Abstract
Recent mass mortality outbreaks around the world in Pacific oysters, Crassostrea gigas, have seriously affected the aquaculture economy. Although the causes for these mortality outbreaks appear complex, infectious agents are involved. Two pathogens are associated with mass mortality outbreaks, the virus ostreid herpesvirus 1 (OsHV-1) and the bacterium Vibrio aestuarianus. Here we describe the interactions between these 2 pathogens and autophagy, a conserved intracellular pathway playing a key role in innate immunity. We show for the first time that autophagy pathway is present and functional in Pacific oysters and plays an important role to protect animals from infections. This study contributes to better understand the innate immune system of Pacific oysters.
Keywords: autophagy, Crassostrea gigas, OsHV-1, Pacific oyster, Vibrio aestuarianus
Abbreviations: ATG, autophagy-related; Atg8–PE, Atg8–phosphatidylethenolamine; DNA, deoxyribonucleic acid; hpi, hours postinfection; LC3-II, cleaved, lipidated and autophagosome-associated form of LC3; MAP1LC3A/B (LC3A/B), microtubule-associated proteins 1 light chain 3 alpha/beta (mammalian orthologs of the predicted Crassostrea gigas LC3 and yeast Atg8); NH4Cl, ammonium chloride; OsHV-1, Ostreid herpesvirus 1; PCR, polymerase chain reaction
Introduction
Since 2008, the oyster aquaculture industry, which mainly relies on the production of the Pacific oyster, Crassostrea gigas, has been affected by mass mortality outbreaks in France and Europe. In 2008, mass mortality of Pacific oysters aged one year or less, occurred simultaneously on the majority of French production sites. These young oysters were decimated by 40% to 100% depending on the site and groups while older animals were less affected.1-3 Mortality outbreaks of French oyster spat then recurred every year. Oysters sampled during mortality events showed high levels of detection of ostreid herpesvirus 1 (OsHV-1) DNA, a virus from malacoherpesviridae family and a particular genotype of OsHV-1 was described for the first time in 2008 and named μVar.2,4,5 This is a major concern for the future of all participants in the oyster industry. In addition, in 2012 and 2013, a bacterium, Vibrio aestuarianus has been reported in French Pacific adult oysters suffering high mortality outbreaks. In experimental conditions, isolates of Vibrio aestuarianus also induced significant mortality in oysters initially healthy.6,7
In the context of mass mortality outbreaks, there is an urgent need to better understand the immune system of oysters in order to potentially find new ways to control OsHV-1 and V. aestuarianus infections. Pacific oysters live in open marine waters and are directly exposed to pathogens. They also lack an adaptive immune system. Conventional vaccination is therefore not an option to protect them against infections. As a consequence, most strategies currently used for other farmed animal species (e.g., cattle, fish) cannot be directly applied to Pacific oysters. The few promising approaches to limit the harmful effect of pathogens in oyster production mainly rely on defining applicable ways to stimulate the oyster immune system and selecting more resistant animals.8
We studied autophagy, a well-conserved pathway, as it plays an important role in innate immunity.9-11 Autophagy is an intracellular degradation pathway that is highly conserved from yeast to plants and animals.12,13 More than 30 genes have been involved in autophagy called ATG for autophagy-related.14,15 In the first steps of autophagosome formation, a portion of cytoplasm is surrounded by a unique double-membraned, cup-shaped structure called the phagophore.13,16 Autophagosomes result from the extension and fusion of the edges of the phagophore.13,16 Ultimately, autophagosomes fuse with lysosomes (in metazoan cells) or vacuoles (in yeast and plant cells). The inner membrane of the autophagosome and the cytoplasm-derived materials contained in the autophagosome are then degraded by lysosomal/vacuolar hydrolases. Autophagy has been extensively studied in higher eukaryotes and its role in pathogen degradation (bacteria and viruses) has been well described.9-11,17 However, little is known about autophagy in molluscs,18-20 and autophagy has never been described in the Pacific oyster, C. gigas. The recent publication of Pacific oyster genome,21 has opened up new possibilities to study innate immunity and particularly autophagy.
In this study, we show that ATG genes are present in the Pacific oyster genome. Oyster ATG genes appear to be closer to human genes than those found in Drosophila or C. elegans. We also show that the autophagy pathway plays a role in protecting Pacific oysters from OsHV-1 and V. aestuarianus infections, and autophagy stimulation is associated with oyster survival. Interestingly, we observe that Pacific oyster families, which have low or high susceptibility to OsHV-1 or V. aestuarianus infections, did not modulate autophagy with the same amplitude. This study shows for the first time a protective role of autophagy against OsHV-1 and V. aestuarianus infections in Pacific oysters in experimental conditions and suggests a potential strategy to fight against these infections by selecting animals that have high autophagy induction potential.
Results
Autophagy in Pacific oyster
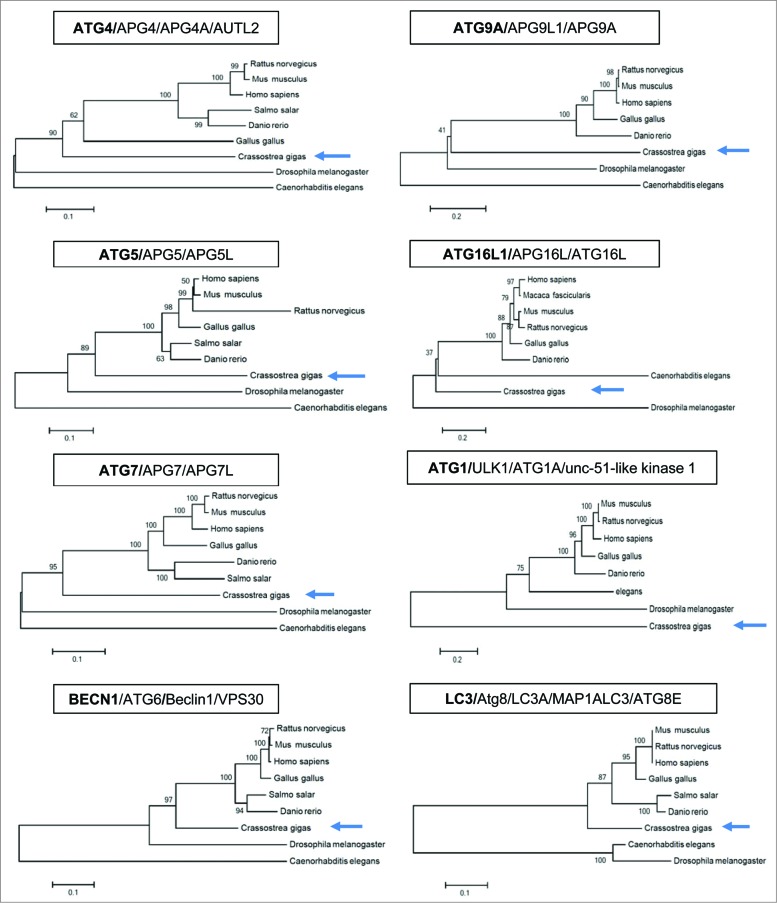
A phylogenetic analysis was carried out and showed that genes involved in autophagosome formation (ATG genes) are present in the Pacific oyster genome (Fig. 1). Many of them share similarities with human autophagy genes (ATG4/APG4/APG4A/AUTL2; ATG5/APG5/APG5L; ATG7/APG7/APG7L; BECN1/ATG6/Beclin1/VPS30; ATG9A/APG9L1/APG9A; ATG16L1/APG16L/ATG16L; ATG1/ULK1/ATG1A/unc-51-like kinase 1; LC3/Atg8/LC3/LC3A/MAP1ALC3/ATG8E; Fig. 1). Most of the oyster ATG genes presented higher homologies to human genes than those of model organisms such as Drosophila melanogaster and Caenorhabditis elegans (Fig. 1). Only ATG1, a serine/threonine-protein kinase, involved in early steps of autophagosome formation, showed divergence compared to human ULK1 (Fig. 1). Among these ATG genes, ATG8/LC3 is important, as its cleaved and lipidated protein product (LC3-II) decorates the autophagosomes and the protein level of lipid-conjugated Atg8/LC3 (LC3-II) correlates with the number of autophagosomes present inside the cells.12 A glycine residue at position 120 in the oyster LC3 protein is present suggesting that the LC3 conjugation system is functional (Fig. 2A).
Figure 1.
Autophagy in Pacific oyster. Phylogenetic trees of Atg/ATG proteins.
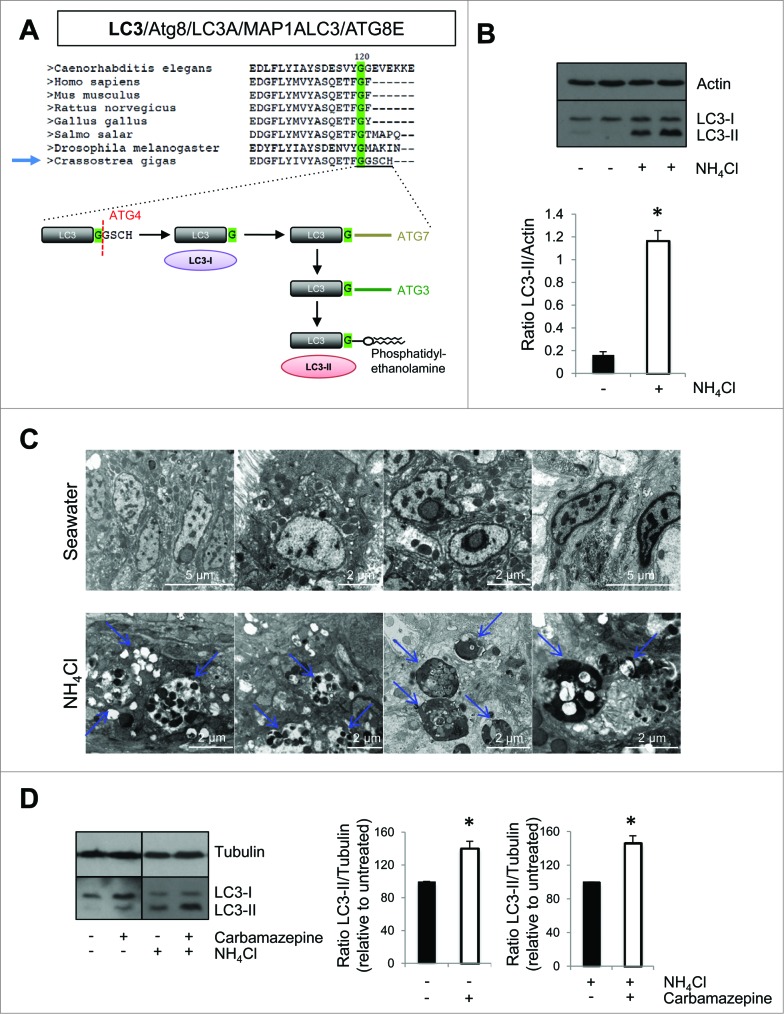
Figure 2.
Autophagy flux in Pacific oyster. (A) Sequence alignement and glycine conservation in position 120; cleavage model of Atg8 and association with a phosphatidylethanolamine. (B) LC3 western blot after contact with NH4Cl, during 20 h. (C) Transmission electron microscopy examination of oyster mantle 20 h post NH4Cl treatment or seawater (control condition). (D) LC3 western blot after contact during 20 h with carbamazepine and NH4Cl+carbamazepine to show autophagy flux in Pacific oysters.
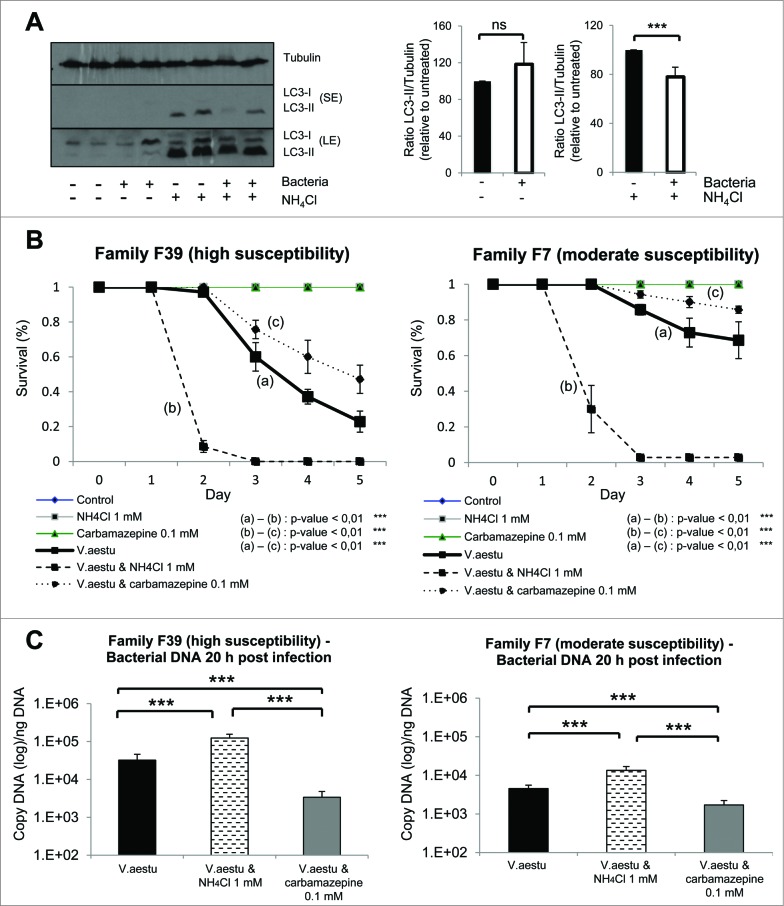
By western blot, we observed the presence of 2 bands when using a specific antibody against LC3, corresponding to LC3-I and LC3-II, as previously described in the literature.12 To confirm the specificity of LC3-II band and to test if autophagy pathway was functional, oysters were treated with a nontoxic concentration of ammonium chloride (NH4Cl), which impairs autophagosome degradation by preventing autophagosome-lysosome fusion (Fig. 2B, S1A and S1B). LC3-II accumulated dramatically in NH4Cl treated conditions (Fig. 2B). To confirm that NH4Cl treatment caused an accumulation of autophagosomes, we performed transmission electron microscopy. We observed a dramatic accumulation of vesicular structures that had the appearance of autophagosomes in NH4Cl treated animals compared to the control (Fig. 2C). These data (western blot and transmission electron microscopy) highly suggested that autophagy flux is present in oysters. Moreover, autophagosome formation was induced in oysters using carbamazepine, which has previously been shown to induce autophagy.22 As seen in Fig. 2D, carbamazepine-treatment caused an accumulation of LC3-II in the presence and absence of NH4Cl, indicating an induction of autophagy.
OsHV-1 infection and autophagy
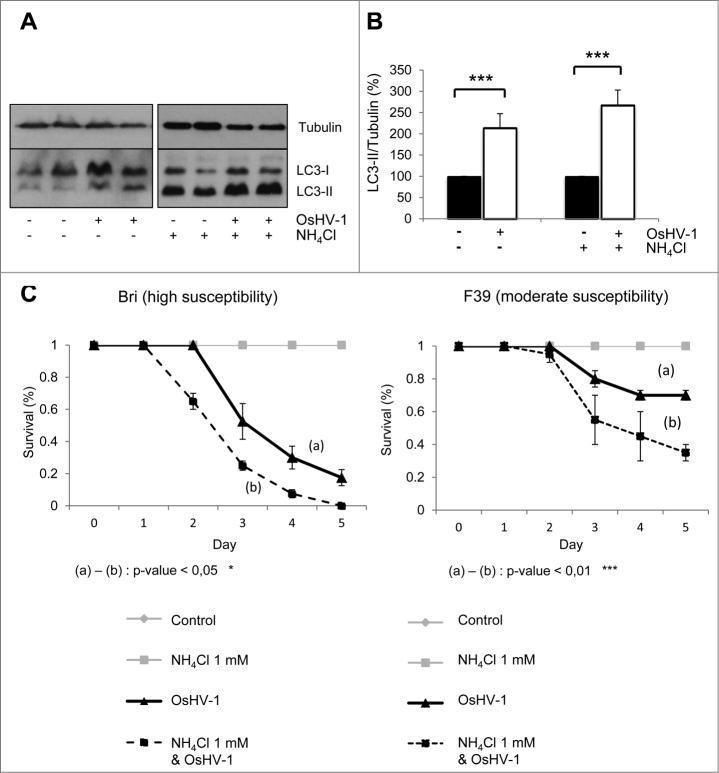
To test the role of autophagy during OsHV-1 infection, different approaches were used. We first analyzed the levels of LC3-II by western blot and observed an increase in LC3-II in experimentally infected oysters (in mantle tissue) in the presence or absence of NH4Cl, suggesting that viral infection triggered autophagy (Fig. 3A and B). As oyster groups with different degrees of mortality after viral infection were available,23 the survival of animals with different susceptibility to OsHV-1 infection was monitored in NH4Cl treated condition (F39; Fig. 3C). NH4Cl treatment in experimentally infected oysters increased mortality in both groups compared to OsHV-1 condition (Fig. 3C). NH4Cl treatment only (without viral infection) did not induce any mortality of oysters. By contrast, animals treated with carbamazepine showed less mortality upon experimental infection in both tested oyster groups (Fig. 4A). Also, animals cultivated without food for 4 wk (starvation, a primordial autophagy stimulus) have better survival upon OsHV-1 infection, compared to fed animals (Fig. S2A and S2B). Importantly, in the presence of NH4Cl, the starved animals have lower survival during OsHV-1 infection, suggesting that the benefits of starvation are autophagy-dependent (Fig. S2B).
Figure 3.
OsHV-1 infection induces autophagy. (A) LC3 western blot 20 h postinfection with different conditions (uninfected and infected without NH4Cl; uninfected and infected with NH4Cl). (B) Quantification of LC3 corresponding to western blot (A). (C) Survival curves of 2 oyster group (Bri and F39) during OsHV-1 infection with or without NH4Cl. Note that no mortality was observed in uninfected animals.
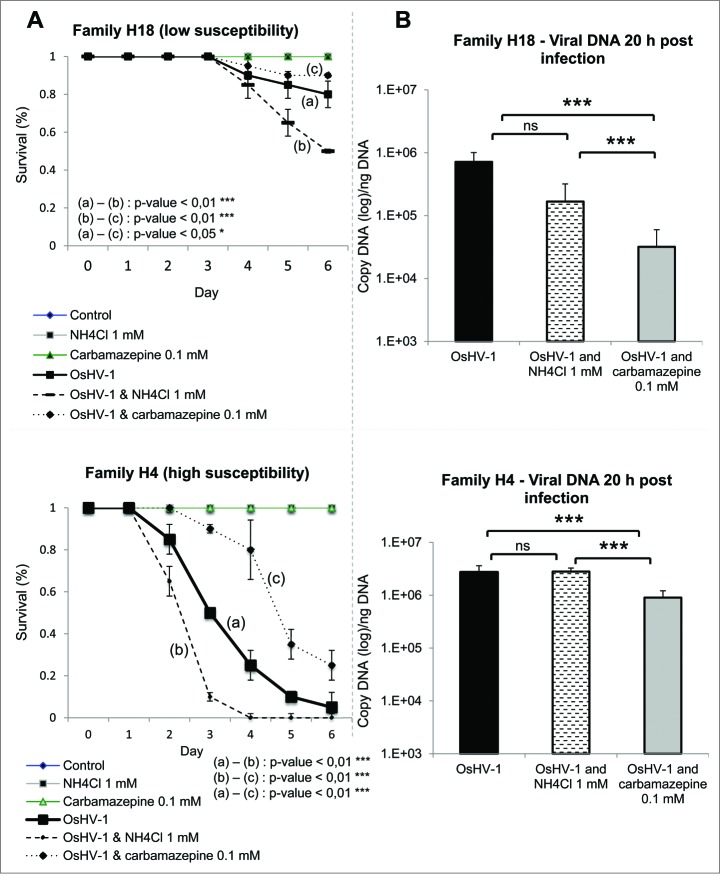
Figure 4.
Autophagy plays protective role during OsHV-1 infection. (A) Survival curves of 2 oyster families (H4 and H18) during OsHV-1 infection with or without NH4Cl and with or without carbamazepine. (B) Viral DNA quantification 20 h postinfection in both families (H4 and H18). Note that no mortality was observed in uninfected animals.
To understand the role of NH4Cl and carbamazepine during experimental viral infection, viral DNA, which correlates with viral replication, was quantified 20 h post-infection. No change was observed in viral DNA levels in NH4Cl treated animals in comparison with control oysters, while a decrease was observed in carbamazepine-treated condition (Fig. 4B). These data suggest that autophagy may participate in virus degradation when stimulated. This has been recently suggested in mammalian cells where autophagy stimulation enhances viral clearance.24,25
ATG genes are expressed differently between different oyster groups challenged with OsHV-1
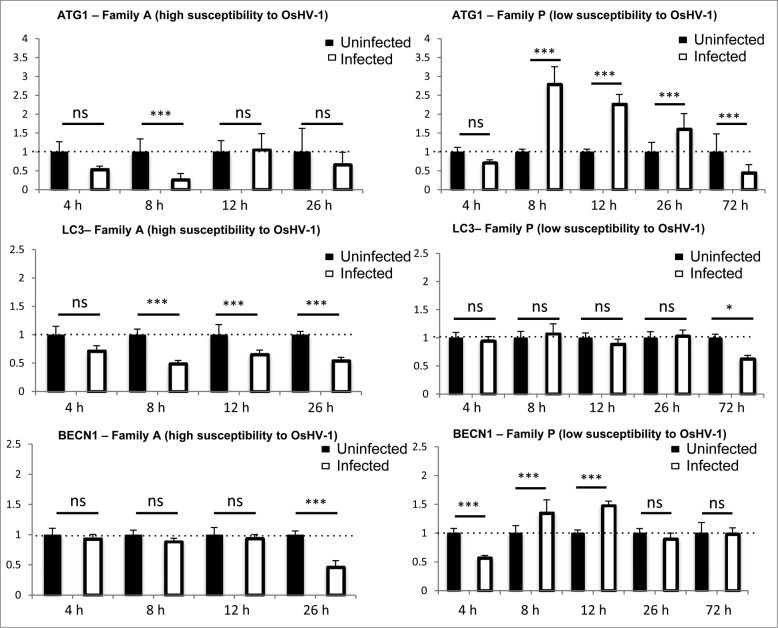
In the same experiment, we performed real-time PCR to analyze the expression of ATG genes (ATG1, ATG8, and BECN1; Fig. 5). Expression of ATG1 and ATG8 were downregulated upon early time points of infection in highly susceptible animals (Fig. 5). BECN1 was downregulated but at a later time point (26 h). In contrast, we observed that ATG1 and BECN1 were upregulated at 8 h and 12 h postinfection in low susceptible animals (Fig. 5). After a phase of induction of ATG1 and BECN1 expression, there was a decrease or a return of expression back to control conditions 72 h postinfection (Fig. 5). These data suggest that sensitive animals were less efficient to induce autophagy when infected by OsHV-1 compared to more resistant animals. Moreover, there may be a feedback regulation of autophagy gene expression in low susceptible animals consistent with decrease of viral DNA during the time course of the experiment.23
Figure 5.
ATG gene expression in different oyster populations challenged with OsHV-1 (Family A and Family P).
Vibrio aestuarianus infection and autophagy
In addition to OsHV-1, V. aestuarianus is associated with mortality outbreaks in Pacific oysters. Thus, the effect of V. aestuarianus infection on autophagy was explored. We observed a decrease of LC3-II in the presence of NH4Cl, suggesting that this bacterium may inhibit autophagy (Fig. 6A). As observed with OsHV-1 infection, Pacific oyster families show different susceptibility to V. aestuarianus infection and for a single family, susceptibility to the viral infection can differ from its susceptibility to the bacterial infection. In the survival assay, we observed that NH4Cl treatment increased oyster mortality upon bacterial infection whereas carbamazepine treatment protected the animals in both high and low susceptible groups in experimental conditions (Fig. 6B). These data suggest that autophagy plays a protective role against both V. aestuarianus and OsHV-1 infections. When measuring bacterial DNA content, we observed a dramatic increase in NH4Cl treated conditions and a clear decrease in carbamazepine conditions 20 h post-infection (Fig. 6C). These data suggest that autophagy could degrade bacteria and that inhibition of autophagy lead to fatal bacterial infection.
Figure 6.
Autophagy protects oysters from Vibrio aestuarianus infection. (A) LC3 western blot 20 h postinfection with different conditions (uninfected and infected without NH4Cl; uninfected and infected with NH4Cl), also quantification of LC3 protein corresponding at the different conditions of western blot. (B) Survival curves of 2 oysters families (F39 and F7) during bacterial infection with or without NH4Cl and with or without carbamazepine. (C) Bacterial DNA quantification 20 h postinfection in both families (F39 and F7). Note that no mortality was observed in uninfected animals.
Discussion
Autophagy is an important pathway involved in the maintenance of cellular homeostasis. Perturbations of autophagy have been associated with several diseases, such as neurodegenerative diseases, diabetes, cancers, and infectious diseases.12,13,26 Autophagy is an important regulator of innate immunity as it helps clear pathogens and control the inflammatory response.9-11,17 As current information indicates that oysters do not develop acquired immunity,27 we decided to study autophagy as it might represent a major defense against pathogens. The recent publication of the Pacific oyster genome allowed us to observe that ATG genes are conserved in oysters.21 Using phylogenetic tools, ATG genes from Pacific oysters showed greater similarities to their human orthologs compared to those in Drosophila melanogaster or Caenorhabditis elegans. We also observed that the autophagy pathway appears functional in oysters, as seen by the presence of the membrane-bound form of LC3 (LC3-II) by western blot, which is associated with autophagosomes, and also as seen by the accumulation of LC3-II when oysters were treated with NH4Cl that inhibits autophagosome-lysosome fusion, therefore accumulating autophagosomes. LC3 is primarily a cytosolic protein (called LC3-I). During autophagosome formation, LC3-I is cleaved by ATG4 to expose a glycine residue at its C terminus at position 120, which allows its conjugation to phosphatidylethanolamine (PE), which is mediated by ATG7 and ATG3 (Fig 2A).13,16 The LC3 moiety is conjugated to PE on autophagosomal precursor membranes. Moreover, autophagy can be stimulated in oysters by a treament with carbamazepine, a known autophagy inducer.22 These data highly suggest that there is an autophagy flux in oysters, meaning that autophagosomes can be formed and fuse with lysosomes where their content can be degraded. Transmission electron microscopy examination confirmed these data as we observed a dramatic accumulation of vesicular structures that look like autophagosomes in NH4Cl treated animals compared to the control. The 2 autophagy modulators used in this study appeared to inhibit (NH4Cl) or induce (carbamazepine) autophagy so we used these compounds for the rest of our analysis to modulate autophagy.
We examined the role of autophagy during experimental infections, using 2 pathogens that have been associated with high mortalities during the last decade, a virus from malacoherpesviridae family called OsHV-1 and a bacterium belonging to the Vibrio genus, Vibrio aestuarianus. Our data suggest that autophagy may play a protective role in oysters against OsHV-1 and Vibrio aestuarianus infections as seen by survival assay when autophagy was inhibited by NH4Cl treatment or induced by carbamazepine or by starvation. These data need to be interpreted with the caveat that these compounds have autophagy-independent actions via multiple targets. However, the protection observed when we used starvation or carbamazepine treatment was abolished when we treated the animals with NH4Cl, suggesting that the effects of these treatments were autophagy-dependent.
Results obtained by western blot highly suggest that autophagy is induced during OsHV-1 infection and inhibited (NH4Cl condition) during Vibrio aestuarianus infection. More work is required to understand the different interactions between OsHV-1 and Vibrio aestuarianus with autophagy. However, this will be limited by the lack of classical tools. There is no oyster cell line that can be used to produce OsHV-1 in vitro. However, we observed a clear decrease of bacterial DNA when autophagy was stimulated and an increase when autophagy was inhibited, suggesting that bacteria are cleared by autophagosomes. In the context of OsHV-1 infection, we observed a decrease of viral DNA when autophagy was stimulated, but no increase when autophagy was inhibited, suggesting that viral particles may be cleared when autophagy is induced. These data illustrate the different relationships that exist between pathogens and the autophagy pathway that has been extensively studied in mammalian cells.9-11,17 For example, pathogens such as viruses and bacteria use the autophagy pathway for their own benefit (replication niche, nutrient supply). This is the case for viruses like Influenza A virus, coxsackievirus B3, dengue virus, and bacteria such as Yersinia, Coxiella.10,28–30 Conversely, autophagy can clear viruses and bacteria and are an important part of the immune response. This is the case for viruses such as Sindbis virus, α-herpesvirus, and vesicular stomatitis virus and bacteria such as Shigella, Salmonella, and Mycobacteria.10,28-30
Finally, mortality outbreaks are often observed during spring and summer where concentrations of algae, which represent the main source of energy for oysters, are elevated. It is well known that starvation is a potent stimulus of autophagy. Thus, it would of interest to analyze if there is a correlation between food supply, autophagy, and mortality outbreaks knowing that starvation protects oysters from OsHV-1 infection.
In conclusion, we show for the first time that autophagy is conserved and functional in Pacific oysters and autophagy is an important defense mechanism against 2 pathogens, OsHV-1 and Vibrio aestuarianus, associated with mortality outbreaks that affect oyster economy.
Materials and Methods
Pacific oysters
Within the framework of the European project Bivalife (FP7, 2011–2014), 45 biparental families of Pacific oysters, C. gigas, were produced at Ifremer's facilities (LGPMM, La Tremblade) in order to obtain biological material presenting contrasted susceptibilities to different pathogens, including OsHV-1 and V. aestuarianus. Six of the 45 biparental families were selected on the basis of their susceptibility to the viral infection in experimental conditions, as well as their susceptibilities to the bacterial infection. Four families (F39, H4, F22, and H44) had moderate mortality rates and one family (H18) presented high mortality rates after experimental OsHV-1 infection. Family F39 presented high mortality rates and family F7 presented moderate mortality rates after experimental V. aestuarianus infection. cDNA from 2 other families (Family A with low mortality rates; Family P with high mortality rates after experimental OsHV-1 infection) already used in a previous study,23 were used for ATG gene expression.
Another group of animals was also used. They were produced at Ifremer's facilities located in Argenton (Brittany). This oyster group (Bri) presented high mortality rates after experimental OsHV-1 infection.
Phylogenetic analysis
Phylogenetic analysis was performed on ATG protein sequences derived from the Pacific oyster genome and genomes of model organisms using 3 computational approaches (Neighbor-Joining (NJ) method; Maximum Likelihood; Maximum Parsimony). Bootstrap data sets (1000 replicates) were generated. All approaches were implemented using the MEGA5 program.
Antibodies and reagents
Antibodies used in this study include rabbit anti-LC3B (Cell Signaling Technology, LC3A/B 4108) for western blotting, rabbit anti-actin (Sigma-Aldrich; A4700) and mouse anti-α-tubulin (Sigma-Aldrich, T9026). Reagents also include ammonium chloride (NH4Cl) (Sigma-Aldrich, 09718) and carbamazepine (Sigma-Aldrich, 94496 Fluka).
Modulation of autophagy
To inhibit autophagosome degradation, oysters were treated with NH4Cl at 1 mM during 20 h in aquariums (3 L). To induce autophagy, oysters were treated with carbamazepine at 0.1 mM during 20 h in aquariums (3 L). For starvation experiment, oysters were put in aquariums (50 L of seawater) without food during 4 wk (Fig. S2).
Transmission electron microscopy
After fixation in 3% glutaraldehyde in 0.2 M cacodylate buffer (pH 7.2), mantle pieces of oysters were treated as previously described.31 Ultrathin sections stained with uranyl acetate and lead citrate were examined with a JEOL JEM 1200EX transmission electron microscope (http://www.lehigh.edu/∼inmicro/1200EX.html) at 80 kV.
Viral infection: intramuscular injection of OsHV-1 suspension
Two hundred and 10 oysters were “anesthetized” during 4 h in a solution containing magnesium chloride (MgCl2, 50 g/L) in seawater (1 v)/distilled water (4 v).3 One hundred μL of a OsHV-1 (μVar genotype,2,4) suspension at 1 × 104 copies of viral DNA/μL were injected into the adductor muscle and oysters placed in tanks containing 3 L of filtered seawater (1 μm) at 22°C without food supply (10 oysters per tank). Survival was monitored during 5 d (experiment with F39 and Bri) or 6 d (experiment with H4 and H18) after injection. Percentages of cumulative survival were defined daily for the different conditions (1) oysters injected with OsHV-1 suspension (2 tanks with seawater, 2 tanks with seawater supplemented with NH4Cl at 1 mM, and 2 tanks with seawater supplemented with carbamazepine at 0.1 mM), or (2) injected with sterile artificial seawater (2 tanks with seawater, 2 tanks with seawater supplemented with NH4Cl at 1 mM, and 2 tanks with seawater supplemented with carbamazepine at 0.1 mM). Dead oysters were removed from tanks during the time course of the experiment. The experiment was performed 3 times for all experiment with OsHV-1 (experiment with F39 and Bri), except for experiments with families H4 and H18 (1 time). Oysters were less than 1-y-old for all experiments.
The same protocol was used to test the effect of starvation on oysters: percentages of cumulative survival were defined daily for the different conditions (1) oysters injected with OsHV-1 suspension (2 tanks with seawater, 2 tanks with seawater supplemented with NH4Cl at 1 mM, 2 tanks with seawater after starvation treatement and 2 tanks with seawater supplemented with NH4Cl at 1 mM after starvation treatement), or (2) injected with sterile artificial seawater (2 tanks with seawater, 2 tanks with seawater supplemented with NH4Cl at 1 mM, 2 tanks with seawater after starvation treatement and 2 tanks with seawater supplemented with NH4Cl at 1 mM after starvation treatment). Dead oysters were removed from tanks during the time course of the experiment. The experiment was performed one time for family H44 (by duplicate for each conditions), and 2 times for family F22 (by triplicate for each conditions). Oysters were less than 1-y-old for all experiments.
Bacterial infection: intramuscular injection of Vibrio aestuarianus
Two hundred and 10 oysters were “anaesthetized” during 4 h as described above. Fifty μL of a bacterial (V. aestuarianus 02/041; purity and concentration suspension checked by plating) suspension at 1 × 104 bacterium/μL were injected into the adductor muscle and oysters placed in tanks containing 3 L of filtered seawater (1 μm) at 22°C without food supply (10 oysters per tank). Survival was monitored during 6 d after injection and percentages of cumulative survival were assessed daily for the different conditions (1) oysters injected with bacteria (2 tanks with seawater, 2 tanks with seawater supplemented with NH4Cl at 1 mM, and 2 tanks of seawater with carbamazepine at 0.1 mM) or (2) injected with sterile artificial seawater (2 tanks of seawater, 2 tanks of seawater with NH4Cl at 1 mM, and 2 tanks of seawater supplemented with carbamazepine at 0.1 mM). Dead oysters were removed from tanks during the time course of the experiment. The experiment was performed 3 times with V. aestuarianus with families F39 and F7. Oysters were more than 1-y-old for all assays.
Western blotting
A mantle piece (15 to 20 mg) was collected from each oyster for all tested conditions 20 h postinfection. For each condition, 2 pools of 5 samples were analyzed. For each pool, a protein extraction was carried out in 100 μL of cell extraction buffer (Invitrogen, FNN0011). Initially, mechanical manual grinding using pellet piston was performed and lysis on ice for 30 min in cell extraction buffer with 1 mM PMSF and a protease inhibitor cocktail (Sigma, P-2714). Lysates were centrifuged at 16,000 g for 10 min at 4°C, and supernatant fractions were resolved by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with TBST (TBS, 0.1% Tween-20 [Sigma, P2287]) containing 1% nonfat dry milk and were then incubated overnight at 4°C with primary antibodies diluted in TBST. Membranes were washed with TBST, incubated for 1 h at room temperature with 2,500x dilutions of HRP-conjugated secondary antibodies (GE Healthcare Bioscience; NA934 and NA931) in TBST containing 1% nonfat dry milk, and washed. Immunoreactive bands were then detected using enhanced chemiluminescence (GE Healthcare Bioscience).
DNA extraction
Total DNA was extracted from tissue fragments (mantle) using QiAamp tissue mini kit® (QIAgen, 51306) combined with the use of the QIAcube automate, according to the manufacturer's protocol. Elution was performed in 100 μL of AE buffer provided in the kit. The DNA quality and quantity were determined using a NanoDrop 2000 instrument (Thermo Scientific, http://www.nanodrop.com/Productnd2000overview.aspx). Extracted DNA was stored at −20°C or 4°C prior OsHV-1 detection and quantification by real-time PCR.
Relative ATG genes expression from oysters
Relative expression of 3 ATG genes from C. gigas spat was studied during experimental OsHV-1 infection at 4, 8, 12, 26 h postinfection (hpi) for families A and P,23 and 72 and 144 hpi for family P.23 The relative quantification value (ratio R) was calculated using the method described by Pfaffl: R = [(Etarget)ΔCTtarget(control-sample)]/ [(Eref)ΔCTref(control-sample)].32 The efficiency of each primer pair was determined by constructing a standard curve from serial dilutions (Table 1). These 3 genes from Pacific oyster were (i) Serine/threonine-protein kinase atg1 (ATG1), (ii) The α and β isoforms of microtubule-associated protein 1 light chain 3 (LC3A/B or ATG8), (iii) Beclin-1 (VPS30/BECN1) (Table 1). Host gene expression was normalized to the elongation factor 1-α (EF1-α), as no significant differences of Ct values were observed for this housekeeping gene between several conditions during the course time.23 The calibrator used for the experiment were individuals sampled at time 0 hpi from each family.23
Table 1.
List of oyster genes targeted by real-time PCR
| GenBank | Gene name | Abbreviation | Forward primer Reverse primer |
Efficiency (%) |
|---|---|---|---|---|
| EKC36832.1 | Serine/threonine-protein kinase atg1 | ATG1 | CAATGCGTGCGAAGAAGATG GCCGTTCATTGTTGGGTGAT |
97,7 |
| EKC40439.1 | Microtubule-associated proteins 1A/1B light chain 3A | ATG8/LC3 | CCGATGCTTGACAAGACCAA CCGTCCTCGTCTTTCTCCTG |
98,2 |
| EKC28450.1 | Beclin-1 | BECN1/ATG6 | AAATGCTGCTTGGGGTCAGA CGGAATCCACCAGACCCATA |
102,2 |
| AB122066.1 | Elongation Factor 1-α | EF1-α | AGTCACCAAGGCTGCACAGAAAG TCCGACGTATTTCTTTGCGATGT |
98,8 |
OsHV-1 DNA quantification by real-time PCR
OsHV-1 DNA quantification was carried out using a real-time PCR protocol.33 Real-time PCR was performed in duplicate using a Mx3000p Thermocycler sequence detector (Agilent, 401512). Amplification reactions were performed in a total volume of 20 μL. Each well contained 5 μL of genomic DNA (5 ng/μL), 10 μL of Brillant III Ultra-Fast SYBR® Green Master Mix (Agilent), 2 μL of each primer (5 μM: OsHVDPFor 5′ATTGATGATGTGGATAATCTGTG3′; 34
Five μM OsHVDPRev 5′GGTAAATACCATTGGTCTT-GTTCC3′),34 and 1 μL of distilled water. Real-time PCR cycling conditions were as follow: 3 min at 95°C, followed by 40 cycles of amplification at 95°C for 5 s, 60°C for 20 s. The results were expressed as a log10 of the virus DNA copy number per ng of total DNA.
Vibrio aestuarianus DNA quantification by real-time PCR
OsHV-1 DNA quantification was carried out using a real-time PCR protocol.35 Real-time PCR TaqMan was performed in duplicate using a Mx3000p Thermocycler sequence detector (Agilent, 401512). Amplification reactions were performed in a total volume of 20 μL. Each well contained 5 μL of genomic DNA (5 ng/μL), 10 μL of 2X Mastermix Ultra-Fast Brillant III (Agilent, 600880), 0.06 μL of each primer (100 μM: DNAj-F and DNAj-R), 0.04 μL of DNAj probe (100 μM) and 4.84 μL of distilled water. Real-time PCR cycling conditions were as follow: 3 min at 95°C, followed by 40 cycles of amplification at 95°C for 10 s, 60°C for 20 s. The results were expressed as a log10 of the bacterial DNA copy number per ng of total DNA.
Statistical analysis
Statistical analysis was performed using the Wilcoxon-Mann Whitney test by statistical software R, to compare 2 groups. The null hypothesis (H0) corresponding to the distribution of the quantitative variable is the same in the groups. Significance was set at P ≤ 0.05 (*) and at P ≤ 0.01 (***).
Significance levels for comparisons between groups were determined with Student t tests, repeated-measure, factorial ANOVA and LSD test, Test log Rank and/or Mann-Whitney using the STATVIEW software, version 4.53 (Abacus Concepts, Berkeley, CA, USA).
Acknowledgments
The authors wish to thank the Ifremer's hatchery team (LGPMM; Patrick Azema and Lionel Degremont for families H) in La Tremblade and the nursery team (LSPC) in Bouin for the production of Pacific oysters. We thank Anna Albecka-Moreau for critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was partially funded through the EU project Bivalife (FP7 KBBE, contract n°266157), the Poitou Charentes Region and DPMA (Direction des pêches maritimes et de l’aquaculture, AESTU project). David Rubinsztein is a Wellcome Trust Prinicipal Research Fellow.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. EFSA J. Scientific opinion of the Panel on Animal Health and Welfare on a request from the European Commission on the increased mortality events in Pacific oysters Crassostrea gigas. 2010; 1894-953. [Google Scholar]
- 2. Segarra A, Pépin JF, Arzul I, Morga B, Faury N, Renault T. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res 2010; 153:92-9; PMID:20638433; http://dx.doi.org/ 10.1016/j.virusres.2010.07.011 [DOI] [PubMed] [Google Scholar]
- 3. Schikorski D, Renault T, Saulnier D, Faury N, Moreau P, Pépin J-F. Experimental infection of Pacific oyster Crassostrea gigas spat by ostreid herpesvirus 1: demonstration of oyster spat susceptibility. Vet Res 2011; 42:27; PMID:21314910; http://dx.doi.org/ 10.1186/1297-9716-42-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Renault T, Moreau P, Faury N, Pepin J-F, Segarra A, Webb S. Analysis of clinical ostreid herpesvirus 1 (Malacoherpesviridae) specimens by sequencing amplified fragments from three virus genome areas. J Virol 2012; 86:5942-7; PMID:22419803; http://dx.doi.org/ 10.1128/JVI.06534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jenkins C, Hick P, Gabor M, Spiers Z, Fell SA, Gu X, Read A, Go J, Dove M, O’Connor W, et al. Identification and characterisation of an ostreid herpesvirus-1 microvariant (OsHV-1 μ-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis Aquat Organ 2013; 105:109-26; PMID:23872855; http://dx.doi.org/ 10.3354/dao02623 [DOI] [PubMed] [Google Scholar]
- 6. Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas J-L. Evidence for the involvement of pathogenic bacteria in summer mortalities of the pacific oyster Crassostrea gigas. Microb Ecol 2007; 53:187-96; PMID:17245611; http://dx.doi.org/ 10.1007/s00248-006-9061-9 [DOI] [PubMed] [Google Scholar]
- 7. De Decker S, Normand J, Saulnier D, Pernet F, Castagnet S, Boudry P. Responses of diploid and triploid Pacific oysters Crassostrea gigas to Vibrio infection in relation to their reproductive status. J Invertebr Pathol 2011; 106:179-91; PMID:20833182; http://dx.doi.org/ 10.1016/j.jip.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 8. Dégremont L, Bédier E, Boudry P. Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). II. Response to selection for survival and its influence on growth and yield. Aquaculture 2010; 299:21-9; http://dx.doi.org/ 10.1016/j.aquaculture.2009.11.017 [DOI] [Google Scholar]
- 9. Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol 2006; 18:375-82; PMID:16782319; http://dx.doi.org/ 10.1016/j.coi.2006.05.019 [DOI] [PubMed] [Google Scholar]
- 10. Deretic V, Levine B. Autophagy, Immunity, and Microbial Adaptations. Cell Host Microbe 2009; 5:527-49; PMID:19527881; http://dx.doi.org/ 10.1016/j.chom.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 2007; 7:767-77; PMID:17767194; http://dx.doi.org/ 10.1038/nri2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/ 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- 14. Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 2010; 6:438-48; PMID:20484971; http://dx.doi.org/ 10.4161/auto.6.4.12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jordan TX, Randall G. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect Inst Pasteur 2012; 14:126-39; PMID:22051604; http://dx.doi.org/ 10.1016/j.micinf.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102-9; PMID:17909521; http://dx.doi.org/ 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 17. Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151-75; PMID:18188003; http://dx.doi.org/ 10.4161/auto.5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore MN, Viarengo A, Donkin P, Hawkins AJS. Autophagic and lysosomal reactions to stress in the hepatopancreas of blue mussels. Aquat Toxicol Amst Neth 2007; 84:80-91; PMID:17659356; http://dx.doi.org/ 10.1016/j.aquatox.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 19. Moore MN. Autophagy as a second level protective process in conferring resistance to environmentally-induced oxidative stress. Autophagy 2008; 4:254-6; PMID:18196967; http://dx.doi.org/ 10.4161/auto.5528 [DOI] [PubMed] [Google Scholar]
- 20. Bai R, You W, Chen J, Huang H, Ke C. Molecular cloning and expression analysis of GABAA receptor-associated protein (GABARAP) from small abalone, Haliotis diversicolor. Fish Shellfish Immunol 2012; 33:675-82; PMID:22771962; http://dx.doi.org/ 10.1016/j.fsi.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 21. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012; 490:49-54; PMID:22992520; http://dx.doi.org/ 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- 22. Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. An autophagy-enhancing drug promotes degradation of mutant 1-antitrypsin Z and reduces hepatic fibrosis. Science 2010; 329:229-32; PMID:20522742; http://dx.doi.org/ 10.1126/science.1190354 [DOI] [PubMed] [Google Scholar]
- 23. Segarra A, Mauduit F, Faury N, Trancart S, Dégremont L, Tourbiez D, Haffner P, Barbosa-Solomieu V, Pépin J-F, Travers M-A, et al. Dual transcriptomics of virus-host interactions: comparing two Pacific oyster families presenting contrasted susceptibility to ostreid herpesvirus 1. BMC Genomics 2014; 15:580; PMID:25012085; http://dx.doi.org/ 10.1186/1471-2164-15-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201-6; PMID:23364696; http://dx.doi.org/ 10.1038/nature11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sumpter R, Levine B. Selective autophagy and viruses. Autophagy 2011; 7:260-5; PMID:21150267; http://dx.doi.org/ 10.4161/auto.7.3.14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709-30; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng TC. Bivalves. Invertebr Blood Cells 1981; 233-300 [Google Scholar]
- 28. Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science 2005; 307:727-31; PMID:15576571; http://dx.doi.org/ 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- 29. Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137-49; PMID:19683680; http://dx.doi.org/ 10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 30. Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G. Matrix protein 2 of influenza a virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009; 6:367-80; PMID:19837376; http://dx.doi.org/ 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Renault T, Le Deuff RM, Cochennec N, Chollet B, Maffart P. Herpes-like viruses associated with high mortality levels in larvae and spat of Pacific oysters, Crassostrea gigas: a comparative study, the thermal effects on virus detection in hatchery-reared larvae, reproduction of the disease in axenic larvae. Vet Res 1995; 26:539-43; PMID:8581037 [PubMed] [Google Scholar]
- 32. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/; http://dx.doi.org/ 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pepin JF, Riou A, Renault T. Rapid and sensitive detection of ostreid herpesvirus 1 in oyster samples by real-time PCR. J Virol Methods 2008; 149:269-76; PMID:18342377; http://dx.doi.org/ 10.1016/j.jviromet.2008.01.022 [DOI] [PubMed] [Google Scholar]
- 34. Webb SC, Fidler A, Renault T. Primers for PCR-based detection of ostreid herpes virus-1 (OsHV-1): application in a survey of New Zealand molluscs. Aquaculture 2007; 272:126-39; http://dx.doi.org/ 10.1016/j.aquaculture.2007.07.224 [DOI] [Google Scholar]
- 35. Saulnier D, De Decker S, Haffner P. Real-time PCR assay for rapid detection and quantification of Vibrio aestuarianus in oyster and seawater: a useful tool for epidemiologic studies. J Microbiol Methods 2009; 77:191-7; PMID:19318049; http://dx.doi.org/ 10.1016/j.mimet.2009.01.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.