Abstract
The P140 peptide, a 21-mer linear peptide (sequence 131–151) generated from the spliceosomal SNRNP70/U1–70K protein, contains a phosphoserine residue at position 140. It significantly ameliorates clinical manifestations in autoimmune patients with systemic lupus erythematosus and enhances survival in MRL/lpr lupus-prone mice. Previous studies showed that after P140 treatment, there is an accumulation of autophagy markers sequestosome 1/p62 and MAP1LC3-II in MRL/lpr B cells, consistent with a downregulation of autophagic flux. We now identify chaperone-mediated autophagy (CMA) as a target of P140 and demonstrate that its inhibitory effect on CMA is likely tied to its ability to alter the composition of HSPA8/HSC70 heterocomplexes. As in the case of HSPA8, expression of the limiting CMA component LAMP2A, which is increased in MRL/lpr B cells, is downregulated after P140 treatment. We also show that P140, but not the unphosphorylated peptide, uses the clathrin-dependent endo-lysosomal pathway to enter into MRL/lpr B lymphocytes and accumulates in the lysosomal lumen where it may directly hamper lysosomal HSPA8 chaperoning functions, and also destabilize LAMP2A in lysosomes as a result of its effect on HSP90AA1. This dual effect may interfere with the endogenous autoantigen processing and loading to major histocompatibility complex class II molecules and as a consequence, lead to lower activation of autoreactive T cells. These results shed light on mechanisms by which P140 can modulate lupus disease and exert its tolerogenic activity in patients. The unique selective inhibitory effect of the P140 peptide on CMA may be harnessed in other pathological conditions in which reduction of CMA activity would be desired.
Keywords: antigen-presenting cells, autophagy, B lymphocytes, chaperone-mediated autophagy, class II MHC molecules, heat shock proteins, HSPA8/HSC70, lupus, lysosomal chaperones, lysosomes
Abbreviations: ALF, artificial lysosomal fluid; APC, antigen-presenting cell; bodipy: BODIPY FL C5 Lactosylceramide/bovine serum albumin; CMA, chaperone-mediated autophagy; CoIP, coimmunoprecipitation; CPZ: chlorpromazine; CTSD, cathepsin D; DAPI, 4′, 6-diamidino-2-phenylindole; ELISA, enzyme-linked immunosorbent assay; FCS, fetal calf serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCQ, hydroxychloroquine; iv, intravenous; LAMP2A, lysosomal-associated membrane protein 2A; LC3-II, MAP1LC3-II; LC-MS, liquid chromatography-mass spectrometry; MHCII, major histocompatibility complex class II; NBD, nucleotide binding domain; paraquat, 1, 1′-dimethyl-4, 4′-bipyridyldinium dichloride; PBS, phosphate-buffered saline; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; RP-HPLC, reversed-phase high-performance liquid chromatography; RPL5, ribosomal protein L5; SBD, substrate binding domain; SD, standard deviation; SEM, standard error of the mean; SLE, systemic lupus erythematosus; SNRNP70/U170K: small nuclear ribonucleoprotein 70kDa; SQSTM1/p62, sequestosome 1; TF, transferrin; TFA, trifluoroacetic acid
Introduction
The 21-mer peptide P140 displays protective properties in MRL/lpr lupus-prone mice1 and most importantly in lupus patients, as shown in phase II clinical trials.2,3 This peptide corresponds to the fragment 131–151 of the spliceosomal SNRNP70/U1–70K (small nuclear ribonucleoprotein 70kDa [U1]) and contains a phosphoserine residue at position 140, which was found to represent a natural apoptosis-specific post-translational modification of the protein.4 P140 binds both the HSPA8/HSC70/HSP73 chaperone and major histocompatibility complex class II (MHCII) molecules, and colocalizes with both molecules intracellularly and at the cell surface in splenic MRL/lpr B cells.5-7 Administered intravenously to MRL/lpr mice, P140 decreases the HSPA8 overexpression observed in this murine strain, and increases the accumulation of the autophagy markers SQSTM1/p62 (sequestosome 1) and LC3-II, consistent with a downregulation of lysosomal degradation at the autolysosome stage or with lysosomal dysfunction.7 P140 also decreases MHCII overexpression and attenuates biological and clinical signs of systemic autoimmunity (i.e. proteinuria, vasculitis, dermatitis, anti-DNA antibody production). Thus, in MRL/lpr mice, P140 displays a unique mode of action in altering the autophagy pathway, which is involved in endogenous autoantigen processing and presentation.8-10 This phenomenon could be associated with the decreased response of autoreactive CD4+ T cells observed in P140-treated MRL/lpr mice.11
P140 bioavailability was previously studied using real-time biodistribution experiments. We observed that fluorochrome-labeled P140 especially accumulates in the lungs and the spleen of MRL/lpr mice.7 In the latter, P140 fluorescence was mainly observed in the lymphoid white pulp, mostly in the mantle and marginal zones. Apparently, P140 homing is essentially detectable in organs enriched in antigen-presenting cells (APCs), including activated B cells. Yet, key questions remained regarding the intracellular trafficking of P140 and its possible location within specific cell compartments or organelles where it generates its effects.
In the present study, we characterized the biochemical effects exerted by P140 on the HSPA8-containing heterocomplexes integrity and on HSPA8 intrinsic functions, and showed in vitro that P140 does interfere with the binding to HSPA8 of cochaperones, ligands, and client proteins. We then examined both in vitro and in vivo, how P140 is internalized from the cell surface and in which compartment it homes. We found that P140 uses the clathrin-mediated internalization pathway in B cells and circulates via early endosomes before ending up into lysosomes. Based on established data indicating that HSPA8 protein resident in the lysosomal lumen is crucial for chaperone-mediated autophagy (CMA), studies were conducted to address whether P140 specially interferes with this selective type of autophagy, which is essential for the degradation of a subset of cytosolic soluble proteins in lysosomes.12 Using 2 independent fluorescent assays, we found that P140 readily decreases levels of CMA activity in living cells. Furthermore, using an in vitro system that reconstitutes CMA in isolated lysosomes, we confirmed that the P140 effect on CMA is targeted on the lumenal chaperones. Interestingly, our studies also indicate that LAMP2A (lysosomal-associated membrane protein 2A), limiting on substrate binding and translocation through CMA13 is overexpressed in MRL/lpr B cells and that in P140-treated lupus mice its overexpression is reduced. In all these experimental settings, and particularly in vivo, the influence of the phosphoryl moiety occurring at serine residue 140 was crucial. Our findings provide the first evidence showing that the P140 peptide acts directly on the CMA pathway.
Results
Effect of the P140 peptide on the HSPA8-HSP90AA1 complex
P140 is a phosphorylated 21-mer peptide of sequence 131RIHMVYSKRpSGKPRGYAFIEY151 (mass = 2640; calculated isoelectric point = 10.6). The sequence from which it is issued (residues 131 to 151 of the SNRNP70 spliceosomal protein) is highly conserved across numerous species (100% homology between human, monkey, mouse, rat) and is exposed to the solvent on the U1 snRNP particle.14 Its structure in aqueous solution has been determined by 1H-NMR spectrometry and molecular dynamics calculation.6 We have previously shown that P140 readily binds to HSPA8.6,15 Our present data based on recombinant HSPA8 fragments show that P140 interacts with the N-terminal HSPA8 nucleotide binding domain (NBD) and not with the C-terminal substrate binding domain (SBD), which encompasses the client protein-binding region (Fig. S1). This result indicates that P140 rather behaves as a ligand and not as a potential substrate of HSPA8.
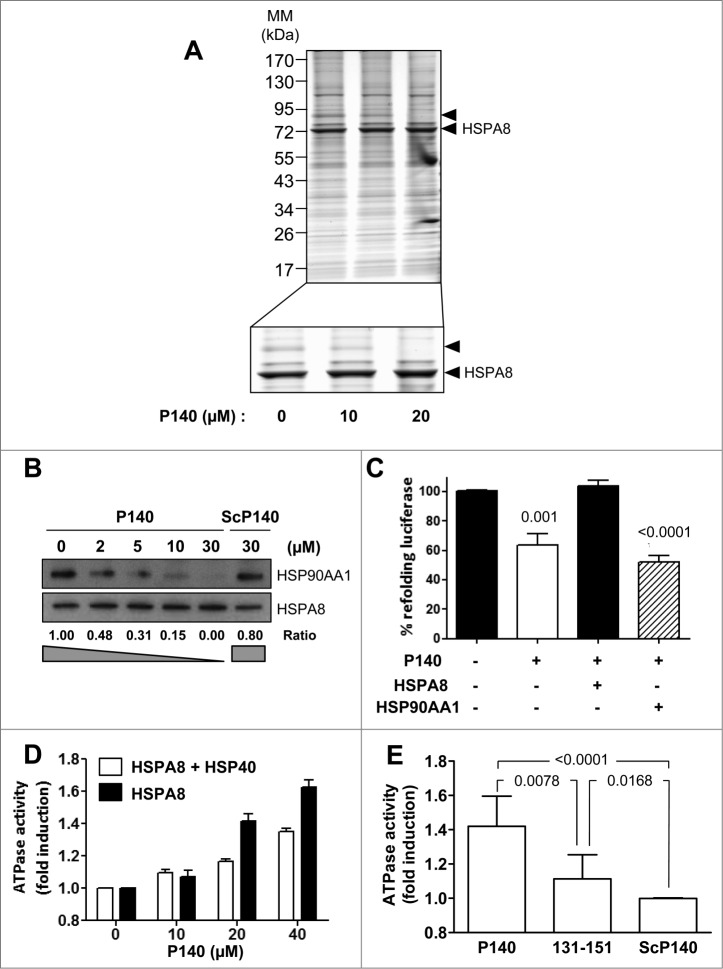
Knowing that in vitro P140 alters folding properties of HSPA8,7 we sought to determine the potential effect of the P140 peptide binding on the integrity of HSPA8-cochaperone heterocomplexes. Using Raji cell lysates incubated with the P140 peptide and analyzed by electrophoresis in native conditions followed by immunoblotting with anti-HSPA8 antibodies, we observed that HSPA8-containing heterocomplexes were partially destabilized by P140 in a dose-dependent manner (Fig. S2). To identify partners of the HSPA8-cochaperone complex that are underrepresented after incubating cells with increasing concentrations of the P140 peptide, Raji cells were preincubated at 37°C for 24 h with the peptide, lysed, and immunoprecipitated with anti-HSPA8 monoclonal antibody. SDS-PAGE analysis revealed that most bands were recovered in the pulldowns but, strikingly, a band migrating at ∼90 kDa, which was not visible in the heterocomplex isolated from the P140-treated cells (arrow; Fig. 1A). Mass spectrometry analysis of the corresponding band in the untreated extract revealed the presence of nearly 2 dozen proteins including the ATPase WRNIP1, helicases, HSP90AA1, ACTA/α-actin, BAG5, and heterogeneous nuclear RNPs. Using coimmunoprecipitation (CoIP) experiments for HSPA8 and immunoblotting with specific antibodies, we confirmed that P140, but not a scrambled form of the peptide (ScP140), diminished the recruitment of HSP90AA1 in a peptide dose-dependent manner (Fig. 1B).
Figure 1.
The P140 peptide disrupts HSPA8 heterocomplex assembly in vitro and alters functional HSPA8 properties. (A) Native protein extracts from P140-treated Raji cells were subjected to CoIP with anti-HSPA8 antibody 1B5. Gel staining was assessed by colloïdal blue. The arrow at ∼90 kDa (see enlargement) indicates a band that disappeared after P140 treatment. (B) Raji cells were treated with increasing concentrations of the P140 peptide or ScP140 peptide as control. Cell lysates were immunoprecipitated with anti-HSPA8 antibody 1B5 and western immunoblotting was performed for detecting HSP90AA1 and HSPA8. HSP90AA1 protein levels were normalized by densitometry to HSPA8 level using ImageJ Software. The ratios are indicated. (C) Luciferase was denatured at 40°C for 30 min and then refolded during 1 h at 30°C in rabbit reticulocyte lysate in the presence of 40 μM P140 peptide. The results (expressed in %) compare the capacity of exogenous recombinant HSPA8 (0.5 μM) or HSP90AA1 (0.5 μM) to counter-balance P140 inhibition of luciferase refolding. The values are the mean + standard deviation (SD) of 3 independent measurements. P values are indicated (Student t test). (D) Increasing concentrations of the P140 peptide were incubated for 3 h at 37°C with HSPA8 (500 nM) with or without HSP40 (500 nM), and the amount of hydrolyzed ATP was measured by luminescence detection assay. ATPase activity measured in the presence of increasing concentrations of the P140 peptide was expressed in fold induction compared to conditions in the absence of peptide. The spontaneous hydrolysis observed with the peptide alone was deduced from the values. Bars represent averaged values from 3 independent experiments + SD. (E) HSPA8 (500 nM) and HSP40 (500 nM) were incubated with 40 μM of either P140 or unphosphorylated peptide 131–151 or ScP140 peptides, and HSPA8 ATP-hydrolytic activity was assayed as described in (D). The values are the mean + SD of 6 independent experiments. The P value is indicated (Student t test).
Numerous regulatory functions have been assigned to HSPA8, mostly related to its chaperoning activity.16 We have previously reported that P140 impairs the folding properties of chaperone HSPA8.7 Here we show that, in contrast to the effect observed after adding exogenous HSPA8, exogenous HSP90AA1 supplementation was unable to reverse the inhibitory effect of P140 on the folding ability of HSPA8 (Fig. 1C).
Housekeeping functions of intracellular HSPA8 are based on the ability of this chaperone to interact with hydrophobic peptide substrates in an ATP-controlled fashion.17,18 Consequently, we next assessed in vitro the effect of P140 on HSPA8 ATPase activity. We found that P140 was capable to effectively increase both the endogenous and HSP40-stimulated ATPase activities of HSPA8 in a peptide dose-dependent manner (Fig. 1D). The effect of the unphosphorylated peptide 131–151 was less pronounced and virtually, the ScP140 peptide had no effect on HSP40-stimulated HSPA8 ATPase activity (Fig. 1E).
Overall these results substantiate the fact that the P140 peptide alters the integrity of HSPA8-HSP90AA1-bearing heterocomplexes by a mechanism that most likely involves HSPA8 ATPase activity.
Endosomal trafficking of the P140 peptide
Given the above results generated in vitro, we next asked whether in cellulo, P140 resides in a particular cell compartment where it may target chaperones and alter their functions. Immunofluorescence and immunoelectron microscopy experiments showed previously that P140 is detectable in the cytoplasm of MRL/lpr peripheral blood lymphocytes and splenocytes 1h after intravenous administration of peptide in saline.6,7 Fluorescent staining of Alexa Fluor 488-labeled P140 was found in the cytoplasm of nonpermeabilized purified B cells incubated at 37°C with the peptide in phosphate-buffered saline (PBS), while the labeling was localized at the membrane at 4°C (Fig. S3). These data confirm that translocation of P140 across biological membranes is direct via an energy-dependent cellular process. No fluorescence staining was visualized when the labeled ScP140 analog was assayed (Fig. S3).
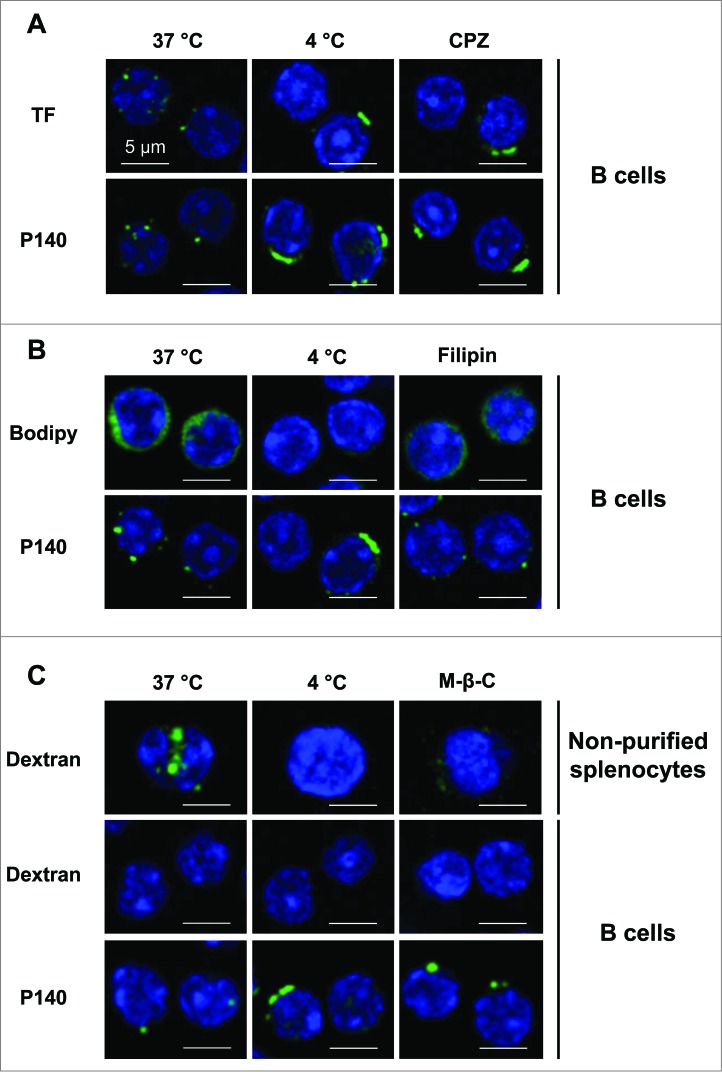
To characterize the P140 entry pathway into B cells we followed the localization of peptide and marker molecules in purified B lymphocytes by confocal microscopy in the absence or presence of selective inhibitors. We determined that P140, like TF (transferrin) used as a positive marker, was endocytosed by a clathrin-dependent mechanism in B lymphocytes purified from 12-wk-old MRL/lpr mice (Fig. 2A). Internalization of both P140 and TF that colocalize in MRL/lpr B lymphocytes (Fig. S4), was inhibited by incubation at low temperature (4°C) or by incubation with chlopromazine (CPZ) (Fig. 2A). To address possible contribution of other entry pathways we used BODIPY FL C5 Lactosylceramide/bovine serum albumin (bodipy) as a control marker of caveolin-dependent endocytosis and filipin was applied as selective inhibitor of this pathway. Filipin was effective at blocking bodipy internalization but devoid of effect on P140 entry into purified MRL/lpr B cells (Fig. 2B). Finally, we analyzed the contribution of macropinocytosis, mostly described as a constitutive pathway for dendritic cells and macrophages and poorly used by B cells19 (see dextran entry in B cells and unpurified splenocytes in Fig. 2C). In agreement with the low rate of macropinocytosis in B cells, we found that P140 did not use macropinocytosis to enter into purified B cells from MRL/lpr mice (Fig. 2C).
Figure 2.
B cells internalize the P140 peptide by clathrin-mediated endocytosis. B cells from MRL/lpr mice were pretreated 30 min at 37°C with the following endocytosis inhibitors: chlorpromazine (CPZ) for clathrin-mediated endocytosis (A), filipin for caveolin-mediated endocytosis (B), and methyl-β-cyclodextrin for macropinocytosis (C), followed by the addition of Alexa Fluor 488-labeled P140 peptide for 30 min at 37°C. TF, bodipy, and dextran were used as respective markers of each pathway. As a control, cells were incubated at 4°C to inhibit endocytosis. For macropinocytosis analysis, unpurified splenocytes were also used since dextran was not detected into B cells. Representative results of 2 different experiments are shown.
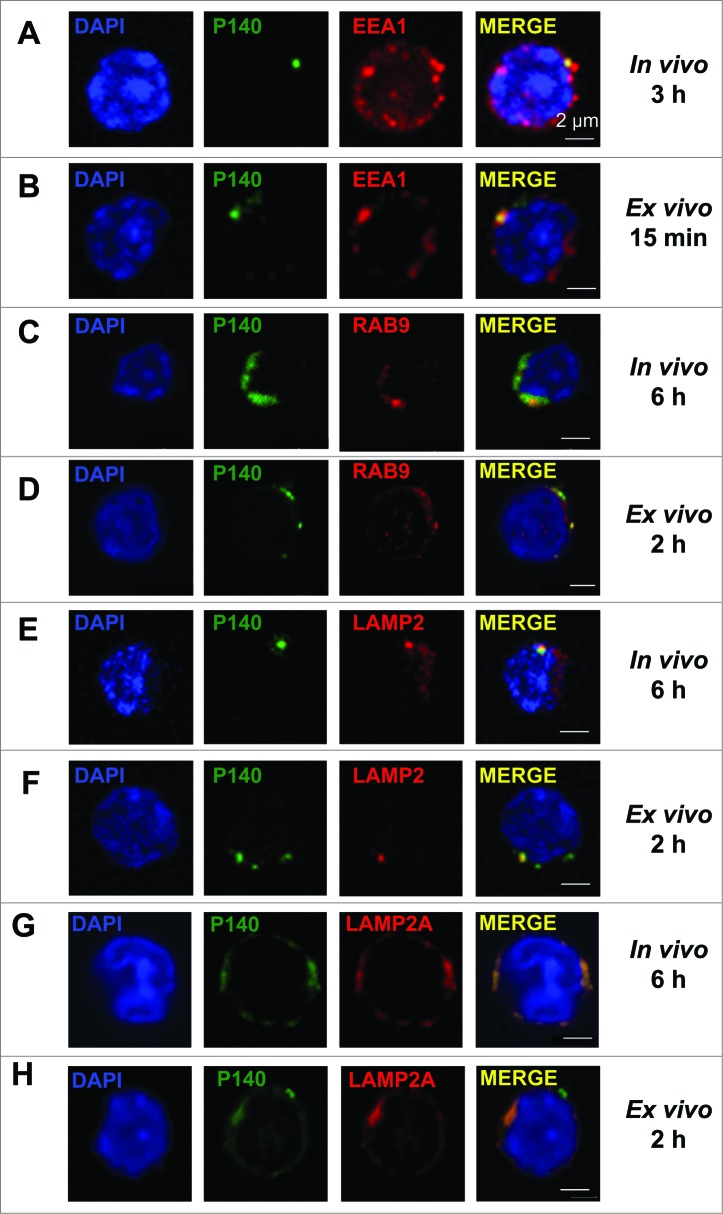
We next examined the lymphocyte entry pathway in vivo using P140 peptide labeled with Alexa Fluor 488 administrated to 10–12-wk-old MRL/lpr mice by intravenous (iv) injection (the route used in our therapeutic protocols1). We found that from 3 h onward, the peptide was recovered in splenocyte early endosomes where it colocalizes with the early endosomal marker EEA1 (Fig. 3A), whereas after 6 h, P140 was detected in late endosomes and lysosomes where it colocalizes with RAB9, LAMP2/CD107b, and LAMP2A (Fig. 3C, E, G). Counterstaining with LysoTracker Red lysosomal and MitoTracker Deep Red mitochondrial markers confirmed that 1 h after iv administration, P140 accumulated into lysosomes (Fig. S5).
Figure 3.
Colocalization of the P140 peptide with endolysosomal markers and components. (A, C, E, G) MRL/lpr mice received Alexa Fluor 488 labeled-P140 peptide (100 μg/mouse) by the iv route. Splenocytes were collected 3 h later and stained for the early endosomal marker EEA1 (A), or 6 h later and stained for the late endosomal marker RAB9 (C), lysosomal component LAMP2 (E) or LAMP2A isoform (G). Representative results of 3 independent experiments are shown. (B, D, F, H) B cells from MRL/lpr mice were incubated for 15 min (B) or 2 h (D, F, H) in the presence of Alexa Fluor 488 labeled-P140 peptide and stained for EEA1 (B), RAB9 (D) LAMP2 (F) or LAMP2A isoform (H). Representative results of 2 to 4 independent experiments are shown (2 different experimenters).
Similar internalization, although with shorter kinetics (15 min onward and 2 h, respectively) was found when Alexa Fluor 488-labeled P140 was incubated ex vivo with purified MRL/lpr B cells (Fig. 3B, D, F, H).
When the same experiments were performed with the unphosphorylated peptide 131–151, striking differences, notably in vivo, were observed (Fig. S6). The Alexa Fluor 488-labeled unphosphorylated peptide could not be visualized in early or late endosomes (Fig. S6A, D, F), indicating either that the unphosphorylated peptide administrated into MRL/lpr mice by the iv route uses another cellular uptake pathway, that the kinetics was radically different, or that this peptide was rapidly degraded (see below) or released. In ex vivo experiments, the unphosphorylated peptide 131–151 was visualized in late endosomes and lysosomes (Fig. S6E, G), but colocalization with EEA1 was hardly seen, even when different times points were used for microscopy observation (Fig. S6B, C).
The beneficial influence of phosphorylation on P140 behavior was further substantiated by showing that the phosphorylated form of the peptide is significantly more resistant to proteases contained in the serum from sick MRL/lpr mice (Fig. S7A). Using a reconstituted medium containing a combination of CTSD/cathepsin D and CTSL/cathepsin L, thereafter named artificial lysosomal fluid (ALF)20 (Fig. S7B) or the complete matrices of isolated lysosomes21,22 (Fig. S7C), no substantial change between phosphorylated and unphosphorylated peptides could be noted at pH 4.2, 5.3 and 7.2. Furthermore, even at pH 4.2 in ALF, and for about 2 h, the phosphoryl moiety was preserved in the P140 peptide (Fig. S8), a result that is most satisfactory in the present context.
Of note, however, we observed marked differences in the stability at low pH of P140 when incubated with the matrices from different lysosomal subpopulations (Fig. S7C). In fact, P140 stability at the acid pH remained much higher upon incubation with matrices from lysosomes active for CMA, than from lysosomes inactive for this autophagic pathway. Since the enzymatic load and proteolytic activity in both subpopulations have been extensively characterized to be comparable in both groups,21 we speculate that interaction with components present in one lysosomal lumen but not in the other could be behind the stabilizing effect observed for P140. The best-characterized difference between the group of CMA-competent and -incompetent lysosomes is actually the presence of HSPA8 only in the lumen of the active group. These findings suggest that the interaction of P140 with lumenal HSPA8 could be behind the stability of this peptide in the lumen of a specific subpopulation of lysosomes.
Overall, these results confirm that P140 is mainly internalized in MRL/lpr B cells via clathrin-mediated endocytosis and that most of the internalized peptide reaches the lysosomal compartment. The critical influence of phosphorylation at residue 140 is highlighted here by the observation that in contrast to the P140 peptide, the unphosphorylated peptide analog appears to use an alternative cellular uptake route to reach late endosomes and lysosomes in MRL/lpr B cells, a pathway that might affect its in vivo protective efficacy.
The P140 peptide interferes with CMA
The fact that we observed P140 accumulation in lysosomes, the higher stability of the peptide in those lysosomes containing HSPA8 and the ability of P140 to alter the integrity of HSPA8-HSP90AA1 heterocomplexes in vitro, led us to analyze the possible effect of P140 on known functions of this chaperone complex in the lysosomal-endosomal compartment. Among those, we focused on CMA where the involvement of HSPA8 and HSP90AA1 forms that are resident in the lysosomal lumen has been demonstrated.21,23,24 In CMA, the substrate proteins, known to contain a motif biochemically related to the pentapeptide KFERQ, are selectively recognized by cytosolic HSPA8 and targeted to the lysosomal membrane. There the substrate-chaperone complex binds to LAMP2A, which acts as a receptor for this pathway.12 Lumenal HSPA8 is also required for the complete translocation of the substrate protein across the membrane into the lysosomal lumen where it is completely degraded by the lysosomal proteases.12 Lumenal levels of HSPA8 determine the efficiency of CMA and blockage of this lumenal chaperone abrogates CMA.22,23
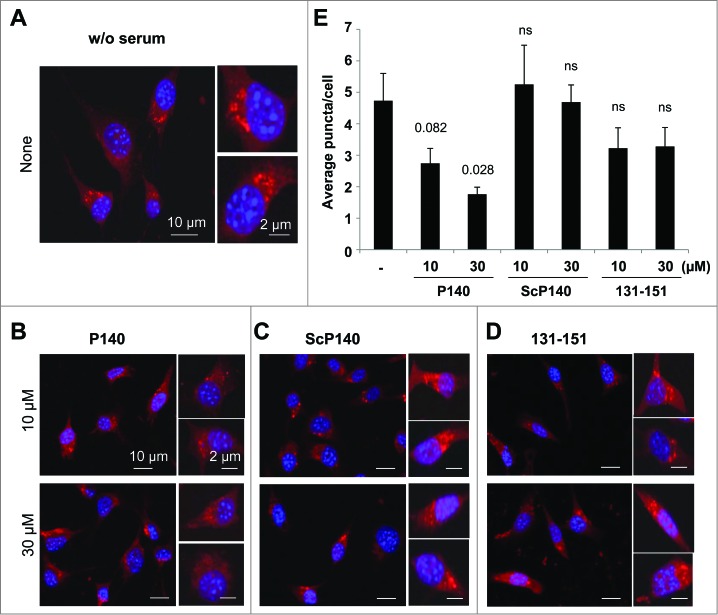
To determine whether the P140 peptide may directly affect CMA, we measured the activity of this pathway in P140-treated fibroblasts using a novel photoactivable reporter. This artificial CMA substrate was constructed by inserting the CMA-targeting motif KFERQ in the N terminus of a photoactivable form of mCherry 1 protein.25 As this cytosolic protein is delivered to lysosomes for degradation, the fluorescence pattern shifts from diffuse to punctate. CMA activity can be measured in cells stably expressing this construct as the number of fluorescent puncta per cell. Using these procedures, we did not observe any changes in CMA activity in cells treated with the ScP140 peptide or the unphosphorylated (nonprotective) peptide 131–151 and maintained in serum-free conditions to maximally activate CMA. In contrast, treatment with P140 clearly diminished the average number of puncta per cell in a dose-dependent manner (Fig. 4). These results were further validated using a different paradigm of maximal activation of CMA, oxidative stress26 and a second independent fluorescent reporter where the CMA targeting motif was inserted in frame with the photoswitchable fluorescent protein Dendra2 (KFERQ-PS-Dendra; unpublished). These experiments were also designed to show a more detailed P140 dose-dependence effect (6 P140 concentrations), in 3 test conditions, namely in the presence of serum, under a starvation stress and under a mild oxidative stress generated by 1,1′-dimethyl-4,4′-bipyridyldinium dichloride (paraquat).26 The brighter fluorescence of this reporter makes it suitable for high-content microscopy allowing for analysis in more than 700 cells per condition. Compared to an inactive P140 analog described earlier,15 P140 was found to significantly diminish CMA activity in a peptide concentration-dependent manner upon maximally activation of CMA through 2 different stimuli (Fig. S9). It should be pointed out that the fact that P140 was capable to inhibit CMA in both conditions, despite the different mechanisms of CMA activation occurring during starvation and oxidative stress,26 is in agreement with the proposed direct effect of P140 in the CMA-active lysosomal compartment that is shared in both cases. Taken together, the foregoing results support the idea that treatment of certain cell populations with P140, which remains stable on a large range of pH in CMA+ lysosomal extracts, may lead to a downregulation of CMA activity.
Figure 4.
Effect of the P140 peptide on CMA activity on mouse fibroblasts. NIH3T3 cells stably expressing the photoactivable CMA reporter KFERQ-PA-mCherry1 were photoactivated and maintained in serum-free medium supplemented with or without (w/o) with the indicated peptides for 12 h. (A to D) Representative images, low magnification field and higher magnification single cell inserts of cells that were kept untreated (A), or treated with the P140 peptide (B), scrambled peptide ScP140 (C) or unphosphorylated peptide 131 to 151 used as control (D). Nuclei were highlighted by DAPI. (E) Quantification of the average number of fluorescent puncta per cell. Values are mean + standard error of the mean (SEM). N > 50 cells. P values are indicated (Student t test).
To further explore the effect of P140 on CMA, we attempted to discriminate if inhibition of CMA by P140 was exerted on the chaperones located in the cytosol or at lumenal side of the lysosomal membrane (since both forms participate on CMA).21,23,24 We used a standardized assay with isolated lysosomes that recapitulates CMA in vitro.21 In this assay, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), a well-characterized CMA substrate, is incubated with lysosomes pretreated with protease inhibitors to prevent the degradation of any GAPDH internalized into the lumen, and CMA of the protein is measured as the amount of GAPDH associated with lysosomes recovered by centrifugation (that represents GAPDH bound to the lysosomal membrane and internalized into the lumen). We found that addition of increasing concentrations of P140 did not modify the amount of GAPDH recovered in lysosomes (Fig. S10A), not even when exogenous HSPA8 was added into the incubation media to increase the dependence on this chaperone (Fig. S10B). Similar results were observed when lysosomal binding and uptake of GAPDH were analyzed separately. Thus, incubation of GAPDH with lysosomes in the absence of protease inhibitors permits to measure the binding step, since all internalized GAPDH will be readily degraded by the lysosomal proteases. In this assay, uptake can be calculated as the difference of GAPDH associated to lysosomes treated with or without protease inhibitors. Using this paradigm, we confirmed that addition of P140 or ScP140 to the incubation media did not significantly modify binding or uptake of GAPDH by CMA (Fig. S10C, D). These results support a mechanism by which the inhibitory effect of P140 on CMA observed in cultured cells is exerted on the lumenal chaperones.
Lumenal HSPA8 has been proposed to interact with substrate proteins as they are being internalized through the CMA translocation complex to facilitate their complete translocation. Based on the results described above, we anticipate that P140 could inhibit CMA by interfering with the binding of lumenal HSPA8 to GAPDH. In the enzyme-linked immunosorbent assay (ELISA) test format we developed, while ovalbumin used as negative control was inactive (Fig. S11A), the CMA substrate GAPDH (a protein multimer in its native state; Fig. S11B) was highly efficient at inhibiting HSPA8 binding to plastic plate-immobilized P140. In this assay, the ribosomal protein L5 (RPL5) that bears a regulatable CMA targeting motif FKRYQ in its sequence (residues 12–16) displayed some activity, which was however much weaker than that of GAPDH (not shown). Taken together, our results are consistent with an inhibitory effect of the P140 peptide on CMA at the level of substrate-chaperone binding in the lysosomal lumen.
Lysosomal abnormalities in MRL/lpr B cells
We have shown previously that peripheral T lymphocytes from 2 distinct lupus-prone mouse models (MRL/lpr and [NZBxNZW]F1 mice) and also patients suffering from lupus exhibit a deregulated macroautophagy.27 We thus decided to characterize the lysosomal system in B cells from MRL/lpr lupus-prone mice to determine if lysosomal abnormalities may underlie their phenotype and to further typify the basis for the inhibitory effect of P140 on CMA.
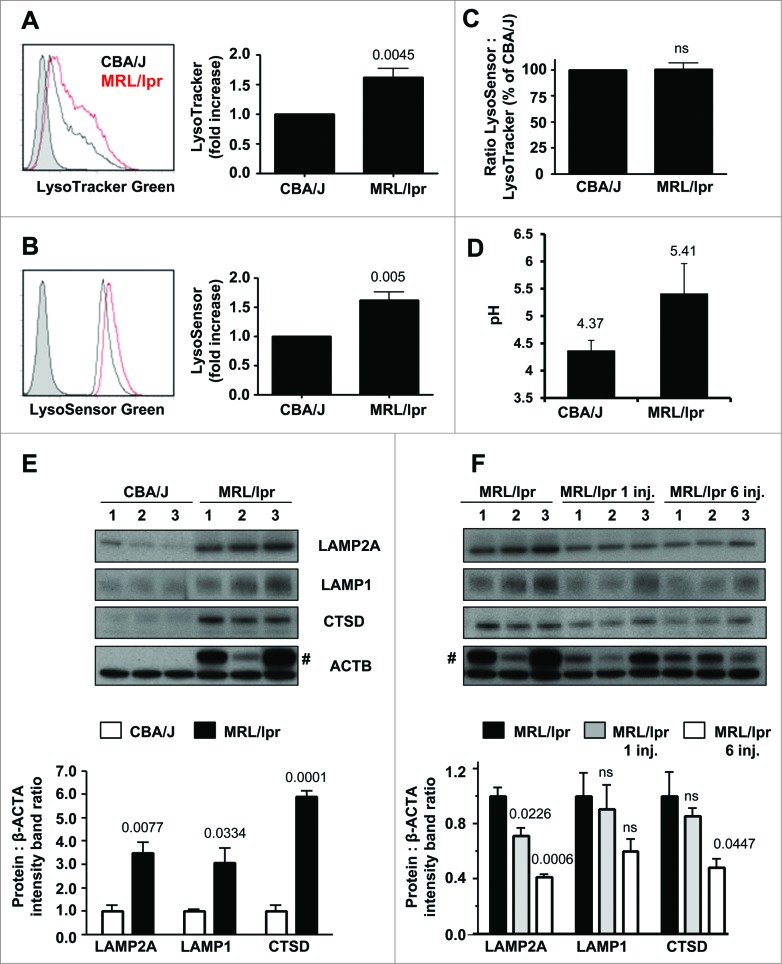
FACS analysis after LysoTracker Green staining of splenocytes revealed a marked increase in the amount of lysosomal material in B cells from 10–12-wk-old MRL/lpr mice when compared to B cells purified from age-matched CBA/J mice (Fig. 5A; effect not detectable at 5 wk old). This increase of lysosomal number and/or volume in living MRL/lpr B cells was correlated with an increase of pH as shown after staining cells with LysoSensor Green (Fig. 5B). The fluorescence ratio of LysoSensor Green:LysoTracker Green staining did not reveal significant differences (Fig. 5C). Fluorescence measurements with the LysoSensor Yellow/Blue revealed that vesicles in CBA/J splenocytes consistently acidified to a pH of 4.4 in average, while those within MRL/lpr splenocytes never reached this value and was rather around 5.4 in average (Fig. 5C; see the calibration tests and the data of individual mice in Fig. S12 A to C).
Figure 5.
(See previous page). Expression of LAMP2A in MRL/lpr B cells and effect of the P140 peptide. (A to C) Splenic cells from 10–12-wk-old CBA/J and MRL/lpr mice (4 mice of each strain) were stained with LysoTracker Green, LysoSensor Green or LysoSensor Yellow/Blue and acquired by flow cytometry. Representative flow cytometry images (filled histograms, unstained cells; empty histograms, stained cells) and relative fluorescence values of LysoTracker Green (A) and LysoSensor Green (B). In (C), the ratio LysoSensor Green over LysoTracker Green fluorescence values was calculated and compared to control CBA/J mice. (D) Lysosomal pH measurement of splenocytes isolated from CBA/J and MRL/lpr mice with ratiometric dye LysoSensor Yellow/Blue DND-160. The pH values of lysosomes were calculated from the linear standard curve established in each individual assay (see Fig. S12A for this experiment). The values are the mean + SD of measurements performed in 3 mice with 3 replicates each. The error bars are the SD values of 3 replicates (the results of 3 individual mice are shown in Fig. S12C). (E, F) western immunoblotting experiments were performed for detecting the expression of LAMP2A (antibody ab18528), LAMP1 (antibody ab24170) and CTSD (antibody ab75852) in purified B cells. Comparison was made between CBA/J mice, MRL/lpr mice and P140-treated MRL/lpr mice (3 10–13 wk-old mice/group). MRL/lpr mice received 100 μg P140/mouse/injection either once (and cells were collected 5 d later) or 6 times at daily intervals, and cells were collected 1 d after the last injection. #, bands corresponding to the immunoglobulin heavy chain level visible in MRL/lpr extracts due to the presence of important amounts of remaining autoantibodies. The histograms represent the mean ratio of 3 mice + SD between band intensity corresponding to LAMP2A, LAMP1 and CTSD over β-ACTA in the different study groups tested in at least 2 independent experiments (quantification using ImageJ software). Ratios have been arbitrarily normalized to normal mice (D) or untreated MRL/lpr mice (E). P values are indicated (Student t test).
To date, the best method to directly assess CMA activity in vivo is by substrate uptake experiments with lysosomes isolated from the organ of interest.12 Because this type of lysosome isolation was not feasible in primary B cells, we analyzed instead possible changes in levels of LAMP2A as an indirect indication of CMA activity. Lysosomal levels of LAMP2A are limiting for CMA and often correlate with changes in the activity of this pathway.13,28 Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analyses showed that in B cells from 5-wk-old or 9–12-wk-old MRL/lpr mice, the relative expression of Lamp2a (and Lamp2) was unchanged or marginally enhanced (fold change ≤ 2) with regard to CBA/J mice (Fig. S13). However, immunoblot analysis for LAMP2A protein, revealed a consistent and significant increase in LAMP2A levels in purified MRL/lpr B cells compared to B cells from healthy age-matched CBA/J mice (Fig. 5E). A similar trend was observed for LAMP1 expression while expression of the lysosomal protease CTSD was strongly augmented.
Our findings generated using several independent approaches strongly support the view that at least a pool of lysosomes are defective in MRL/lpr B cells. At this point, it is not known if all B cells subsets are affected and if the same observation could be made in all B cell compartments. It is not known either if other APCs are also affected in MRL/lpr mice and other lupus mouse models.
P140 induces changes in lysosomes
Finally, we examined whether the P140 peptide influences lysosomal dysfunctions observed in MRL/lpr mice. Remarkably, in P140-treated MRL/lpr mice that received the peptide once, and even more strikingly when they received the peptide 6 times at daily intervals, we observed a highly significant reduction of the LAMP2A (and also CTSD) expression in purified B cells (Fig. 5F). A similar trend was observed for LAMP1 expression. The lack of change at the mRNA level (Fig. S13) suggests that the decrease in LAMP2A levels upon treatment with P140 must be due, for the most part, to changes in the stability of LAMP2A, which incidentally has been shown to be dependent on lysosomal lumenal chaperones.24
Overall, our results strongly suggest a negative impact of the P140 peptide on the LAMP2A-HSPA8 lysosomal axis and lead us to conclude that downregulation of CMA activity is an important component of the therapeutic effect of this peptide in lupus.
Discussion
In this work, we have identified CMA as a target of the P140 peptide, which was previously shown beneficial in the treatment of lupus in human and in mouse models of this disease. The inhibitory effect of P140 on CMA seems to result, for the most part, from the effect of this peptide on the HSPA8 heterocomplex of lysosomal chaperones. The marked expansion in MRL/lpr B cells of the lysosomal compartment, along with a measurable raise of the average pH in acidic vesicles in splenocytes, and increased levels of CMA proteins observed in the lupus mouse model along with the efficient reduction of these markers by the P140 peptide provide rationale to further explore the potential therapeutic value of CMA downregulation.
HSPA8 and HSP90AA1 localize both at the lumenal and cytosolic lysosomal membrane sides, from where they modulate different steps of CMA.12 Cytosolic HSPA8 is responsible for substrate targeting and for regulation of the dynamics of the CMA translocation complex.24,29 The role of cytosolic HSP90AA1 in CMA is still unclear but has been proposed to contribute, in coordination with HSPA8, to substrate unfolding, which is required before translocation into the lumen.30 Further studies support a role for lumenal form of HSP90AA1 in the stabilization of LAMP2A as it organizes into the CMA translocation complex.24 The role of lumenal HSPA8 has been more extensively characterized as the presence of this lumenal form of HSPA8 is an essential requirement for CMA.21,23 In fact, only the subset of cellular lysosomes containing HSPA8 in their lumen are competent for CMA activity21 and blockage of this chaperone with specific antibodies delivered by endocytosis abolished internalization of substrates.23 Levels of lysosomal HSPA8 increase proportionally to the rise of CMA activity and the lumenal fraction of this chaperone becomes limiting when LAMP2A is in excess.21,26 The overall expansion of the lysosomal compartment in the splenic B cells of the MRL/lpr murine model of lupus and the increase in the 2 key CMA components LAMP2A (in this work) and HSPA8 (our previous studies7) provide strong evidence for modifications in CMA activity in this disease. Interestingly, the mechanisms that modulate these changes in these essential CMA components are different. Whereas HSPA8 protein expression correlates with its increased mRNA expression in MRL/lpr splenocytes,7 variations in mRNA are not observed for Lamp2a. Levels of LAMP2A in lysosomes are modulated in part by changes in the degradation of this protein in lysosomes,28 making possible that modification in the stability of LAMP2A occurs in the lupus model. Interestingly, inhibition of lysosomal HSP90AA1 leads to rapid destabilization of LAMP2A,24 suggesting that the low levels of LAMP2A observed upon P140 peptide treatment could indeed be a result of the interference of this peptide with the HSPA8 heterocomplex containing HSP90AA1.
These alterations in the lysosomal compartment (size and acidity) and the enhancement of CMA components observed in the B cells of the strain of lupus mice used in this study may favor an overdelivery to or a qualitative change of autoantigens presented in the context of MHCII molecules (most lysosomal hydrolytic enzymes such as cathepsin proteases are exquisitely pH sensitive). In this respect, although CMA has been shown to mainly lead to complete degradation of the proteins internalized by this pathway, studies using professional APCs have proposed that CMA may contribute to antigen presentation as overexpression of LAMP2A in these cells increases MHCII loading.31 At this stage it is not known whether this augmentation is directly linked to the activated status of B cells or, for instance, to an effect of internal pH of (some) MRL/lpr B cell lysosomes and if the same process also occurs in other APCs, i.e., dendritic cells, macrophages, or certain epithelial cells. It is worth mentioning that Kupffer cells (macrophages present in the liver) in MRL/lpr mice exhibit an increased number of lysosomes.32
The P140 peptide, by acting as a ligand of HSPA8, impairs its chaperoning functions in part by altering the interaction of HSPA8 with HSP90AA1. Although the HSPA8 and HSP90AA1 chaperone system can be detected in the cytosol where it contributes to modulate the protein folding stage, both proteins are also present in the lysosomal lumen.24 The fact that P140 accumulates in the lysosomal lumen and that the inhibitory effect of P140 on lysosomes could only be attained when P140 is delivered by endocytosis and not if the peptide is presented from the cytosolic side of lysosomes, strongly suggest that P140 mediates disruption of HSPA8-HSP90AA1 heterocomplexes that may be present in the lysosomal lumen. Dysfunction of lumenal HSPA8 may thus directly compromise CMA by interfering with the function of this chaperone in internalization of substrate proteins, whereas loss of lysosomal HSP90AA1 may be behind the destabilization of LAMP2A in lysosomes. We propose that this dual effect of P140 on CMA interferes with loading of cellular autoantigens to MHCII molecules leading to a weaker activation of autoreactive T cells.5,11,33 Interestingly, the disruptive effect of P140 on the ability of HSPA8 to bind substrate proteins may be selective for a subset of CMA substrates. For example, our ELISA analysis reveals strong competition by GAPDH (that contains an atypical CMA targeting motif where Q is replaced by N) while, by comparison, the efficacy of RPL5 (that contains a CMA targeting motif that forms upon phosphorylation of a Y residue) in this assay was lower. Future studies on the structural basis of HSPA8 binding to different CMA targeting motifs are required to clarify these inhibitory differences of P140, but overall they support the above mentioned possible selectivity of the effect inhibitory effect of the peptide toward a subset of autoantigens.
The fact that the P140 peptide enters MRL/lpr B lymphocytes via a clathrin-dependent endo-lysosomal pathway supports this mechanism. Yet it is not known whether P140 uses the same entry pathway in other (normal and autoimmune) cell types and if it recirculates at the membrane. The receptor and the adaptor or accessory molecules used by the P140 peptide to be endocytosed remain also unknown. Although most of the effect of P140 on CMA seems to be a consequence of its accumulation into the lysosomal lumen, we do not know if some of the P140 peptide could reach the cytosol and interfere there with other HSPA8 functions such as clathrin disassembly or the recently described targeting of cytosolic proteins to multivesicular bodies by endosomal microautophagy.34 The changes of lysosomal pH we discovered in lupus splenocytes compared to normal splenocytes, might critically affect the coordinated biochemical reactions occurring along both the endocytic and secretory pathways. To this regard, the fact that the P140 peptide seems to be more stable at higher pH (5.3 and 7.2 compared to 4.2) is certainly important in its mode of action and might play a crucial role in its selective effect toward some altered lysosomes naturally occurring with time in diseased lupus mice. These differences in pH may work favorably for the effect of P140 on lysosomal HSPA8 since increase in lumenal pH has been previously shown to markedly affect the stability of this chaperone. It is thus possible that pH-induced changes in lysosomal HSPA8 conformation in lupus cells make them particularly amenable for the interaction with P140.
The selective inhibitory effect of the P140 peptide on CMA described in this study opens now the possibility of using this peptide to modulate other conditions in which CMA downregulation may be desirable. For example, we described that CMA is constitutively upregulated in many cancer cell types and that inhibition of CMA through genetic manipulation leads to reduced tumorogenesis, metastasis, and shrinkage of the primary tumor.35 Consequently, P140 may have potential antioncogenic applications in some of these instances. In fact, until now, the few compounds for which an inhibitory effect on CMA has been proposed lack selectivity.36,37 For example the protein synthesis inhibitors anisomycin and cycloheximide can inhibit CMA but also affect macroautophagy and many other cellular processes. Although inhibition of both autophagic pathways may have additive effect on these cells, from the therapeutic point of view it could be less desirable. The antimalarial drug hydroxychloroquine (HCQ) prescribed for SLE also exerts its effects via multiple (nonspecific) molecular pathways that differ (and are sometimes opposite) according to cell subsets,38 and nowadays it is not clear which one prevails in its beneficial action. As HCQ is a weak base, it has been notably claimed to operate as an alkalinizing lysomotropic agent, which acts as inhibitor of both macroautophagy and CMA pathways by interfering with lysosomal degradation, a step shared by these 2 pathways. While it is useful in treating patients, HCQ possesses some toxicity and can induce deleterious side effects, making compounds that only act in one of these pathways more attractive therapeutically.
In conclusion, we propose that the P140 peptide, which ameliorates the course of the lupus disease in patients and mice, enters B cells via a clathrin-dependent endosomal pathway and that its accumulation in lysosomes compromises CMA, at least in part, by disruption of the lysosomal lumenal HSPA8 heterocomplexes containing HSP90AA1. The consequence of the inhibitory effect on CMA may be a slowing down or a qualitative change of cellular autoantigen loading to MHCII molecules and as a result a weaker priming of autoreactive T cells. This will potentially end up by a beneficial reduction of autoreactive B cell proliferation and differentiation into deleterious autoantibody-secreting plasma cells, leading to an improvement of the autoimmune status observed in patients with SLE.
Materials and Methods
Synthetic peptides and anti-peptide antibodies
The P140 peptide as well as analogs ScP140, 131–148 P140 and 131–151 were synthesized using classical N-(9-fluorenyl) methoxycarbonyl solid-phase chemistry and purified by reversed-phase high-performance liquid chromatography (RP-HPLC).1,6 Their homogeneity was checked by analytical HPLC, and their identity was assessed by liquid chromatography-mass spectrometry (LC-MS) on a Finnigan LCQ Advantage Max system (Thermo Fischer Scientific, Courtaboeuf, France). For labeling purposes, the P140 and ScP140 peptides were also synthesized with a cysteine residue added at their N terminus (Cys-P140, Cys-ScP140). Alexa Fluor 488 Cys-P140, Cys-131–151 and Cys-ScP140 peptides were prepared by reacting equimolar amounts of the maleimide derivatives (Life technologies, 1–10254) and peptides in dimethylformamide. The conjugates were purified by HPLC and lyophilized in the dark.
Stability of P140 and the P140 phosphoryl moiety in different media at different pH
The stability of peptides was tested in vitro as described previously.39,40 Peptides (1 mg/mL) were incubated at 37°C with pooled fresh serum from MRL/lpr mice. The reaction was stopped at different intervals by adding trifluoroacetic acid (TFA; 10% v/v of the final volume) and the samples were diluted 5 times in water. After centrifugation, the supernatants were collected and the extent of peptide degradation was estimated by RP-HPLCanal on a Beckman Coulter gold 126 instrument (Villepinte, France) equipped with a ProStar 410 autosampler (Beckman Coulter, Villepinte, France) and a Beckman Coulter gold 166 NMP detector (Villepinte, France), and using a Macherey-Nagel Nucleodur 100–3 C18 column (720120.46) (gradient: 5 to 65% B in 20 min at 1.2 mL/min flow rate; λ = 220 nm). Eluents for analytical RP-HPLC were (A) H2O + 0.1% TFA and (B) CH3CN + 0.08% TFA. The resistance of peptides to enzymatic degradation was evaluated from the area of the peak corresponding to the intact peptide remaining at several time intervals. The stability of peptides was also studied in ALF20 at different pH (4.2, 5.3, and 7.2), supplemented with either CTSD and CTSL or extracted lysosomal matrix. The choice of CTS introduced in ALF (D and L in this study) was predetermined according to the amino acid composition of the P140 peptide. Briefly, 40 μM of peptide was incubated in the corresponding ALFs at 37°C for indicated time points followed by addition of TFA to prevent further degradation of the peptides by lysosomal proteases. The supernatant was subjected to RP-HPLC and mass analysis using LC-MS. Eluents for analytical RP-HPLC were (A) H2O + 0.1% formic acid and (B) CH3CN + 0.08% formic acid. ESI mass spectra were recorded on Thermo Fisher Finnigan LCQ Advantage Max instrument.
Cloning, expression, and purification of the recombinant human HSPA8 and HSPA8 fragments
Applying the retrotranscription PCR experiment, the full-length as well as the N-terminal fragment 1–386 (NBD) and the C-terminal fragment 387–646 (SBD) were amplified from Raji cells with synthetic primers. The full-length HSPA8 forward primer (GCCATATGTCCAAGGGACCTGC) was identical to the coding sequence of human HSPA8 except that a restriction site NdeI was added at the 5′ end of the primer. Full-length HSPA8 reverse primer (GCGGATCCTTAATCAACCTCTTCAATGG) was complementary to the coding sequence around the termination codon, except that a BamHI site was added at the 3′ end of the coding strand. The NBD fragment for the forward primer was the same as that used for full-length HSPA8, the NBD fragment for the reverse primer (GCGGATCCCTAAGACTTGTCTCCAGAC) was complementary to the coding sequence around the codon 386 of HSPA8 with a BamHI site added at the 3′ end of the coding strand. The SBD reverse primer was the same as the one used for full-length HSPA8 and the SBD forward primer (GCCATATGAATGTCCAAGATTTGCTGC) was complementary to the coding sequence around the codon 387, except that an NdeI site was added at the 5′ end of the coding strand. The PCR products were then cloned into a pCR®TOPO 2.1 vector (Life technologies, K4500–01) and sequenced. The digested cDNA fragment of HSPA8 was ligated in pET-15b vector (Novagen, 69661) linearized with both NdeI and BamHI. The resulting plasmids contained a sequence coding for a tag of 6 histidine residues at the N terminus of HSPA8 fragments. The recombinant proteins were expressed in Origami rosetta B (Novagen, 71137) and purified using Qiagen Ni-NTA Superflow resin (Invitrogen, R901–01). The purity of 3 proteins (full length, NBD and SBD) was checked by SDS-PAGE and colloidal blue staining.
Immunocytochemical analyses
B cells were purified from splenocytes using Pan B Cell Isolation kit (Miltenyi, 130–090–862) according to the manufacturer's protocol. Their purity was verified by flow cytometry. For endocytosis inhibition studies, total splenocytes or purified B cells were pretreated with endocytosis inhibitors for 30 min at 37°C, and then loaded with Alexa Fluor 488-labeled P140 peptide at 10 μM for 30 min at 37°C. In some experiments, Alexa Fluor 488-labeled ScP140 control peptide was used at the same concentration. All these experiments were conducted in PBS (Lonza, BE17–516F) supplemented with 2% (v/v) fetal calf serum (FCS; Dominique Dutscher, S1800–500) and the inhibitors were kept during the entire duration of the experiments. Ten μg/mL CPZ (Sigma-Aldrich, C-8138) was used to inhibit clathrin-mediated endocytosis, 5 μg/mL filipin (Sigma-Aldrich, F-4767) to disrupt caveolae-dependent endocytosis, and 5 mM methyl-β-cyclodextrin (Sigma-Aldrich, C-4555) to block macropinocytosis. As a positive control, cells were also incubated at 4°C to inhibit internalization. The inhibitory effect of the compounds was verified with well-known fluorescent substrates for each endocytosis pathway, namely 100 μg/mL TF/transferrin (Life technologies, T13342) for CPZ, 0.5 μM BODIPY FL C5- Lactosylceramide/bovine serum albumin (Life Technologies, I34402) for filipin and 250 μg/mL dextran (Life Technologies, D1820) for methyl-β-cyclodextrin. After incubation, cells were washed twice with PBS-2% FCS and fixed with 4% paraformaldehyde. Cells were then permeabilized with 0.05% (v/v) of Triton X-100 (Pharmacia Biotech, 17–1315–01), stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, D1306), and finally mounted on glass slides with fluorescent mounting medium (DAKO, S302380). Images were acquired with a Zeiss Axiovert LSM700 confocal microscope (Jena, Germany) with a 63× oil immersion objective.
For in vitro colocalization studies, B cells were incubated in the presence of 10 μM Alexa Fluor 488-labeled P140 peptide or Alexa Fluor 488-labeled peptide 131–151, for 15 min (for colocalization with EEA1) or 2 h (for colocalization with RAB9 or LAMP2/2A) at 37°C in PBS-2% FCS. Cells were washed, fixed, permeabilized as described above, and then incubated with polyclonal antibodies to EEA1, RAB9, and LAMP2 (Santa Cruz Biotechnology, sc-6415, sc-28573 and sc-8100, respectively), or LAMP2A. For EEA1, RAB9, and LAMP2 staining, cells were then successively incubated with biotin-coupled anti-goat antibody (Jackson Immunoresearch, 705–065–147), Alexa Fluor 546-labeled streptavidin (Molecular Probes, S11225) and DAPI. For LAMP2A staining, cells were incubated with Alexa Fluor 647-coupled antibody (Molecular Probes, A31573) and DAPI. They were finally mounted on glass slides and images were acquired as described above. For colocalization of the P140 peptide with TF, B cells were incubated in the presence of 10 μM Alexa Fluor 488-labeled P140 peptide and 100 μg/mL Alexa Fluor 546-labeled TF (Life Technologies, T-23364) for 30 min at 37°C in PBS-2% FCS. Cells were washed, fixed and permeabilized. They were then stained with DAPI, mounted on a glass slides, and images were acquired as above.
For immunofluorescence studies after in vivo administration, 100 μg of Alex Fluor 488-labeled peptides were injected iv and splenocytes were collected 3 h or 6 h later and stained as described above with anti-EEA1, RAB9 or LAMP2 antibodies, respectively. For lysosomal and mitochondrial staining, 1 h after injection, splenocytes were incubated with 100 nM LysoTracker Red-DND 99 or 500 nM MitoTracker Deep Red FM (Life Technologies, L-7528 and M22426, respectively) for 30 min at 37°C in complete RPMI medium. After washings in the same medium, images were immediately acquired on the confocal microscope as described above.
Coimmunoprecipitation
HSPA8 CoIP experiments were done using solubilized P140-treated Raji cells in CoIP buffer corresponding to complete Lysis-M buffer supplemented with protease inhibitors cocktail (Roche, 04 693 124 001). Briefly, anti-HSPA8 antibody (clone 1B5; Abcam, 19136) was added for 2 h on P140-treated Raji cell lysates, and agarose-protein G beads (Millipore, 16–266) were added subsequently for 2 h. Beads were washed 3 times in CoIP buffer and proteins were eluted in Laemmli buffer. HSPA8-interacting proteins were then subjected to either denaturing SDS-PAGE or blue native electrophoresis followed by western immunoblotting analysis with antibodies to HSP90AA1 (clone 16F1; Enzo Life Science, ADI-SPA-835-D) or to HSPA8 (clone 1B5).
Luciferase renaturation assay
The test was carried out essentially as previously described41 using either intact or heat-denatured luciferase (Sigma Aldrich, L4899; 30 min at 40°C) and rabbit reticulocyte lysate (Promega, L4960) in the presence of increasing concentrations of the P140 peptide (0 to 40 μM). Luciferase activity was determined using Bright-glo reagent (Promega, E2610).
Measurement of ATPase activity
ATPase activity measurements were performed as described.42 Briefly, HSPA8 (0.5 μM; Enzo Life Sciences, ADI-SPP-751-D) and in some experiments HSP40 (0.5 μM; Enzo Life Sciences, ADI-SPP-400-D) were suspended in assay buffer (0.02% Triton X-100, 100 mM Tris-HCl, 20 mM KCl, and 6 mM MgCl2, pH 7.4). This solution was distributed into a 96-well plate with 100 μM ATP and incubated with increasing concentrations of different peptides used as competitors. After 3 h at 37°C, ATPlite 1-step reagent (PerkinElmer, 6016736) was added to detect ATP in assay buffer, and luminescence signal was measured on a Victor2 reader (Wallac, Turku, Finland).
Measurement of CMA activity in culture cells
CMA activity was assessed in NIH3T3 cells stably expressing the photoactivable CMA reporter KFERQ-PA-mCherry1.25 Cells were photoactivated as described previously and maintained in serum-free medium to maximally activate CMA with or without supplementation of 10 and 30 μM peptides P140, ScP140 or 131–151 (unphosphorylated) for 12 h. Quantification of the average number of fluorescence puncta per cell was done in a minimum of 50 cells using ImageJ software (NIH). In some experiments, a second reporter based on a variation of the previously published photoswitchable25 version of the CMA reporter was also used. Briefly, NIH3T3 cells stably expressing a construct with a KFERQ-containing region in frame with the photoswitchable fluorescent protein Dendra2, were exposed to a 405 nm light-emitting diode lamp for photoswitching and the association of the red-converted protein to lysosomes was monitored as formation of fluorescent red puncta. In these studies, cells were plated in glass-bottom 96-well plates and image analysis was performed using the high-content microscopy Operetta system (PerkinElmer, Waltham, Massachusetts). The average number of puncta per cell was calculated in 9 different fields for a total of > 700 cells. Where indicated CMA activity was upregulated by addition of paraquat (50 μM; Sigma, P-856177) directly to the culture media.
Measurement of CMA activity in vitro
Direct binding and uptake of CMA substrates by lysosomes were measured using a previously standardized in vitro assay.20 Briefly, lysosomes (75 μg protein) isolated from mouse liver, and treated with or without a cocktail of protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (100 μM; Fisher Bioreagent, BP2644), leupeptin (100 μM; Fisher Bioreagent, BP2662), pepstatin (10 μM; Sigma, P-4265), and ethylenediaminetetraacetic acid (100 μM) upon isolation, were incubated in an isotonic medium [10 mM MOPS (Sigma-Aldrich, M3183), 0.25 M sucrose (American Bioanalytical, C12H22011), pH 7.3] with GAPDH (25 μg; Sigma-Aldrich, G2267) for 20 min at 37°C. At the end of the incubation, lysosomes were recovered by centrifugation and the amount of GAPDH associated to the organelles was assessed by SDS-PAGE and immunoblot for GAPDH (Abcam, 8245). Quantification was performed by densitometric analysis of the immunobloted membranes using the square method of ImageJ. GAPDH associated to untreated lysosomes represents protein bound to the lysosomal membrane, whereas uptake can be calculated as the difference in the amount of GAPDH between lysosomes treated with or without protease inhibitors.
Quantification of LAMP2A expression
For measuring mRNA expression levels, total RNA was extracted from purified B cells using RNeasy Mini kit (Qiagen, 74704) according to the manufacturer's protocol. After treatment with DNase (Qiagen, 79254) to degrade residual genomic DNA, the first strand cDNA was synthetized from 200 ng of RNA using ImProm-II Reverse Transcriptase and oligo(dT)15 (Promega, A3802 and C1101, respectively). Ten ng of newly cDNA templates were then amplified using RT2 SYBR Green Rox qPCR Mastermix (Qiagen, 330520) with primers specific for exon 9 of mouse Lamp2a and housekeeping genes Actb, Ppia, and Rpl13a. Amplification of DNA products was measured on Step One apparatus (Applied Biosystems). The following primers, designed to span 2 exons, were used: Lamp2a 5′-AAAAGGACAGTATTCTA CAGCTCAAGACT-3′ and 5′-AATAAAATAAGCCAGCAACACTAGAATAAG-3′; β-ACTA 5′-ACGGCCAGGTCATCACTATTG-3′and 5′-CACAGGATTCCATACCCAAGA AG-3′; Ppia 5′-GTGCCAGGGTGGTGACTTTAC-3′and 5′-TGCCAGGACCTG TATGCTTTAG-3′; Rpl13a 5′-GGGCAGGTTCTGGTATTGGA-3′ and 5′-GAAGTACCTTGGCCACAA-3′. The expression levels of Lamp2a were normalized to those of housekeeping genes.
For measuring LAMP2A protein expression levels, purified B cells were lysed in complete Lysis-M buffer supplemented with protease inhibitors cocktail (Roche, 04693124001), and LAMP2A expression levels in the proteins extracts were measured by western immunoblotting using LAMP2A specific antibody (Abcam, 18528). LAMP1 and CTSD were revealed with specific antibodies from Abcam (24170 and 75852, respectively). The expression of ACTB protein was used as standard in each blot.
Amount and pH of lysosomes
Splenocytes were loaded with 100 nM of LysoTracker Green DND-26 or 1 μM of LysoSensor Green DND-189 (Life Technologies, L-7526 and L-7535, respectively) for 15 min at 37°C in prewarmed complete RPMI medium. Stained cells were washed twice in the same medium and stained with allophycocyanin-labeled anti-CD19 antibody (BD Biosciences, 561738). Data were collected on a Gallios flow cytometer (Beckman Coulter) and analyzed using FlowJo software (Tree Star). Differences in lysosome amounts between CBA/J and MRL/lpr B cells were assessed by normalizing the LysoTracker Green staining of CD19+ cells from MRL/lpr mice to the one with CD19+ cells from CBA/J mice. For lysosomal pH comparison, we normalized the LysoSensor Green staining of CD19+ cells to the LysoTracker Green one, and the ratio calculated for MRL/lpr cells was compared to the one calculated of CBA/J cells. Alternatively, lysosomal pH measurements were also performed using splenocytes collected from CBA/J and MRL/lpr mice and stained for 5 min at 37°C with 5 μM LysoSensor Yellow/Blue DND-160 (Molecular Probes, L7545) in RPMI medium. Stained cells were washed twice in cold PBS, followed by fluorescence measurement in a fluorescence spectrophotometer (Mithras) at an excitation wavelength of 360 nm and at 2 emission wavelengths, namely 460 nm (blue) and 535 nm (yellow). The blue/yellow fluorescence ratio was calculated after subtracting the background fluorescence. In situ pH calibration was performed in Raji cells to relate the blue/yellow fluorescent ratio to lysosomal pH according to the protocol described previously.43,44 Briefly, lysosomes were stained with LysoSensor Yellow/Blue DND-160 as above, and lysosomal pH was equilibrated in a series of 2-(N-morpholino)ethanesulfonic acid buffers containing ionophores [5 mM NaCl, 115 mM KCl, 1.2 mM MgSO4,7H2O, 25 mM 2-(N-morpholino)ethanesulfonic acid (Sigma, M2933), 10 μM monensin (Sigma, M5273), 10 μM nigericin (Sigma, N7143)] over the pH range of 4.01 to 6.51. NH4Cl was used as a positive control to increase the lysosomal pH.45,46 The pH values of lysosomes was calculated from the linear standard curve.
Competitive ELISA
For competitive ELISA assays using different proteins in solution as inhibitor, increasing amounts of these proteins (and P140 as internal control) were first incubated 1 h at 37°C with a constant concentration of HSPA8 (25 ng/mL) in PBS containing 0.05% (v/v) Tween 20 (Sigma, P1379) (PBS-T). The mixtures were subsequently transferred to polyvinyl microtiter plates (Falcon, 3912) precoated overnight at 37°C with 0.1 μM P140 in 0.05 M carbonate buffer, pH 9.6, and postcoated with PBS-T containing 1% (w/v) BSA (Aurion, 900.011) (PBS-T-BSA). After a 1 h-incubation at 37°C and washing, bound HSPA8 was detected using rabbit antibodies (Ig fraction, 60 μg/mL) generated against the C terminus of HSPA8 and then a peroxidase-conjugated secondary antibody to rabbit IgG (Fc specific) diluted in PBS-T (Jackson ImmunoResearch Laboratories, 111–035–008; dilution 1/20,000). Certain tests used 12.5 ng/mL HSPA8 and 2 μM immobilized P140. Final revelation was made with H2O2 as substrate and 3,3′,5,5′-tetramethyl benzidine as chromogen. Absorbance was measured at 450 nm. Proteins used as inhibitors were GAPDH, ovalbumin (Sigma-Aldrich, S7951) and RPL5 (Abnova, H00006125-P01). For the test used to measure the binding of the P140 peptide to HSPA8 fragments, increasing concentrations of recombinant HSPA8 proteins containing a polyhistidine-tag at their N terminus were added for 1 h at 37°C to P140 peptide-coated plates (2 μM peptide) and their binding was revealed with peroxidase-conjugated anti-His tag antibodies (Sigma-Aldrich, H1929; 1:10,000 in PBS-T). The subsequent steps of the test were as described above.
Statistical analysis
Statistical tests were performed using GraphPad Prism version 5.0. Statistical significance was assessed using the Student t test. P values less than 0.05 were considered significant.
Acknowledgments
We gratefully acknowledge Frédéric Gros for valuable discussions, and Olivier Chaloin, Hayet Dali, Monique Duval and Jean-Daniel Fauny for expert technical help.
Disclosure of Potential Conflicts of Interest
CM and NP were supported in part by an ImmuPharma France doctoral fellowship. NS is a salaried employee of ImmuPharma. SM and JPB are consultants of ImmuPharma. The other authors have no conflicting financial interest.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Ethics Statement
All experimental protocols were carried out with the approval of the local Institutional Animal Care and Use Committee (CREMEAS).
Funding
This research was funded by the French Center National de la Recherche Scientifique and the Laboratory of Excellence Medalis, Initiative of Excellence (IdEx), Strasbourg University and the National Institute on Aging of the National Institute of Health. CM was supported by a Region Alsace and ImmuPharma France doctoral fellowship. FW was supported by a post-doctoral grant from the University of Strasbourg Institute for Advanced Study (USIAS). IT was supported by a Fulbright fellowship from the Ministerio de Educacion y Ciencia.
References
- 1. Monneaux F, Lozano JM, Patarroyo ME, Briand JP, Muller S. T cell recognition and therapeutic effects of a phosphorylated synthetic peptide of the 70K snRNP protein administered in MRL/lpr lupus mice. Eur J Immunol 2003; 33:287-96; PMID:12548559; http://dx.doi.org/ 10.1002/immu.200310002 [DOI] [PubMed] [Google Scholar]
- 2. Muller S, Monneaux F, Schall N, Rashkov RK, Oparanov BA, Wiesel P, Geiger JM, Zimmer R. Spliceosomal peptide P140 for immunotherapy of systemic lupus erythematosus. results of an early phase II clinical trial. Arthritis Rheum 2008; 58:3873-83; PMID:19035498; http://dx.doi.org/ 10.1002/art.24027 [DOI] [PubMed] [Google Scholar]
- 3. Zimmer R, Scherbarth HR, Rillo OL, Gomez-Reino J, Muller S. Lupuzor/P140 peptide in patients with systemic lupus erythematosus: a randomised, double-blind, placebo-controlled phase IIb clinical trial. Ann Rheum Dis 2013; 72:1830-5; PMID:23172751; http://dx.doi.org/ 10.1136/annrheumdis-2012-202460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dieker J, Cisterna B, Monneaux F, Décossas M, van der Vlag J, Biggiogera M, Muller S. Apoptosis changes the phosphorylation status and subcellular localization of the spliceosomal autoantigen U1-70K. Cell Death Diff 2008; 15:793-804; PMID:18202700; http://dx.doi.org/ 10.1038/sj.cdd.4402312 [DOI] [PubMed] [Google Scholar]
- 5. Monneaux F, Hoebeke J, Sordet C, Nonn C, Briand JP, Maillère B, Sibillia J, Muller S. Selective modulation of CD4+ T cells from lupus patients by a promiscuous, protective peptide analogue. J Immunol 2005; 175:5839-47; PMID:16237076; http://dx.doi.org/ 10.4049/jimmunol.175.9.5839 [DOI] [PubMed] [Google Scholar]
- 6. Page N, Schall N, Strub JM, Quinternet M, Chaloin O, Décossas M, Cung MT, Van Dorsselaer A, Briand JP, Muller S. The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by γδ T cells. PloS ONE 2009; 4:e5273; PMID:19390596; http://dx.doi.org/ 10.1371/journal.pone.0005273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Page N, Gros F, Schall N, Décossas M, Bagnard D, Briand JP, Muller S. HSC70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHCII presentation in murine lupus. Ann Rheum Dis 2011; 70:837-43; PMID:21173017; http://dx.doi.org/ 10.1136/ard.2010.139832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H., et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA 2005; 102:7922-7; PMID:15894616; http://dx.doi.org/ 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/ 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Randow F, Münz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol 2012; 33:475-87; PMID:22796170; http://dx.doi.org/ 10.1016/j.it.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monneaux F, Parietti V, Briand JP, Muller S. Importance of spliceosomal RNP1 motif for intermolecular T-B cell spreading and tolerance restoration in lupus. Arthritis Res Ther 2007; 9:R111; PMID:17963484; http://dx.doi.org/ 10.1186/ar2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 2012; 22:407-17; PMID:22748206; http://dx.doi.org/ 10.1016/j.tcb.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996; 273:501-3; PMID:8662539; http://dx.doi.org/ 10.1126/science.273.5274.501 [DOI] [PubMed] [Google Scholar]
- 14. Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 2009; 458:475-80; PMID:19325628; http://dx.doi.org/ 10.1038/nature07851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schall N, Page N, Macri M, Chaloin O, Briand JP, Muller S. Peptide-based approaches to treat lupus and other autoimmune diseases. J Autoimmun 2012; 39:143-53; PMID:22727561; http://dx.doi.org/ 10.1016/j.jaut.2012.05.016 [DOI] [PubMed] [Google Scholar]
- 16. Stricher F, Macri C, Ruff M, Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy 2013; 9:1937-54; PMID:24121476; http://dx.doi.org/ 10.4161/auto.26448 [DOI] [PubMed] [Google Scholar]
- 17. Takeda S, McKay DB. Kinetics of peptide binding to the bovine 70 kDa heat shock cognate protein, a molecular chaperone. Biochem 1996; 35:4636-44; PMID:8605215; http://dx.doi.org/ 10.1021/bi952903o [DOI] [PubMed] [Google Scholar]
- 18. Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Molec Cell Biol 2010; 11:579-92; PMID:20651708; http://dx.doi.org/ 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rosales-Reyes R, Pérez-López A, Sánchez-Gómez C, Hernández-Mote RR, Castro-Eguiluz D. Ortiz-Navarrete V, Alpuche-Aranda CM. Salmonella infects B cells by macropinocytosis and formation of spacious phagosomes but does not induce pyroptosis in favor of its survival. Microb Pathog 2012; 52:367-74; PMID:22475626; http://dx.doi.org/ 10.1016/j.micpath.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 20. Beeston MP, van Elteren JT, Selih VS, Fairhurst R. Characterization of artificially generated PbS aerosols and their use within a respiratory bioaccessibility test. Analyst 2010; 135:351-7; PMID:20098770; http://dx.doi.org/ 10.1039/b910429a [DOI] [PubMed] [Google Scholar]
- 21. Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem 1997; 272:5606-15; PMID:9038169; http://dx.doi.org/ 10.1074/jbc.272.9.5606 [DOI] [PubMed] [Google Scholar]
- 22. Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell 2010;39:535-47; PMID:20797626; http://dx.doi.org/ 10.1016/j.molcel.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 1997;137:825-34; PMID:9151685; http://dx.doi.org/ 10.1083/jcb.137.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 2008; 28:5747-63; PMID:18644871; http://dx.doi.org/ 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun 2011; 2:386; PMID:21750540; http://dx.doi.org/ 10.1038/ncomms1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 2004; 15:4829-40; PMID:15331765; http://dx.doi.org/ 10.1091/mbc.E04-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gros F, Arnold J, Page N, Décossas M, Korganow AS, Martin T, Muller S. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy 2012; 8:1113-23; PMID:22522825; http://dx.doi.org/ 10.4161/auto.20275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 2000;113:4441-50; PMID:11082038 [DOI] [PubMed] [Google Scholar]
- 29. Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kgdalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989; 246:382-5; PMID:2799391; http://dx.doi.org/ 10.1126/science.2799391 [DOI] [PubMed] [Google Scholar]
- 30. Salvador N, Aguado C, Horst M, Knecht E. A role for a 70-kgdalton heat shock protein in lysosomal degradation of intracellular proteins. J Biol Chem 2000; 275:27447-56; PMID:10862611 [DOI] [PubMed] [Google Scholar]
- 31. Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. LAMP2A facilitates MHC Class II presentation of cytoplasmic antigens. Immunity 2005; 22:571-81; PMID:15894275; http://dx.doi.org/ 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 32. Cho K, Seki S, Nakatani K, Kobayashi K, Kaneda K. Kupffer cell activation and hematopoiesis in the liver of autoimmune MRL-lpr/lpr mice. Arch Histol Cytol 2000; 63:473-83; PMID:11201206; http://dx.doi.org/ 10.1679/aohc.63.473 [DOI] [PubMed] [Google Scholar]
- 33. Monneaux F, Parietti V, Briand JP, Muller S. Intramolecular T cell spreading in unprimed MRL/lpr mice: importance of the U1-70K protein sequence 131-151. Arthritis Rheum 2004; 50:3232-8; PMID:15476231; http://dx.doi.org/ 10.1002/art.20510 [DOI] [PubMed] [Google Scholar]
- 34. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 2011; 20:131-9; PMID:21238931; http://dx.doi.org/ 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett 2010; 584:1399-404; PMID:20026330; http://dx.doi.org/ 10.1016/j.febslet.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gros F, Muller S. Pharmacological regulators of autophagy and their link with modulators of lupus disease. Brit J Pharmacol 2014; 171:4337-4359; PMID:24902607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709-30; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallace DJ, Gudsoorkar VS, Weisman MH, Venuturupalli SR. New insights into mechanisms of therapeutic effects of antimalarial agents in SLE. Nat Rev Rheumatol 2012; 8:522-33; PMID:22801982; http://dx.doi.org/ 10.1038/nrrheum.2012.106 [DOI] [PubMed] [Google Scholar]
- 39. Stemmer C, Quesnel A, Prévost-Blondel A, Zimmermann C, Muller S, Briand JP, Pircher H. Protection against lymphocytic choriomeningitis virus infection induced by a reduced peptide bond analogue of the H-2Db-restricted CD8+ T cell epitope GP33. J Biol Chem 1999; 274:5550-6; PMID:10026170; http://dx.doi.org/ 10.1074/jbc.274.9.5550 [DOI] [PubMed] [Google Scholar]
- 40. Briand J-P, Benkirane N, Guichard G, Newman JFE, Van Regenmortel MHV, Brown F, Muller S. A retro-inverso peptide corresponding to the GH loop of foot-and-mouth disease virus elicits high levels of long lasting protective neutralizing antibodies. Proc Natl Acad Sci USA 1997; 94:12545-50; PMID:9356486; http://dx.doi.org/ 10.1073/pnas.94.23.12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J 1995; 14:2281-92; PMID:7774586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O'Leary J, Morgan D, Lee DC, Shults CL. Chemical manipulation of Hsp70 ATPase activity regulates tau stability. J Neuroscience 2009; 29:12079-88; PMID:19793966; http://dx.doi.org/ 10.1523/JNEUROSCI.3345-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diwu Z, Chen CS, Zhang C, Klaubert DH, Haugland RP. A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem Biol 1999; 6:411-8; PMID:10381401; http://dx.doi.org/ 10.1016/S1074-5521(99)80059-3 [DOI] [PubMed] [Google Scholar]
- 44. Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G., et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 2010; 141:1146-58; PMID:20541250; http://dx.doi.org/ 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA 1978; 75:3327-31; PMID:28524; http://dx.doi.org/ 10.1073/pnas.75.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu J, Lu W, Reigada D, Nguyen J, Laties AM, Mitchell CH. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4−/− mice: Pharmacologic approaches and functional recovery. Invest Ophthalmol Vis Sci 2008; 49:772-80; PMID:18235027; http://dx.doi.org/ 10.1167/iovs.07-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.