5-GTP, not RAB5-GDP (Fig. 4A).44 Consistently, RAB5A knockdown markedly decreased vacuolin-1-induced accumulation of both LC3B-II and SQSTM1 (Fig. 4B–D). Similarly, overexpression of a dominant-negative form of RAB5A (RAB5A-DN) blocked vacuolin-1-induced autophagy arrest and homotypic fusion, whereas overexpression of a constitutive active form of RAB5A (RAB5A-CA)45 augmented the ability of vacuolin-1 to induce autophagy arrest and homotypic fusion (Fig. 4E and F). Interestingly, RAB5A-CA overexpression alone also inhibited the autophagosome-lysosome fusion, thereby inducing the accumulation of LC3B-II and SQSTM1 (Fig. 4G; Figs. S10 and S13). Notably, RAB5A-DN expression or RAB5A knockdown did not affect starvation-induced autophagy (Fig. 4C and D; Figs. S11 and S12). Taken together, these data indicate that RAB5A is required for vacuolin-1-induced autophagy arrest.
Figure 4.
RAB5A is required for vacuolin-1-induced autophagy arrest and homotypic fusion in HeLa cells. (A) Vacuolin-1 (1 μM) activated RAB5A in HeLa cells. The active RAB5A in cells treated with vacuoin-1 or transfected with RAB5A-CA or RAB5A-DN were examined by a GST-tagged RABEP1 affinity isolation assay. Quantification of RAB5A-GTP/total RAB5A (relative fold) is expressed mean ± S.D., n = 3. (B) RAB5A knockdown blocked vacuolin-1-induced accumulation of LC3B-II and SQSTM1 in HeLa cells. Quantifications of LC3B-II/GAPDH and SQSTM1/GAPDH (relative fold) are expressed mean ± S.D., n = 4. (C) RAB5A knockdown blocked GFP-LC3 puncta induced by vacuolin-1 (1 μM) but not by starvation in HeLa cells. (D) Quantification of GFP-LC3B puncta/per cell in (C) is expressed as mean ± S.E., n = ∼50 to 60 cells of 3 independent experiments. The * symbols indicate the results of the Student t test analysis, P < 0.05. (E) Expression of RAB5A-CA enhanced vacuolin-1-induced autophagy arrest, whereas expression of RAB5A-DN blocked it in HeLa cells. (F) Data quantifications of GFP-LC3B puncta/per cell in (E) are expressed as mean ± S.E., n = ∼80 cells of 3 independent experiments. (G) Expression of RAB5A-CA alone induced the accumulation of LC3-II as assessed by western blot analyses (representative of 3 independent experiments).
Discussion
Here we found that vacuolin-1 potently and reversibly inhibited autophagosome-lysosome fusion, thereby accumulating autophagosomes (Fig. 1; Figs. S1–S6). Although vacuolin-1 is at least 10 times more potent than CQ in inhibiting autophagy (Fig. 2A; Fig. S7), vacuolin-1 showed much less cell toxicity than CQ (Fig. 2B; Fig. S8). Vacuolin-1 alkalized lysosomal pH (Fig. 2D and E) and decreased lysosomal Ca2+ content (Fig. 2F), but vacuolin-1 only weakly and nonspecifically inhibited V-ATPase (Fig. 3E). On the other hand, vacuolin-1 markedly activated RAB5 (Fig. 4A), and expression of RAB5A-DN or RAB5A knockdown significantly inhibited vacuolin-1-induced autophagosome accumulation (Fig. 4B–F). We speculate that RAB5A activation by vacuolin-1 indirectly contributes to the increase of lysosomal pH, thereby compromising normal lysosomal functions, including autophagasome-lysosome fusion and endosomal-lysosomal degradation.
Vacuolin-1 was originally found to induce homotypic fusion of endosomes or lysosomes, thereby forming large vacuoles. Yet, it does not alter other cell structures and membrane-trafficking functions.33-35 It remains controversial whether vacuolin-1 blocks the Ca2+-dependent exocytosis of lysosomes. One study suggests that the effects of vacuolin-1 on lysosomal exocytosis depend on the cell types and mode of activation.33 Interestingly, Cerny et al. have found that some enlarged vacuoles induced by vacuolin-1 contain amorphous content,35 which is conspicuously similar to the morphology of autophagosomes (Fig. 1D). Indeed, here we found that vacuolin-1 potently inhibited the fusion between autophagosomes and lysosomes, thereby accumulating autophagosomes (Fig. 1; Figs. S1–S6). Although vacuolin-1 did not stop genesis of early endosomes, vacuolin-1 also arrested the fusion of endosomes and lysosomes, thus stopping the general endosomal-lysosomal degradation (Fig. 3B–D). In summary, vacuolin-1 not only induces homotypic fusion to form enlarge vacuoles but also disrupts general lysosomal functions, e.g., autophagy and endocytosis.
RAB5 is required for biogenesis of endolysosomal system. Here, we found that vacuolin-1 markedly activated RAB5A, while inhibiting RAB5A activity reversed the effects of vacuolin-1 (Fig. 4A–F). How vacuolin-1 activates RAB5A remains elusive. RAB5 is a small monomeric GTPase. The conversion of GDP-bound RAB5 (inactive form) into the GTP-bound active form is catalyzed by a guanine nucleotide exchange factor (GEF). The RAB5-GTP form is subsequently recognized by multiple effectors for its function. The switch of RAB5-GTP back to the GDP-bound form is not only driven by the intrinsic GTPase activity of RAB5 but also stimulated by a GTPase-activating protein (GAP). Multiple RAB5 GEF and GAP have been identified.46 We speculate that vacuolin-1 could either directly inhibit RAB5 intrinsic GTPase activity, or activate a RAB5 GEF, or inhibit a RAB5 GAP, thus keeping RAB5 in a GTP-bound active state.46 We are currently examining these possibilities.
RAB5A activation is required for vacuolin-1-induced autophagosome-lysosome fusion blockage (Fig. 4B–F). However, overexpression of RAB5A-CA more potently activated RAB5A (Fig. 4A) but less efficiently inhibited autophagy compared with vacuolin-1 (Fig. 4E and F; Fig. S13), hinting that vacuolin-1 regulates other factor besides RAB5A to affect autophagy.
Indeed, vacuolin-1 treatment alkalized lysosomal pH (Fig. 2D and E) and alkalized pH generally leads to autophagy arrest.36 Notably, we have recently found that alkalized lysosomal pH by NAADP-TPCN2-Ca2+ signaling prevents the recruitment of RAB7A to autophagosomes, which might be one of the causes of the blockage of autophagosome-lysosome fusion.36 Likewise, few RFP-RAB7A puncta were colocalized with GFP-LC3B in vacuolin-1 treated cells, whereas rapamycin markedly induced the colocalization of RAB7A with GFP-LC3B in control cells (Fig. S14).
Regarding how vacuolin-1 alkalizes lysosomal pH, obviously, the nonspecific inhibition of V-ATPase by vacuolin-1 (Fig. 3D) could directly lead to the increase of lysosomal pH. In addition, endosomal traffic is important for the function and biogenesis of lysosome because lysosomes depend on the influx of new components. Without incoming endosomal traffic, lysosomes lose their intact morphology, contents of protons and other ions, and perinuclear localization.47,48 Endosome maturation requires a conversion from RAB5 to RAB7, and expressing a constitutively active mutant of RAB5A actually blocks endosome maturation.10 Thus, besides inhibiting V-ATPase, vacoulin-1 could activate RAB5A to stop endosome maturation, which subsequently compromises the biogenesis and function of lysosomes, e.g., pH and Ca2+ content. In addition, many enlarged lysosomes were found in vacuolin-1 treated cells (Fig. S15),35 which is likely due to RAB5A activation because expression of RAB5A-DN abolished it (data not shown). Therefore, it is reasonable to speculate that V-ATPases might not be able to pump enough protons into the enlarged lysosomes to maintain the acidic pH; if so, this should lead to increased lysosomal pH as well. Other than RAB5A and V-ATPase, whether vacuolin-1 regulates other factors to inhibit autophagosome-lysosome fusion remains to be determined as well.
RAB5 family has 3 isoforms, A, B, and C, which are more than 90% homologous.49 Here, we found that RAB5A knockdown or expression of RAB5A-DN markedly inhibited vacuolin-1-induced autophagy arrest (Fig. 4B–F), but had little effect on starvation-induced autophagy (Figs. S11 and S12). Consistently, expression of RAB5A-CA also arrested autophagic flux (Fig. 4E–G; Fig. S13). These data indicate that RAB5A is not required for autophagy induction. Notably, several studies report that RAB5 is required for autophagy induction.15,16 The study by Dou et al. also suggests that RAB5 is only involved in growth factor starvation, not glucose or amino acid limitation, induced autophagy.14 It is possible that RAB5 isoforms function differently in distinct cellular processes, including autophagy.50-53 Further investigations are needed to resolve the controversy.
Materials and Methods
Antibodies and reagents
The antibodies used were as follows: LC3B (Novus, NB100-2220), SQSTM1 (Novus, NBP1-48320), CTSL (BD Bioscience, 611084), CTSB (Santa Cruz Biotechnology, SC-13985), EGFR (Santa Cruz Biotechnology, SC-03), RAB5A (Cell Signaling Technology, 3547), RAB7 (Cell Signaling Technology, 2094), LAMP1 (Cell Signaling Technology, 9091), GAPDH antibodys (Sigma, G8795). Vacuolin-1 (Santa Cruz Biotechnology, SC-216045), glycyl-L-phenyl-alanine-β-naphthylamide (GPN, Santa Cruz Biotechnology, SC-252858), BAF (Sigma-Aldrich, B1793), CQ (Sigma-Aldrich, C6628), rapamycin (Sigma-Aldrich, R8781), Leup (Sigma-Aldrich, L2884), Fura-2 AM (Invitrogen, F1221), LysoSensor Green DND-189 (Invitrogen, L7535), LysoSensor Yellow/Blue DND-160 (Invitrogen, L7545), DQ green-BSA (Invitrogen, D12050) and Alexa Fluor 488-EGF (Invitrogen, E13345).
Cell culture
HeLa, MCF-7, A549, and HepG2 cells (ATCC) were maintained in DMEM (Invitrogen, 12800-017) plus 10% fetal bovine serum (Invitrogen, 10270-106) and 100 units/ml of penicillin/streptomycin (Invitrogen, 15140-122) at 5% CO2 and 37°C. PC3 cells were maintained in RPMI (Invitrogen, 31800-022) plus 10% fetal bovine serum (Invitrogen, 10270-106) and 100 units/ml of penicillin/streptomycin (Invitrogen, 15140-122) at 5% CO2 and 37°C.
shRNA and lentivirus production and infection
Two optimal 21-mers were selected in the human RAB5A gene (Table S1). One 21-mer was selected in the GFP nucleotide sequence as a control. These sequences were then cloned into the pLKO.1 vector for expressing shRNA. The shRNA lentivirus production and infection were performed as described previously.36
Intracellular Ca2+ measurement
Cells were cultured in 24-well plates at the density of 7 × 104 cells/well in regular medium overnight and were labeled with 4 μM Fura-2 AM in regular HBSS (Invitrogen, 14175-059) at room temperature for 30 min. The cells were then washed with calcium-free HBSS containing 2 mM EGTA (Sigma, 3889) 3 times and incubated in the presence or absence of vacuolin-1 at room temperature for another 10 min. Cells were put on the stage of an Olympus inverted epifluorescence microscope (Telefon, Germany) and visualized using a 20× objective. Fluorescence images were obtained by alternate excitation at 340 nm and 380 nm with emission set at 510 nm. Images were collected by a CCD camera every 3 or 6 s and analyzed by Cell R imaging software.
Western blot and immunofluorescence staining analyses
Both assays were performed as described previously.36 The colocalization coefficients of images were quantified by the Ziess LSM 710 Software, a quantitative colocalization analysis of multichannel images, as following: (a) correct background (background correction settings are constant for all images), (b) select region of interest (ROI), (c) generate histogram of ROI, and (d) obtain the table of results, including colocalized area, the degree of colocalization, and colocalization coefficient.
Cell preparation for transmission electron microscopy (TEM)
Cell preparation for TEM was performed as described previously.54
MTT cell proliferation assay
Cells were treated in 4 replicates and seeded into 96-well plates. Following drug treatment, MTT solution (USB Corporation, 19265) of 20 μl for every 100 μl medium was added to wells and incubated for 2 h, followed by the addition of 150 μL of the DMSO solution to each well. The final reaction product, a purple formazan solution, was detected by a microplate reader (Techan infinite M200, Männedorf, Switzerland) for absorbance at a wavelength of 570 nm and a reference wavelength of 630 nm.
Lysosomal pH measurement
LysoSensor Green DND-189 is commonly used to measure the pH of acidic organelles, such as lysosomes, which become more fluorescent in acidic environments. Briefly, cells were loaded with 1 μM LysoSensor Green DND-189 in prewarmed regular medium for 20 min at 37°C. Then the cells were washed twice with PBS (Invitrogen, 10010-023) and immediately analyzed by flow cytometry (collecting FL1 fluorescence and 10,000 cells were collected for each sample) or in a microplate reader (excitation/emission = 485/530 nm).
Quantification of lysosomal pH was performed using a ratiometric lysosomal pH dye, LysoSensor Yellow/Blue DND-160. The pH calibration curve was generated as described previously.55 Briefly, cells were trypsinized and labeled with 2 μM Lysosensor Yellow/Blue DND-160 for 30 min at 37°C in regular medium, and excessive dye was washed out using PBS. The labeled cells were treated for 10 min with 10 μM monensin (Sigma, M5273) and 10 M nigericin (Sigma, N7143) in 25 mM 2-(N-morpholino) ethanesulfonic acid (MES) calibration buffer, pH 3.5–6.0, containing 5 mM NaCl, 115 mM KCl and 1.2 mM MgSO4. Quantitative comparisons were performed in a 96-well plate, and the fluorescence was measured with a microplate reader at 37°C. Light emitted at 440 and 535 nm in response to excitation at 340 and 380 nm were measured, respectively. The ratio of light emitted with 340 and 380 nm excitation was plotted against the pH values in MES buffer, and the pH calibration curve for the fluorescence probe was generated from the plot using Microsoft Excel.
V-ATPase assay
Fifth instar larvae of M. sexta (Lepidoptera, Sphingidae), weighing 6 to 8 g, were reared under long-day conditions (16 h of light) at 27°C using the gypsy moth diet (MP Biomedicals, 02960292). The purification of the V1VO holoenzyme was performed as previously described.56 Activity assays were performed in triplicate in a final volume of 160 μL and at a pH of 8.1. Assays contained 3 μg of purified V1V0 holoenzyme, 50 mM Tris-MOPS, 3 mM 2-mercaptoethanol, 1 mM MgCl2, 20 mM KCl, 0.003% C12E10, 20 mM NaCl, and 3 mM TRIS-HCl. After 10 min of preincubation at 30°C, with or without inhibitors, 1 mM Tris-ATP was added and after an incubation for 2 min at 30°C the reaction was stopped by freezing the samples in liquid nitrogen. Inorganic phosphate was determined as previously described.57
Glutathione S-Transferase (GST) Affinity Isolation Assay
Cells were collected and lyzed in an ice-cold EBC lysis buffer described above. Lysates were clarified by centrifugation at 13,000 g for 10 min at 4°C, and equal amount of protein (500 μg) from each supernatant fraction was incubated with 30 μl of GST-R5BD bound to the 30 μl glutathione-Sepharose beads (GE Life Sciences, 17-0756-01) for 1 h at 4°C on a rotating mixer. The beads were subsequently washed and resuspended in the standard SDS-sample buffer, boiled, and subjected to SDS-PAGE (15% gel), followed by immunoblot analysis with the anti-RAB5 monoclonal antibody.
Statistical analysis
Data were presented as mean ± s.e.m. the statistical significance of differences was estimated by one-way anova or the Student t test. P < 0.05 was considered significant.
Acknowledgments
We thank Prof Guangpu Li for GST-tagged RABEP1/RABaptin5, and Richard Graeff and members of the Yue lab for advice on the manuscript. FACS analysis and Confocal imaging were performed in the Faculty of Medicine Core Facility at the University of Hong Kong.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Research Grant Council (RGC) grants (782709M, 785911M, 769912M, 785213M, and 17126614M) to JY and the Deutsche Forschungsgemeinschaft (SFB 944) to HM.
Supplemental Material
Supplemental data for the article can be accessed on the publisher's website.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/ 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nature reviews. Clin Oncol 2011; 8:528-39. [DOI] [PubMed] [Google Scholar]
- 3.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12:401-10; PMID:22534666; http://dx.doi.org/ 10.1038/nrc3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy 2005; 1:1-10; PMID:16874026; http://dx.doi.org/ 10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- 5.Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5:886-97; PMID:16239905; http://dx.doi.org/ 10.1038/nrc1738 [DOI] [PubMed] [Google Scholar]
- 6.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. . Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/ 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- 7.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741-52; PMID:17717517; http://dx.doi.org/ 10.1038/nrm2239 [DOI] [PubMed] [Google Scholar]
- 8.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem 2012; 81:637-59; PMID:22463690; http://dx.doi.org/ 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005; 122:735-49; PMID:16143105; http://dx.doi.org/ 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- 11.Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, et al. . Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 2012; 485:465-70; PMID:22622570; http://dx.doi.org/ 10.1038/nature11133 [DOI] [PubMed] [Google Scholar]
- 12.Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci 1999; 112:3667-75; PMID:10523503 [DOI] [PubMed] [Google Scholar]
- 13.Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649-60; PMID:18430781; http://dx.doi.org/ 10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, et al. . Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 2013; 50:29-42; PMID:23434372; http://dx.doi.org/ 10.1016/j.molcel.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 2011; 85:10561-71; PMID:21835792; http://dx.doi.org/ 10.1128/JVI.00173-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chua CE, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 2011; 68:3349-58; PMID:21687989; http://dx.doi.org/ 10.1007/s00018-011-0748-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek KH, Park J, Shin I. Autophagy-regulating small molecules and their therapeutic applications. Chem Soc Rev 2012; 41:3245-63; PMID:22293658; http://dx.doi.org/ 10.1039/c2cs15328a [DOI] [PubMed] [Google Scholar]
- 18.Hanson KK, Ressurreição AS, Buchholz K, Prudêncio M, Herman-Ornelas JD, Rebelo M, Beatty WL, Wirth DF, Hänscheid T, Moreira R, et al. . Torins are potent antimalarials that block replenishment of Plasmodium liver stage parasitophorous vacuole membrane proteins. Proc Natl Acad Sci U S A 2013; 110:E2838-47; PMID:23836641; http://dx.doi.org/ 10.1073/pnas.1306097110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. . An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science 2010; 329:229-32; PMID:20522742; http://dx.doi.org/ 10.1126/science.1190354 [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, et al. . Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 2004; 36:585-95; PMID:15146184; http://dx.doi.org/ 10.1038/ng1362 [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 2005; 170:1101-11; PMID:16186256; http://dx.doi.org/ 10.1083/jcb.200504035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 1982; 79:1889-92; PMID:6952238; http://dx.doi.org/ 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rote KV, Rechsteiner M. Degradation of microinjected proteins: effects of lysosomotropic agents and inhibitors of autophagy. J Cell Physiol 1983; 116:103-10; PMID:6853609; http://dx.doi.org/ 10.1002/jcp.1041160116 [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Wang X, Guo H, Zhang B, Zhang XB, Shi ZJ, Yu L. Synthesis and screening of 3-MA derivatives for autophagy inhibitors. Autophagy 2013; 9:595-603; PMID:23412639; http://dx.doi.org/ 10.4161/auto.23641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10:1533-41; PMID:21878654; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 2013; 73:3-7; PMID:23288916; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2464 [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709-30; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; PMID:17534139; http://dx.doi.org/ 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23:33-42; PMID:9639028; http://dx.doi.org/ 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- 30.Bjørkøy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2006; 2:138-9; PMID:16874037 [DOI] [PubMed] [Google Scholar]
- 31.Hegedűs K, Takáts S, Kovács AL, Juhász G. Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes. Autophagy 2013; 9:1642-6; PMID:24113031; http://dx.doi.org/ 10.4161/auto.25684 [DOI] [PubMed] [Google Scholar]
- 32.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256-69; PMID:23217709; http://dx.doi.org/ 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 33.Shaik GM, Dráberová L, Heneberg P, Dráber P. Vacuolin-1-modulated exocytosis and cell resealing in mast cells. Cell Signal 2009; 21:1337-45; PMID:19376224; http://dx.doi.org/ 10.1016/j.cellsig.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 34.Huynh C, Andrews NW. The small chemical vacuolin-1 alters the morphology of lysosomes without inhibiting Ca2+-regulated exocytosis. EMBO Rep 2005; 6:843-7; PMID:16113649; http://dx.doi.org/ 10.1038/sj.embor.7400495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerny J, Feng Y, Yu A, Miyake K, Borgonovo B, Klumperman J, Meldolesi J, McNeil PL, Kirchhausen T. The small chemical vacuolin-1 inhibits Ca(2+)-dependent lysosomal exocytosis but not cell resealing. EMBO Rep 2004; 5:883-8; PMID:15332114; http://dx.doi.org/ 10.1038/sj.embor.7400243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Hao BX, Graeff R, Wong CW, Wu WT, Yue J. Two pore channel 2 (TPC2) inhibits autophagosomal-lysosomal fusion by alkalinizing lysosomal pH. J Biol Chem 2013; 288:24247-63; PMID:23836916; http://dx.doi.org/ 10.1074/jbc.M113.484253 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Davis-Kaplan SR, Ward DM, Shiflett SL, Kaplan J. Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification. J Biol Chem 2004; 279:4322-9; PMID:14594803; http://dx.doi.org/ 10.1074/jbc.M310680200 [DOI] [PubMed] [Google Scholar]
- 38.DePedro HM, Urayama P. Using LysoSensor Yellow/Blue DND-160 to sense acidic pH under high hydrostatic pressures. Anal Biochem 2009; 384:359-61; PMID:18976626; http://dx.doi.org/ 10.1016/j.ab.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 39.Morgan AJ, Platt FM, Lloyd-Evans E, Galione A. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem J 2011; 439:349-74; PMID:21992097; http://dx.doi.org/ 10.1042/BJ20110949 [DOI] [PubMed] [Google Scholar]
- 40.Jadot M, Andrianaivo F, Dubois F, Wattiaux R. Effects of methylcyclodextrin on lysosomes. Eur J Biochem 2001; 268:1392-9; PMID:11231291; http://dx.doi.org/ 10.1046/j.1432-1327.2001.02006.x [DOI] [PubMed] [Google Scholar]
- 41.Srinivas SP, Ong A, Goon L, Goon L, Bonanno JA. Lysosomal Ca(2+) stores in bovine corneal endothelium. Invest Ophthalmol Vis Sci 2002; 43:2341-50; PMID:12091436 [PubMed] [Google Scholar]
- 42.Katunuma N. Posttranslational processing and modification of cathepsins and cystatins. J Signal Transduct 2010; 2010:375345; PMID:21637353; http://dx.doi.org/ 10.1155/2010/375345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huss M, Wieczorek H. Inhibitors of V-ATPases: old and new players. J Exp Biol 2009; 212:341-6; PMID:19151208; http://dx.doi.org/ 10.1242/jeb.024067 [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Lamb D, Chou MM, Liu YJ, Li G. Nerve growth factor-mediated neurite outgrowth via regulation of Rab5. Mol Biol Cell 2007; 18:1375-84; PMID:17267689; http://dx.doi.org/ 10.1091/mbc.E06-08-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohdanowicz M, Balkin DM, De Camilli P, Grinstein S. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol Biol Cell 2012; 23:176-87; PMID:22072788; http://dx.doi.org/ 10.1091/mbc.E11-06-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513-25; PMID:19603039; http://dx.doi.org/ 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 47.Spang A. On the fate of early endosomes. Biol Chem 2009; 390:753-9; PMID:19361275; http://dx.doi.org/ 10.1515/BC.2009.056 [DOI] [PubMed] [Google Scholar]
- 48.Huotari J, Helenius A. Endosome maturation. EMBO J 2011; 30:3481-500; PMID:21878991; http://dx.doi.org/ 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucci C, Lütcke A, Steele-Mortimer O, Olkkonen VM, Dupree P, Chiariello M, Bruni CB, Simons K, Zerial M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett 1995; 366:65-71; PMID:7789520; http://dx.doi.org/ 10.1016/0014-5793(95)00477-Q [DOI] [PubMed] [Google Scholar]
- 50.Chen PI, Kong C, Su X, Stahl PD. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem 2009; 284:30328-38; PMID:19723633; http://dx.doi.org/ 10.1074/jbc.M109.034546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Dominguez C, Stahl PD. Increased expression of Rab5a correlates directly with accelerated maturation of Listeria monocytogenes phagosomes. J Biol Chem 1999; 274:11459-62; PMID:10206948; http://dx.doi.org/ 10.1074/jbc.274.17.11459 [DOI] [PubMed] [Google Scholar]
- 52.Ulrich F, Krieg M, Schötz EM, Link V, Castanon I, Schnabel V, Taubenberger A, Mueller D, Puech PH, Heisenberg CP. Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev Cell 2005; 9:555-64; PMID:16198297; http://dx.doi.org/ 10.1016/j.devcel.2005.08.011 [DOI] [PubMed] [Google Scholar]
- 53.Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell 2005; 16:3847-64; PMID:15944222; http://dx.doi.org/ 10.1091/mbc.E05-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi S, Hu B, Hahm K, Luo Y, Kam Hui ES, Yuan Q, Wong WM, Wang L, Su H, Chu TH, et al. . LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med 2007; 13:1228-33; PMID:17906634; http://dx.doi.org/ 10.1038/nm1664 [DOI] [PubMed] [Google Scholar]
- 55.Bankers-Fulbright JL, Kephart GM, Bartemes KR, Kita H, O’Grady SM. Platelet-activating factor stimulates cytoplasmic alkalinization and granule acidification in human eosinophils. J Cell Sci 2004; 117:5749-57; PMID:15507482; http://dx.doi.org/ 10.1242/jcs.01498 [DOI] [PubMed] [Google Scholar]
- 56.Huss M, Ingenhorst G, König S, Gassel M, Dröse S, Zeeck A, Altendorf K, Wieczorek H. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J Biol Chem 2002; 277:40544-8; PMID:12186879; http://dx.doi.org/ 10.1074/jbc.M207345200 [DOI] [PubMed] [Google Scholar]
- 57.Wieczorek H, Cioffi M, Klein U, Harvey WR, Schweikl H, Wolfersberger MG. Isolation of goblet cell apical membrane from tobacco hornworm midgut and purification of its vacuolar-type ATPase. Methods Enzymol 1990; 192:608-16; PMID:2150092; http://dx.doi.org/ 10.1016/0076-6879(90)92098-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.