Abstract
The endosomal system and autophagy are 2 intertwined pathways that share a number of common protein factors as well as a final destination, the lysosome. Identification of adaptor platforms that can link both pathways are of particular importance, as they serve as common nodes that can coordinate the different trafficking arms of the endolysosomal system. Using a mass spectrometry approach to identify interaction partners of active (GTP-bound) RAB7, the late endosome/lysosome GTPase, and yeast 2-hybrid screening to identify LC3/GABARAP interaction partners we discovered the multivalent adaptor protein PLEKHM1. We discovered a highly conserved LC3-interaction region (LIR) between 2 PH domains of PLEKHM1 that mediated direct binding to all LC3/GABARAP family members. Subsequent mass spectrometry analysis of PLEKHM1 precipitated from cells revealed the HOPS (homotypic fusion and protein sorting) complex as a prominent interaction partner. Functionally, depletion of PLEKHM1, HOPS, or RAB7 results in decreased autophagosome-lysosome fusion. In Plekhm1 knockout (KO) mouse embryonic fibroblasts (MEFs) we observed increased lipidated LC3B, decreased colocalization between LC3B and LAMP1 under amino acid starvation conditions and decreased autolysosome formation. Finally, PLEKHM1 binding to LC3-positive autophagosomes was also essential for selective autophagy pathways, as shown by clearance of puromycin-aggregates, in a PLEKHM1-LIR-dependent manner. Overall, we have identified PLEKHM1 as an endolysosomal adaptor platform that acts as a central hub to integrate endocytic and autophagic pathways at the lysosome.
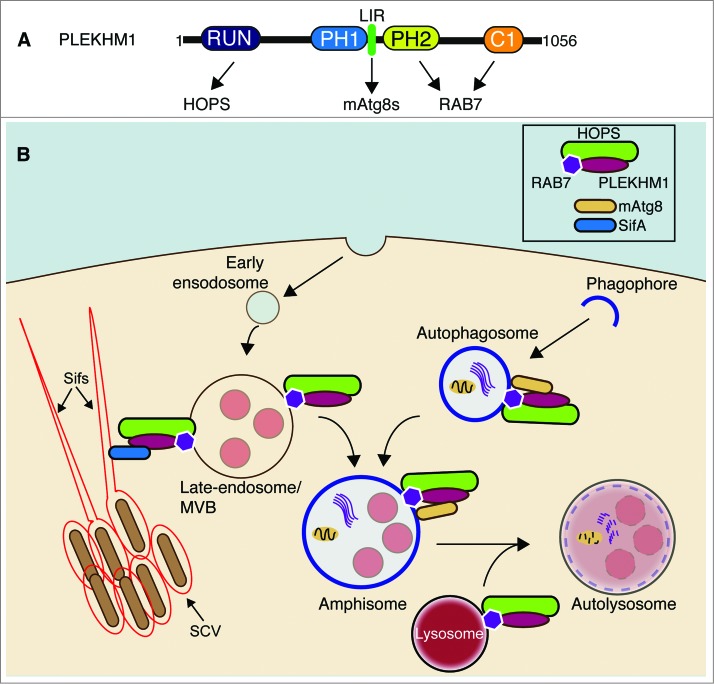
PLEKHM1 (pleckstrin homology domain containing, family M [with RUN domain] member 1) is a ubiquitously expressed protein involved in the regulation of osteoclast function and bone resorption. Recently, it was also described in the context of negatively regulating the endocytic pathway but not autophagy. However, in our recent studies, we show that PLEKHM1 positively regulates the terminal stages of both endocytic and autophagy pathways through direct interaction between PLEKHM1, RAB7, the HOPS complex, and mammalian Atg8 proteins (Fig. 1A). In addition, the PLEKHM1-RAB7-HOPS complex is a direct target for the Salmonella (Salmonella enterica Typhimurium) effector protein SifA (Salmonella-induced filament protein A) that together regulate the Salmonella-containing vacuole (Fig. 1B).
Figure 1.
Model of PLEKHM1 function in the endocytic and autophagic pathways. (A) Domain structure of PLEKHM1 and their interactions. RUN (RUNDC3A/RPIP8, UNC-14 and RUSC1/NESCA); PH1 and PH2 (Pleckstrin homology domain 1 and 2); C1/Zinc finger (C1); HOPS (homotypic fusion and protein sorting). (B) Proposed positioning of PLEKHM1 and its associated complexes in the autophagy and endocytic pathway. PLEKHM1 localizes to late endosomes and lysosomes in an RAB7-dependent manner. The interaction between PLEKHM1, RAB7, and HOPS on vesicles positions these vesicles for tethering and fusion with autophagosomes, through direct interaction with MAP1LC3/GABARAP proteins. The autophagosomes may also fuse with late endosomes/MVBs (multivesicular bodies) in a PLEKHM1-RAB7-HOPS-dependent manner to produce amphisomes, prior to fusion with the lysosome. PLEKHM1-RAB7-HOPS can also be subverted by the Salmonella effector SifA, for the proper maintenance of the Salmonella-containing vacuole (SCV) and Sif (Salmonella-induced filament) formation. mAtg8s, MAP1LC3/GABARAP proteins.
Using a 2-pronged approach, we identified PLEKHM1 as an interaction partner of RAB7 in its GTP-bound active state, RAB7(GTP), and MAP1LC3/GABARAP proteins. PLEKHM1 interacts directly with all MAP1LC3/GABARAP proteins through a highly conserved LC3-interaction motif (LIR) located between the Pleckstrin homology domain 1 (PH1) and PH2 domains of PLEKHM1 (Fig. 1A). Endogenous PLEKHM1 colocalizes with LAMP1 at the cytosolic-facing membrane, but not the lumenal side, of LC3B-containing amphisomes/autolysosomes, indicating that PLEKHM1 is an autophagy adaptor protein rather than a selective cargo receptor.
Using SILAC (stable isotope labeling of cells in culture)-labeled inducible PLEKHM1 cells, we identified the HOPS complex as a significant interaction partner. The hexameric HOPS complex is an essential component of the late endocytic fusion machinery and is required for autolysosome formation. PLEKHM1 interacts directly with the HOPS complex, mediated by the RUN domain of PLEKHM1 and the C terminus of VPS39 (Fig. 1A) Crucially, PLEKHM1 forms an endogenous complex with HOPS. In the context of vesicle fusion, the HOPS complex acts as a tether to anchor and position the vesicles prior to fusion that is driven by SNARE proteins. Multiple SNARE proteins, such as VAMP7, VAMP8, VTI1B, SNAP29, and STX17 have been described to be required for autophagosome-lysosome fusion. Upon autophagy induction, enhanced PLEKHM1 coprecipitation is detected with the HOPS complex and the autophagosome specific SNARE STX17, reinforcing a role for PLEKHM1 in autophagosome-lysosome fusion.
Both RAB7 and the HOPS complex are integral components of the endocytic pathway and, as such, we wanted to test the effect of PLEKHM1 loss on EGFR (epidermal growth factor receptor) degradation. We used 2 epithelial cell lines, HeLa and Hke3. In both instances, loss of PLEKHM1 causes a marked decrease in the rate of EGFR degradation and increases retention in early endosomes. This is in stark contrast to previous reports that used A549 cells and showed that a lack of PLEKHM1 accelerates EGFR degradation. Clearly, cell lines and their background mutations will have to be considered for future studies.
In addition to the endocytic pathway, RAB7 and the HOPS complex are essential for the autophagosome-to-autolysosome transition. Therefore, we also wanted to explore this facet of PLEKHM1 function. We generated Plekhm1 KO MEFs to analyze the effects of autophagy flux in the absence of PLEKHM1. Plekhm1 KO MEFs show a block in autophagy, with the accumulation of SQSTM1/p62 and LC3B-II and, using tandem-fluorescence-LC3B as a marker, a decrease in autolysosome formation. Taken together, these findings suggest that PLEKHM1 functions at the point of autophagosome-lysosome fusion (Fig. 1B).
Finally, we were interested in testing the functional role of PLEKHM1, and in particular the LIR, during selective autophagy of protein aggregates. We treated control and PLEKHM1-depleted cells with puromycin and observed aggregate clearance over time after puromycin removal. Cells lacking PLEKHM1 and those reconstituted with a PLEKHM1-LIR mutant were unable to efficiently remove SQSTM1-ubiquitin-positive aggregates, compared to control or PLEKHM1-wild type reconstituted cells, indicating an important role for the final stages of endosome and autophagosome maturation (Fig. 1B).
“No man is an island, entire of itself” seems of particular prudence when considering the intertwined nature of both autophagic and endocytic pathways. Indeed, it is interesting that there are multiple RAB7 effector proteins functioning at the late endocytic step that also contribute to autophagy, including FYCO1, KIAA0226/Rubicon, UVRAG and now PLEKHM1, where only PLEKHM1 and UVRAG have been shown to interact with the HOPS complex. All of which, when mutated or depleted, have effects on both the endocytic and autophagic pathways. Clearly the roles of these proteins in cell-type and tissue-specific settings have to be determined before we fully comprehend the complexities of how the endosomal and autophagic pathways integrate and communicate with each other.
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed.
Funding
This work is supported by grants from the Deutsche Forschungsgemeinschaft (DI 931/3–1), the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115), LOEWE grant Ub-Net and LOEWE Centrum for Gene and Cell therapy Frankfurt and the European Research Council / ERC grant agreement n° 250241-LineUb to ID.