Abstract
As with the case of the mechanism of autophagosome formation, studies in yeast have taken a leading role in elucidating the molecular basis of target recognition during selective autophagy. Degradation targets are recognized by receptor proteins, which also bind to Atg8 homologs on growing phagophore membranes, leading to the loading of the targets into autophagosomes. However, it remains to be elucidated how these processes are regulated. In yeast, receptors also interact with the scaffold/adaptor protein Atg11, which subsequently recruits core Atg proteins onto receptor-target complexes to initiate autophagosome formation. Recently, we found that Hrr25, a homolog of CSNK1D/casein kinase 1δ, regulates 3 of 4 selective autophagy-related pathways in the budding yeast Saccharomyces cerevisiae by a uniform mechanism: phosphoregulation of the receptor-scaffold interaction.
Keywords: Ams1, Atg19, Atg34, Atg36, casein kinase 1, Cvt pathway, nitrogen starvation, pexophagy, selective autophagy, yeast
To date, 4 receptors, Atg19, Atg32, Atg34, and Atg36, have been identified in S. cerevisiae. In the cytoplasm-to-vacuole targeting (Cvt) pathway, Atg19 recognizes the cytosolic forms of the aminopeptidases Ape1 and Ape4, and the α-mannosidase Ams1, and mediates their transport into the vacuole under nutrient-rich conditions. These targets are not degraded, but function as active enzymes in the vacuole. Atg34 is a homolog of Atg19 and specifically responsible for vacuolar transport of Ams1 under nitrogen starvation conditions. Atg32 and Atg36 localize to mitochondria and peroxisomes and induce autophagic degradation of these organelles, i.e., mitophagy and pexophagy, respectively. Atg11 serves as a common scaffold in all of these pathways. Recent studies revealed that casein kinase 2 (CK2) phosphorylates Atg32 to promote its interaction with Atg11, resulting in the stimulation of mitophagy. However, the regulation of the other pathways was poorly understood.
A previous systematic mass spectrometry analysis identified Atg19 in immunoprecipitates of Hrr25. This prompted us to investigate the involvement of Hrr25 in the Cvt pathway. Indeed, depletion of Hrr25 significantly impairs the Cvt pathway. In Hrr25-depleted cells, the Atg19-prApe1 interaction is normal, but the Atg19-Atg11 interaction is severely impaired. Consistent with this, whereas Atg19 localizes to the Ape1 complex assembly site in these cells, the localization of Atg11 to the Atg19-prApe1 complex is abolished. Consequently, downstream core Atg proteins are not recruited to the complex in Hrr25-depleted cells. We found that Atg19 is a substrate of Hrr25 kinase; depletion of Hrr25 decreases Atg19 phosphorylation in vivo, and Hrr25 directly phosphorylates Atg19 in vitro. While Atg19 is phosphorylated at multiple sites, our results show that phosphorylation of Ser391 by Hrr25 is important for the Atg19-Atg11 interaction and thus for the Cvt pathway. Taken together, we propose that Hrr25 phosphorylates the receptor Atg19 and thereby enhances its interaction with the scaffold Atg11 to activate the Cvt pathway (Fig. 1A).
Figure 1.
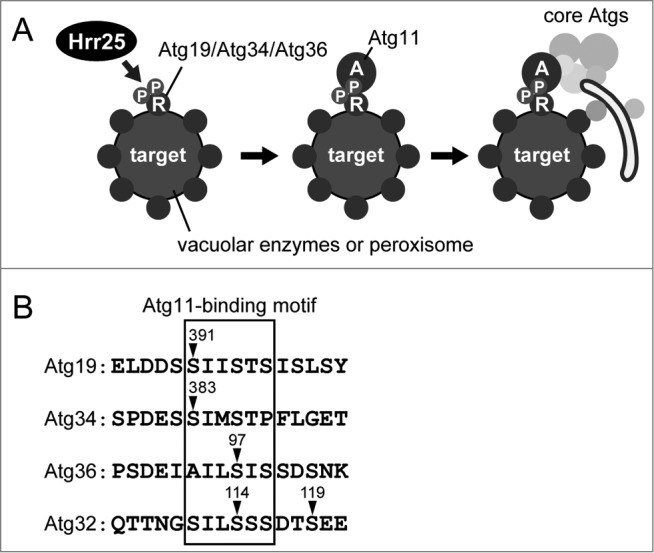
Regulation of selective autophagy-related pathways by Hrr25. (A) Schematic model. Hrr25 phosphorylates autophagy receptors (R) for vacuolar enzymes and peroxisomes. This modification (P) upregulates the interactions of the receptors with the scaffold Atg11 (A), leading to the recruitment of the core Atg proteins that mediate the formation of the autophagosomal membrane. (B) Amino acid sequences of Atg11-binding regions in autophagy receptors in S. cerevisiae. Arrowheads indicate Ser residues phosphorylated to enhance interactions with Atg11.
The Cvt pathway has often been described as constitutively active. However, we found that increased Atg19 phosphorylation by Hrr25 stimulated this pathway when yeast cells are grown to a late growth phase. Our data thus suggest that Hrr25 is important for both basal activity of the Cvt pathway in logarithmically growing cells, and its stimulation in a late growth phase. In the latter situation, cells may require high vacuolar degradation capacity to adapt to environmental changes, and the upregulation of vacuolar transport of degradative enzymes via the Cvt pathway may be important to meet this cellular demand.
We found that Hrr25 depletion also compromises pexophagy, but not mitophagy, ribophagy, the selective autophagic degradation of Ald6, and nonselective autophagy induced by nitrogen starvation. S. cerevisiae proliferates peroxisomes in oleate medium and degrades them via pexophagy when shifted to glucose medium without a nitrogen source. A previous study reported that the pexophagy receptor Atg36 also interacts with Atg11, and is phosphorylated by an unknown kinase under pexophagy-inducing conditions. We hypothesized that as with the case of the Cvt pathway, Hrr25 phosphorylates the receptor Atg36 and enhances its interaction with the scaffold Atg11 to induce pexophagy. Indeed, phosphorylation of Atg36 is attenuated by depletion of Hrr25 in vivo, and Hrr25 directly phosphorylates Atg36 in vitro. Decreased Atg36 phosphorylation by Hrr25 depletion impairs the interaction of Atg36 with Atg11 without affecting its binding to the peroxisomal protein Pex3. Consistent with these results, peroxisomal targeting of Atg11, but not that of Atg36, is abolished by depletion of Hrr25. Thus, our results revealed that Hrr25 regulates pexophagy in a manner similar to that in the Cvt pathway (Fig. 1A).
Furthermore, in a separate paper, we showed that Hrr25 also phosphorylates Atg34 to promote autophagic transport of Ams1 under nitrogen-starvation conditions. Atg34 phosphorylation by Hrr25 also increases its binding to Atg11 (Fig. 1A). Thus, we revealed that Hrr25 regulates 3 of the 4 well-characterized selective autophagy pathways in S. cerevisiae by a uniform mechanism (Fig. 1A). Interestingly, mitophagy is also regulated in a similar manner although the kinase CK2 is employed. In all 4 receptors, phosphorylated Ser residues crucial for Atg11 binding reside within Atg11-binding regions (Fig. 1B). Whereas these regions contain a number of negatively charged residues, a receptor-binding region in Atg11 is enriched in positively charged residues. It is possible that phosphorylation of these Ser residues increases the negative charge in the Atg11-binding region and thereby enhances electrostatic interactions between the receptors and Atg11.
It was surprising that a single kinase regulates 3 different selective autophagy-related pathways. However, this would be reasonable in light of the fact that these pathways are promoted under similar conditions, such as late growth phase and nitrogen starvation. We showed that receptor phosphorylation by Hrr25 is stimulated under these conditions. Further analysis will elucidate this mechanism. It is also interesting to investigate whether Hrr25 homologs are involved in the regulation of selective autophagy in other organisms. Of note, Hrr25 is involved in different cellular processes, including DNA repair, ribosome biogenesis, chromosome segregation, and membrane traffic. Is there any relationship between selective autophagy and these processes? These issues should also be addressed in future studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Our work is supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (25111003 and 25711005).


