Abstract
Primary cilium is an organelle that plays significant roles in a number of cellular functions ranging from cell mechanosensation, proliferation, and differentiation to apoptosis. Autophagy is an evolutionarily conserved cellular function in biology and indispensable for cellular homeostasis. Both cilia and autophagy have been linked to different types of genetic and acquired human diseases. Their interaction has been suggested very recently, but the underlying mechanisms are still not fully understood. We examined autophagy in cells with suppressed cilia and measured cilium length in autophagy-activated or -suppressed cells. It was found that autophagy was repressed in cells with short cilia. Further investigation showed that MTOR activation was enhanced in cilia-suppressed cells and the MTOR inhibitor rapamycin could largely reverse autophagy suppression. In human kidney proximal tubular cells (HK2), autophagy induction was associated with cilium elongation. Conversely, autophagy inhibition by 3-methyladenine (3-MA) and chloroquine (CQ) as well as bafilomycin A1 (Baf) led to short cilia. Cilia were also shorter in cultured atg5-knockout (KO) cells and in atg7-KO kidney proximal tubular cells in mice. MG132, an inhibitor of the proteasome, could significantly restore cilium length in atg5-KO cells, being concomitant with the proteasome activity. Together, the results suggest that cilia and autophagy regulate reciprocally through the MTOR signaling pathway and ubiquitin-proteasome system.
Keywords: cilia, autophagy, MTOR, proteasome, polycystic kidney disease
Abbreviations
- 3-MA
3-methyladenine
- ANKS6
ankyrin repeat and sterile α motif domain containing 6
- ATG/atg
autophagy-related
- Baf
bafilomycin A1
- DAPI
4′-6-diamidino-2-phenylindole
- Ac-TUBA
acetylated-tubulin α
- CQ
chloroquine
- CF
confluence
- FBS
fetal bovine serum
- HK2
human kidney proximal tubular cells
- IFT
intraflagellar transport
- KD
knockdown
- KAP3
kinesin family-associated protein 3
- KIF3A/3B
kinesin family member 3A/3B
- KO
knockout
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- LTA
lotus tetragonolobus agglutinin
- MEF
mouse embryonic fibroblast
- MTOR
mechanistic target of rapamycin
- OFD1
oral-ficial-digital syndrome 1
- PBS
phosphate-buffered saline
- PKD
polycystic kidney disease
- Rapa
rapamycin
- RKRB
Krebs-Henseleit saline containing 25 mM NaHCO3
- RPS6KB1
ribosomal protein S6 kinase
- 70kDa
polypeptide 1
- SD
standard deviation
Introduction
Primary cilia are dynamic organelles protruding from one of the 2 centrioles (mother centriole) in the majority of cells. Structurally, primary cilia are mainly composed of the microtubule-based axonemes, accessorized by motor proteins and intraflagellar transport (IFT) particles.1 IFT is a parallel process of the anterograde transport toward the tip assisted by the kinesin-2 complex (KIF3A, KIF3B, KAP3), and the retrograde transport toward the ciliary base aided by the cytoplasmic motor protein dynein. IFT particles contain at least 20 polypeptides, classified as the complex A and B.2-5 The functions of primary cilia were largely unknown until the landmark research of lov-1 in C. elegans and Ift88 in mice.6,7 It is now recognized that primary cilia not only contribute to cellular sensing but are involved in cell proliferation, differentiation, apoptosis, endocytosis/exocytosis, and planar cell polarity.8-13 Cilium may be a hub for cellular signaling integration, on which certain key molecules from the WNT, notch, and hedgehog pathways have been localized.14 Dysfunction of primary cilia contributes to a large spectrum of human genetic diseases collectively termed ciliopathies, notably including polycystic kidney disease (PKD).15
Autophagy is a fundamental catabolic process whereby many excessive or damaged cytoplasmic components are degraded through lysosomes for the maintenance of cellular homeostasis.16 Three major types of autophagy, i.e, macroautophagy, microautophagy, and chaperone-mediated autophagy, have been described. Macroautophagy (hereafter called autophagy) involves a series of complex steps from the phagophore formation, autophagosome, to autolysosomes. Autolysosomes can move toward the microtubule organizing center with the aid of kinesins and cytoplasmic dyneins that are also the key proteins for ciliogenesis.17,18 Autophagy is evolutionarily essential in many aspects of cellular functions and dysregulation of autophagy is associated with many types of human disorders.19
In 2011, Belibi et al. reported that autophagy is suppressed in Han:SPRD rats and cpk mice, 2 animal models of PKD with ciliary dysfunction.20 Based on this study and our pilot observations, we hypothesized that there may be a regulatory connection between autophagy and ciliogenesis. Very recently, 2 studies have demonstrated the direct interaction between autophagy and ciliary regulation. Tang et al. have discovered that OFD1 at centriolar satellites functions as a general suppressor for ciliogenesis.21 Pampliega et al. further suggest a reciprocal interaction between autophagy and ciliogenesis, whereby ciliary signaling, such as the hedgehog pathway, induces autophagy.22 Our present study has further confirmed the reciprocal interaction between autophagy and cilia. Moreover, we demonstrate the involvement of MTOR signaling and ubiquitin-proteasome system in the reciprocal regulation.
Results
Association of cilium length and autophagy from cell growth to differentiation
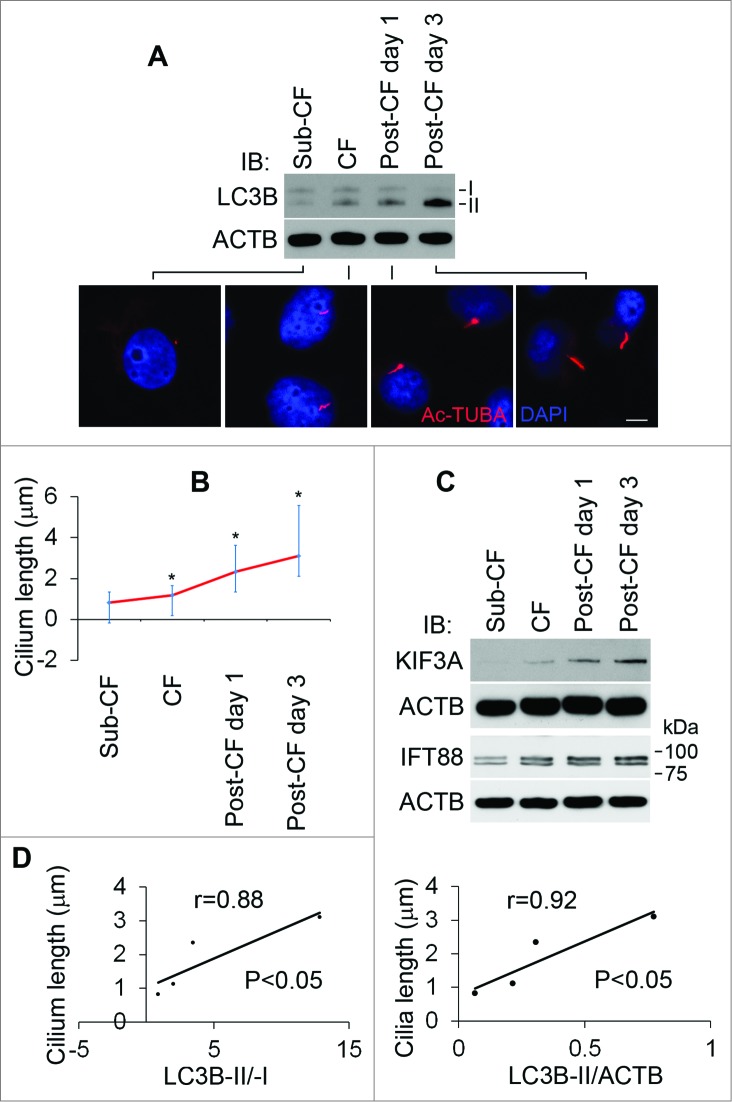
In our initial study, we observed ciliogenesis from cell growth to differentiation in culture dishes. When cells became confluent and entered postconfluence differentiation stage, their cilia grew longer (Fig. 1A, B) and notably, this was accompanied by the accumulation of ciliary IFT proteins, including KIF3A and IFT88 (Fig. 1C). To examine the association between cilia and autophagy, we analyzed LC3B-II/-I and LC3B-II/ACTB ratios in stages of subconfluence, confluence, and postconfluence cells. Cilium length in these cells showed an appreciable correlation with LC3B-II/-I and LC3B-II/ACTB ratios, respectively (Fig. 1D).
Figure 1.
Association of cilium length and autophagy from cell growth to differentiation. To determine the association of cilium length and autophagy, HK2 cells were cultured in DMEM-F12 medium containing 10% FBS at subconfluence (Sub-CF), confluence (CF), or for one or 3 d after reaching confluence (Post-CF d 1, 3). (A) Cells were assayed by immunofluorescence staining with the antibody to Ac-TUBA (red) and DAPI (blue) and cell lysates examined by immunoblot analysis of LC3B and ACTB. Scale bar: 5 5 μ. (B) Cilium length was averaged out of over 50 cilia in each group (Sub-CF, n=52; CF, =56; Post-CF d 1, =51; Post-CF d 3, =60). Cilium length: Post-CF d 3>Post-CF d 1>CF>Sub-CF, * P < 0.05. (C) Expression level of KIF3A and IFT88 from cell growth to differentiation. Please note, as previously reported,7 2 forms of IFT88 were detected. (D) Positive correlation of cilium length with LC3B-II/-I or LC3B-II/ACTB ratio.
Shortening of cilia inhibits autophagy through MTOR signaling
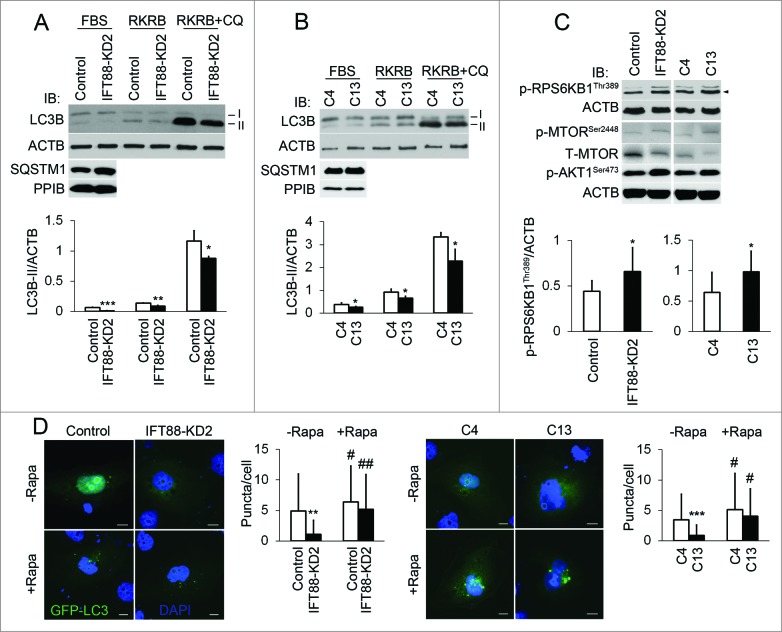
To further study the role of cilia in autophagy regulation, we used 2 cilia-suppressed lines of cells. IFT88 knockdown (IFT88-KD2) cells had short cilia due to knockdown of IFT88, while the C13 cells were selected from heterogeneous kidney epithelial cells for short cilia.23 Immunoblot analysis of LC3B-II/ACTB ratio showed that the basal level of autophagy was suppressed in both IFT88-KD2 and C13 cells, which was also indicated by the accumulation of SQSTM1/p62 (Fig. 2A and B). To further monitor the autophagic flux in these cells, chloroquine was added into the RKRB medium. As shown in Fig. 2A and 2B, chloroquine-induced accumulation of LC3B-II was also partially inhibited in IFT88-KD2 and C13 cells. Moreover, following transfection of GFP-LC3 fewer puncta per cell were detected in IFT88-KD2 and C13 cells in comparison to their respective controls (Fig. 2D). These results together indicate that suppression of cilia leads to autophagy blockade. To determine if MTOR, the major negative regulator, contributed to autophagy inhibition in cilia-suppressed cells, we examined MTOR activity by detecting the phosphorylation of MTOR and its substrate RPS6KB1. Compared with control cells, both IFT88-KD2 and C13 cells showed higher phosphorylation of MTOR at serine 2448 (p-MTOR, Ser2448) and RPS6KB1 at threonine 389 (p-RPS6KB1, Thr389), which were further complicated by activation of MTORC2 indicated by phosphorylation of AKT1 at serine 473 (p-AKT1, Ser473) (Fig. 2C). To further determine whether MTOR activation was responsible for the suppression of autophagy, we tested the effect of rapamycin. As shown in Fig. 2D, rapamycin restored GFP-LC3-puncta in IFT88-KD2 and C13 cells.
Figure 2.
Shortening of cilia suppresses autophagy through the MTOR pathway. (A, B) Inhibition of autophagy in both IFT88-KD2 and C13 cilia-short cells. To assess the effect of cilium length on autophagy, cilia-suppressed cells and their controls (Control, C4) were incubated with FBS-containing culture medium or serum-free RKRB medium without IFT88 or with 25 μM chloroquine (CQ) for 3 h. Cell lysates were collected for immunoblot analysis of LC3B and ACTB. Protein bands were quantified by densitometry. (C) MTOR hyperphosphorylation in IFT88-KD2 and C13 cells. To define MTOR activity, cilia-short cells were cultured in serum-free DMEM-F12 medium. Cell lysates were collected for immunoblot analysis of p-MTOR, Ser2448; p-RPS6KB1, Thr389; p-AKT1, Ser473, and ACTB. Densitometry was conducted to determine the p-RPS6KB1, Thr389/ACTB ratio. (D) Restoration of autophagy by rapamycin (Rapa) in cilia-suppressed cells. GFP-LC3 was transfected into IFT88-KD2 and C13 cilia-short, and their control cells for the analysis of autophagosome puncta. Fewer puncta per cell were seen in IFT88-KD2 cells (n=36) than control cells (n=33). After 24 h treatment of rapamycin (50 nM), the number of puncta per cell increased in both control (n=55) and IFT88-KD2 cells (n=53), and no difference was detected between them. Similar results were shown for cilia-short C13 cells and their control C4 cells. Fifty-two and 53 positive cells, without rapamycin treatment, were counted from C4 and C13 cells; 52 and 54 positive cells, with rapamycin treatment, were counted from C4 and C13 cells. All the experiments were performed 3 times. P*<0.05, **<0.01, ***<0.001 IFT88-KD2 and C13 vs Control and C4 cells; #<0.05, ##<0.01 +rapamycin vs -rapamycin. Scale bar: 10 μm.
Autophagy activation promotes cilium elongation
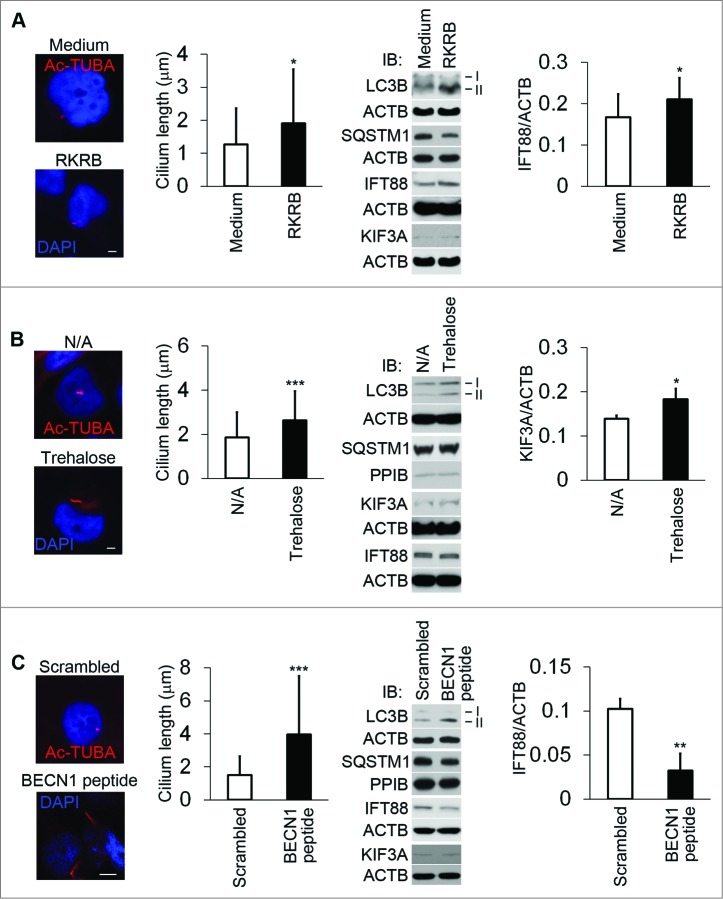
To investigate the effect of autophagy on cilium length, we treated HK2 cells with 3 different approaches respectively. Incubation of cells in a plain buffer (RKRB without serum and glucose) induces autophagy.24 After 3 h of incubation in RKRB medium, cilium length was increased significantly, accompanied by the increment of IFT88 expression. However, the change of KIF3A expression level was minimal (Fig. 3A). After 24 h of incubation with trehalose,25 cilia were significantly elongated. Under this condition, KIF3A (but not IFT88) was accumulated (Fig. 3B). Finally, BECN1/Beclin 1 peptide was used as the third approach to activate autophagy.26 BECN1 peptide lengthened primary cilia after 4 h of treatment, compared to the scrambled peptide-treated controls. Surprisingly, IFT88 was downregulated by BECN1 peptide (Fig. 3C), suggesting that ciliary factor(s) other than IFT88 may be responsible for the observed ciliary lengthening. Obviously, all 3 methods increased LC3B-II levels. SQSTM1 was degraded in RKRB and BECN1 peptide-treated cells.
Figure 3.
Autophagy activation increases cilium length. To determine the effect of autophagy activation on cilium length, 3 different approaches for autophagy induction were used. (A) Increase in cilium length during incubation in serum/glucose-free RKRB medium. HK2 cells were incubated for 3 h in full culture medium (medium) or plain RKRB medium, and then subjected to immunofluorescence analysis of Ac-TUBA or immunoblot analysis of the indicated proteins. Forty-five and 52 cilia were measured, respectively, in culture medium and RKRB-incubated HK2 cells. Densitometry was conducted to determine the IFT88/ACTB ratio. (B) Increase in cilium length induced by trehalose. HK2 cells were incubated in the absence (N/A: no addition) or presence of 100 mM trehalose for 24 h, followed by immunofluorescence analysis of Ac-TUBA or immunoblot analysis of the indicated proteins. Seventy-four and 82 cilia were counted for N/A and trehalose-treated cells. Densitometry was conducted to determine the KIF3A/ACTB ratio. (C) Increase in cilium length induced by BECN1 peptide. HK2 cells were incubated with a scrambled sequence or the BECN1 peptide for 4 h, followed by immunofluorescence analysis of Ac-TUBA or immunoblot analysis of the indicated proteins. Fifty one and 66 cilia were counted for control and BECN1 peptide-treated cells. Densitometry was conducted to determine the IFT88/ACTB ratio. In (A, B, and C), autophagy was confirmed by immunoblotting of LC3B and SQSTM1. The results were the summary of 3 separate experiments. P*<0.05, **<0.01, ***<0.001. Scale bar: 5μm ((A)and B) and 10 μm (C).
Autophagy inhibition decreases cilium length in cultured cells and animal tissues
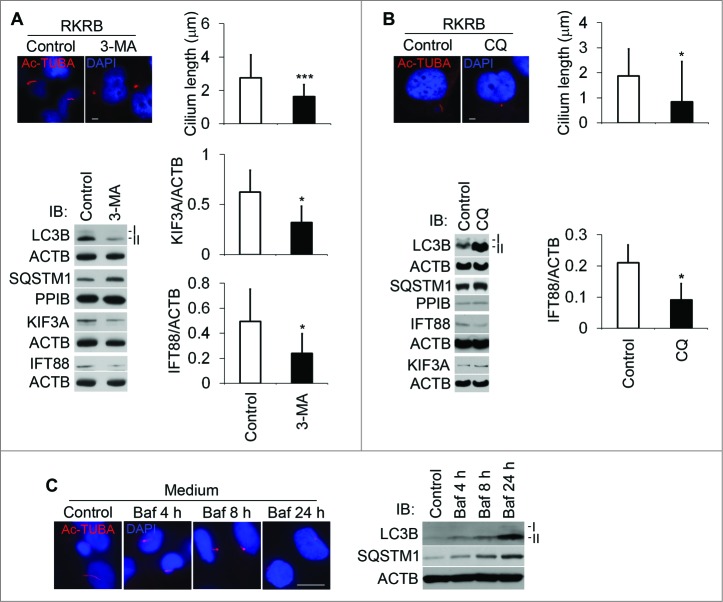
To explore the effect of autophagy inhibition on cilia, we first used 3-MA, an inhibitor at the early stage of autophagy, which inhibits the class III PtdIns3K. After 2 h of incubation of 3-MA, cilia were shortened, accompanied by the decrement of IFT88 and KIF3A expression (Fig. 4A). We then tested the effect of CQ, an inhibitor at the late stage of autophagy. Clearly, CQ shortened cilia after 3 h. CQ inhibition also led to the decrease of IFT88 expression (Fig. 4B). Similarly, bafilomycin A1 (Baf), another inhibitor at a late stage of autophagy, gave results similar to CQ in a time course (Fig. 4C). Not surprisingly, SQSTM1 was accumulated in all 3 conditions of autophagy inhibition. We further examined primary cilia in kidney proximal tubule-specific atg7-KO mice. Cilium length and frequency in kidney proximal tubular cells were significantly reduced compared to those of wild-type tissues (Fig. 5). These data suggest that autophagy inhibition resulted in blockade of ciliogenesis in both cultured cells and tissues.
Figure 4.
Autophagy suppression decreases cilium length. To study the effect of autophagy suppression on cilium length, we used 3 different inhibitors of autophagy. (A) Suppression of cilia by 3-MA. HK2 cells were treated for 2 h with 3-MA and then subjected to immunofluorescence analysis of Ac-TUBA or immunoblot analysis of the indicated proteins. Sixty and 52 cilia were measured for control and 3-MA-treated cells. Densitometry was conducted to determine the KIF3A/ACTB and IFT88/ACTB ratios. (B) Suppression of cilia by chloroquine (CQ). HK2 cells were treated for 2 h with CQ and then subjected to immunofluorescence analysis of Ac-TUBA or immunoblot analysis of the indicated proteins. Fifty-four and 45 cilia were counted for control and CQ-treated cells. Densitometry was conducted to determine the IFT88/ACTB ratio. (C) Suppression of cilia by bafilomycin A1 (Baf) in a time course. HK2 cells were treated with Baf for up to 24 h in a full culture medium and then subjected to immunofluorescence analysis of Ac-TUBA. Autophagy inhibition by 3-MA and CQ and Baf was confirmed by immunoblotting of LC3B and SQSTM1. The results are the summary of 3 separate experiments. P*<0.05, ***<0.001 vs Control. Scale bar: 5 μm ((A) and B) and 10 μm (C).
Figure 5.
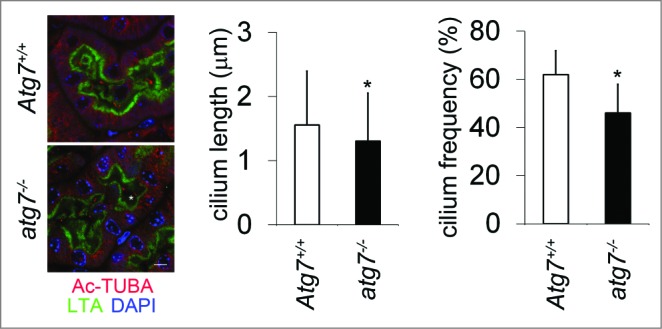
Decrease in cilium length and frequency in proximal tubule-atg7-KO cells. To confirm our in vitro findings, we determined to evaluate cilium length and frequency in proximal tubule-atg7-KO mice. Kidney tissue sections from proximal tubule-atg7-KO mice and wild-type littermate mice were stained for Ac-TUBA and LTA, a proximal tubule marker. One hundred forty seven and 101 cilia were counted for LTA-positive wild-type and atg7-KO cells, respectively. Five different microscope fields of view were used for cilium frequency calculation (cilium number/nucleus number). P*<0.05. Scale bar: 5 μm.
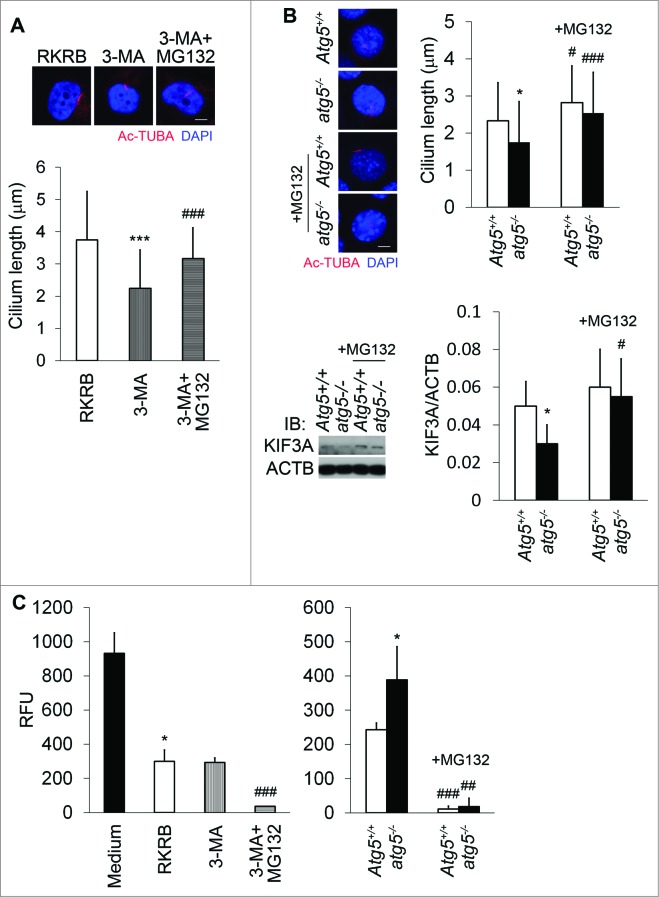
Restoration of cilium length by MG132 in autophagy-defective cells
To understand the mechanism whereby autophagy inhibition blocks ciliogenesis, we hypothesized the involvement of proteasomal degradation of ciliary proteins. Autophagy and the proteasome interact functionally in protein degradation.27 In our study, autophagy inhibition by 3-MA suppressed cilia in HK2 cells, and notably, the proteasomal inhibitor MG132 markedly reversed cilium length caused by 3-MA treatment (Fig. 6A). Similarly, atg5-deficient (atg5−/−) cells exhibited shorter cilia (Fig. 6B). MG132 increased cilium length in wild-type and atg5−/− cells, but it appeared more effective in atg5−/− cells. Consequently, the cilium length difference between wild-type and atg5-deficient cells was reduced significantly after MG132 treatment (Fig. 6B). Interestingly, MG132 induced KIF3A accumulation in wild-type and atg5−/− cells, with noticeably more induction in the latter. To further substantiate the findings, we determined proteasome activity. Clearly, proteasome activity in atg5−/− cells was higher than that in Atg5+/+ cells, both of which were significantly suppressed by MG132 (Fig. 6C). MG132 had a likewise effect in HK2 cells. However, 3-MA did not significantly affect proteasome activity, probably due to its autophagy-independent effects. Compared to cell culture media, RKRB medium activated autophagy and significantly reduced the proteasome activity. These data, in general, suggest that autophagy suppression is associated with an increase of proteasome activity, which may lead to proteosomal degradation of ciliary proteins to attenuate ciliogenesis. Under this condition, MG132, by inhibiting proteasome, can prevent ciliary protein degradation leading to the preservation of cilia.
Figure 6.
Proteasome in cilia shortening in autophagy-suppressed cells. (A) Restoration of cilium length by MG132 in 3-MA-treated cells. HK2 cells were treated for 2 h with 3-MA in the absence or presence of 5 μM MG132, followed by immunofluo-rescence analysis of cilia by Ac-TUBA. Thirty six, 35, and 38 cilia were measured for the control, 3-MA-treated, 3-MA+MG132-treated cells respectively. (B) Effect of MG132 on cilium length in atg5-KO cells and induction of KIF3A by MG132. Atg5+/+ and atg5−/− cells were incubated without or with MG132 for 2 h, followed by immunofluorescence analysis of Ac-TUBA. Thirty three, 48, 46, and 50 cilia were measured for the wild-type and KO cells at the basal condition or with MG132 treatment. atg5-KO cells had shorter cilia at the basal condition. Cell lysates were collected in (B) for immunoblot analysis. Densitometry was conducted to determine the KIF3A/ACTB ratio. The results are the summary of 3 separate experiments. (C) Proteasome activity increment in autophagy-suppressed cells. Compared to Atg5+/+ cells, atg5−/− exhibited enhanced proteasome activity. MG132 significantly suppressed the proteasome activity in HK2 and Atg5+/+ and atg5−/− MEF cells. Compared to culture medium, RKRB medium reduced proteasome activity in HK2 cells. Relative fluorescent units (RFU) were used to indicate the proteasome activity in different culture conditions. Results are the summary of 4 separate experiments. P*<0.05, ***<0.001 vs RKRB (A) or Medium (C), atg5−/− vs Atg5+/+ (B); #<0.05, ##<0.01, ###<0.001 +MG132 vs -MG132 (A, B, and C). Scale bar (A and B): 5 μm.
Discussion
In this study, we show that cilia and autophagy reciprocally affect each other, which is consistent with the latest reports.21,22 Moreover, our results suggest that the MTOR pathway and the proteasome system are involved in these 2 processes respectively. These findings suggest that 2 aspects of cell metabolism, i.e. anabolism (MTOR) and catabolism (proteasome), play distinct roles in the reciprocal regulation of cilia and autophagy.
Thus far, there has been limited knowledge about the effect of cilium length alteration on autophagy, although functional KO of ciliary proteins ANKS6/SamCystin or CYS1/cystin 1 leads to the suppression of autophagy in 2 different animal models.20,28 Pampliega et al. very recently reported that blockage of IFT20 and IFT88 compromises autophagy through the hedgehog signaling pathway, consistent with an early report,22,29 while KO of Ift20 and Ift88 in mice diminishes primary cilia.7,30 In our study, we confirmed that KD of IFT88 shortened primary cilia in kidney tubular epithelial cells; more importantly, the MTOR signaling pathway was activated as previously reported in Ift88-mutated mice.31 Further experiments with rapamycin support that heightened MTOR activity may be responsible for inhibition of autophagy. The recent study by Jimenez-Sanchez et al. shows that cilia-regulated hedgehog signaling modulates autophagy through patched 1 and patched 2 in a GLI2-dependent but MTOR-independent pathway.29 Interestingly, crosstalk between the hedgehog and MTOR pathways has been reported.32 Thus, cilia-regulated autophagy may result from the MTOR and hedgehog pathways independently or combined. On the other hand, some members of the hedgehog pathway are localized in the primary cilia and disruption of cilia leads to the dysfunction of hedgehog signaling.13,33-35 Thus, it is also possible that dysfunction of primary cilia suppresses autophagy through the cilium-hedgehog-autophagy axis. However, further in-depth investigation is needed to delineate how interactions among primary cilia, the hedgehog, and the MTOR signaling pathways are coordinated to regulate autophagy. The research has significant implications for human diseases. For instance, MTOR is also activated in the autosomal dominant and recessive PKD involving the ciliary proteins PKD1/236 and PKHD1,37,38 and rapamycin slows the progression of cystogenesis in animal models.36,39 Hence, shortening of primary cilia may lead to cystogenesis through the inhibition of autophagy. It would be interesting to analyze in detail how autophagy suppression contributes to cystogenesis and progression. Based on the current knowledge about PKD, it seems that no ATG proteins have direct interactions with PKD1/2 or PKHD1. It is tempting to study whether there are individual protein-protein interactions among PKD proteins and ATG proteins and how dysfunction of PKD proteins affects ATG proteins and autophagy regulatory factors. More likely, autophagy suppression is caused by multiple dysfunctional signaling pathways such as MTOR, hedgehog, STAT, and AMPK, which were found abnormal in PKD.40,41 Autophagy abnormality may be a key mechanism for cystogenesis. However, it is noteworthy that proximal tubule-atg7-KO mice do not develop renal cysts during the observation period of 9 mo (unpublished data), suggesting that autophagy defects may need to combine with other signaling abnormalities to promote cystic diseases.
With regard to the effect of autophagy on cilia, our results demonstrate a positive correlation. Cilium length is mainly determined by IFT particles, coordinated by kinesin-2 complex and dynein proteins, although in mammalian cells nonIFT regulators of cilia have been identified.13 KIF3A plays a major part in transporting IFT proteins along the axoneme to elongate the primary cilia while the cytoplasmic dynein is responsible for transporting the disassembled cargos toward the basal body area. In our study, we found that expression levels of IFT88 and KIF3A were not always consistent with changes of the cilium length. Thus, we believe nonIFT regulators may play specific roles in regulating cilium length in the context of autophagy. Tang et al.21 found that ciliogenesis is promoted by autophagy through removal of the OFD1 protein from centriolar satellites, while Pampliega et al.22 support a role of IFT20 in this process. Consistent with these findings, we showed that ciliogenesis was suppressed in atg5-KO MEF cells. Importantly, we showed that cilia were shorter in atg7-KO mouse kidney tissues, further strengthening the concept of dependence of ciliogenesis on autophagy.
Our finding that suppression of proteasome system partially rescued short cilia in autophagy-suppressive cells adds another piece to this complex puzzle of cilia and autophagy interaction. Our data indicate that KIF3A was degraded partially through the ubiquitin-proteasome pathway, which was activated in autophagy-deficient cells. Thus, the cilium length is determined probably by not only the autophagic system but the ubiquitin-proteasome system.21,22 As a matter of fact, the ubiquitin-proteasome system was found around the primary cilia.42 It would be tempting to study whether centrosome-associated proteasomes play a significant role. We are presently not clear about the significance of cilia shortening in cells with autophagy deficiency. It is generally believed that primary cilia act as cellular mechanosensors.10,12,43 For instance, deflection of the primary cilia by fluid shear stress can shorten its length and consequently ameliorate mechanosensitivity.12,44,45 We speculate that shortening of cilia induced by autophagy suppression changes the cilia-sensing ability, and subsequently sensitizes cells to injury and apoptosis as found recently.23
Based upon the fact that some proteins such as IFT20 are found in both secretory vesicles and the primary cilia46 as well as in the urine exosomes,47 it has been suggested that a large number of substances may be excreted from the ciliary membrane through exocytosis or bud-off at the tip.11 A few autophagy proteins are localized in cilia,22 the tiny organelles with diameter of ˜0.25 μm (lysosomes 0.3 to 0.6 μm), suggesting that primary cilia may have a remarkable plasticity. However, other possibilities cannot be completely ruled out. For instance, ATG proteins may reside in the primary cilia as vesicles,22 or ATG proteins are localized in the primary cilia as noncomponents of autophagosomes and autolysosomes. Lin et al. have recently provided evidence that molecules with ˜650 kDa can enter primary cilia, indicating that at least 90% of mammalian proteins can do so.48 Of note, although we showed a general association of cilium length and individual IFT proteins in the context of autophagy change, IFT proteins were not always consistent with the cilium length, probably because IFT proteins are regulated transcriptionally and post-translationally. In addition, ciliary length may also depend on the turnover rate and/or recruitment of IFT.
In summary, together with recent reports,21,22 we have demonstrated the reciprocal positive interaction between cilia and autophagy. Mechanistically, our results suggest that on the one hand, ciliogenesis is associated with the suppression of MTOR and consequent activation of autophagy; on the other hand, autophagy suppresses the proteasome and prevents the degradation of ciliary proteins resulting in ciliogenesis (Fig. 7). Further research should delineate the role and regulation of the crosstalk between cilia and autophagy in physiological and pathophysiological conditions.
Figure 7.
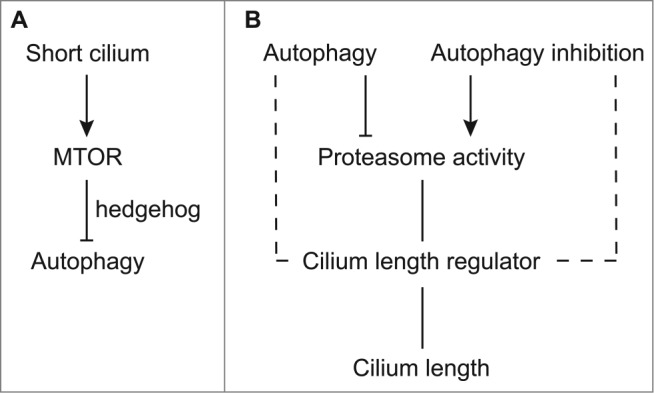
Our working model. Reciprocal interaction between cilia and autophagy. (A) Shortening of cilia activates MTOR, which subsequently inhibits autophagy. (B) Autophagy inhibition or stimulation increases or decreases the proteasome activity, which subsequently affects cilium length regulators. At certain conditions, autophagy may directly regulate cilium- length regulators such as IFT88 in BECN1 peptide-induced cilium elongation. The integrative balance of cilia-suppressing and -elongating factors determines the cilium length.
Materials and Methods
Reagents and antibodies
ACTB (A2228) and acetylated (Ac)-α tubulin/TUBA (T7451) antibodies were purchased from Sigma. IFT88 antibody (13967–1-AP) was acquired from the Proteintech Group. KIF3A monoclonal antibody (611508) was obtained from the BD Transduction Laboratory. Phospho-RPS6KB1 (p-RPS6KB1, Thr389; 9205), phospho-AKT1 (p-AKT1, Ser473; 9271), total MTOR (T-MTOR; 2972) and phospho-MTOR (p-MTOR, Ser2448; 2927) antibodies were purchased from Cell Signaling Technology. LC3B antibody (NB100–2220) was bought from Novus Biologicals. SQSTM1 antibody (PM045) was obtained from MBL. PPIB antibody (ab16045) was purchased from Abcam. Secondary antibodies for immunoblotting were from Pierce (goat antimouse 31430, goat antirabbit 31460) and for immunofluorescence staining from Jackson ImmunoResearch (115–165–062), respectively. Fluorescein-labeled lotus tetragonolobus agglutinin (LTA) was bought from Vector Labs (FL-1321). Three-MA (M9281), rapamycin (R0395), MG132 (M7449), trehalose (T9531), and CQ (C6628) were all obtained from Sigma. BECN1 peptide was commercially synthesized according to the published protocol.26 Other reagents were from Sigma unless specifically indicated.
Cell culture and treatment
HK2 cells obtained from the ATCC (CRL-2190) were routinely cultured in DMEM-F12 medium containing 10% fetal bovine serum (FBS). Isolation of cilia-long and -short HK2 cells was described elsewhere.23 IFT88-KD2 cells were made by transduction of virion particles containing specific IFT88 shRNAs. Control cells were transduced with the Mission Non-Target shRNA Particles (SHC002V, Sigma).23 For the treatment of HK2 cells, 3-MA (5 mM) or CQ (25 μM) was added into RKRB medium (115 mM NaCl, 1 mM KH2PO4, 4 mM KCl, 1 mM MgSO47H2O, 1.25 mM CaCl2, 25 mM NaHCO3, pH 7.4) while Baf (25 nM) was added into culture media for a specified period. Trehalose (100 mM) was added into the cell culture media for 24 h. Six N HCl-treated OPTI-MEM medium (GIBCO, 31985070) containing BECN1 peptide (25 μM) were incubated with HK2 cells for 4 h. MG132 was used at a concentration of 5 μM for 2 h in HK2 and Atg5+/+ and atg5−/− MEF cells.
Immunostaining and confocal microscopy
Tissue and cell treatment as well as staining procedures have been described previously.23 For immunostaining, tissue sections and fixed cells were blocked by bovine serum albumin (Sigma, A9647). After incubation with primary antibodies, samples were washed with phosphate-buffered saline (PBS; 136.89 mM NaCl, 2.68 mM KCl, 8.10 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4), and then incubated with secondary antibodies. Slides were mounted with the Prolong Gold antifade reagent containing DAPI (4′-6-diamidino-2-phenylindole) (P36931, Sigma). All procedures were performed at room temperature. For the image analysis, Zeiss Axio fluorescence and confocal microscopes (Carl Zeiss Inc.. (Thornwood, NY 10594) were used. The confocal microscope was equipped with an LSM Image analysis system and used for measurement of the cilium length.
Immunoblotting
Proteins from cells were extracted with SDS lysis buffer as previously reported.23,49 In brief, protein lysis buffer was added into PBS-washed cells for 5 min at room temperature. Samples were centrifuged at 15000 rcf for 10 min and supernatant fractions were used for protein analysis. Protein samples were electrophoresed in a mini-PROTEAN TGX gel and transferred to PVDF nylon membranes (Bio-Rad, 162–0177). After blocking with 5% nonfat dry milk (Lab Scientific, M0841) in PBS, the membrane was incubated with the primary antibody and washed with PBS containing 0.1% Tween 20 (MP Biomedicals, 11TWEEN201), followed by incubation with the secondary immunoglobulins conjugated with horseradish peroxidase. The bound secondary antibodies were detected with the SuperSignal™ West Pico Chemiluminescent Substrate (Pierce, 34080).
GFP-LC3 transfection
HK2 cells were placed into 6-well plates containing sterile coverslips. On the second d, cells were transfected with GFP-LC3 constructs by the Lipofectomine 2000 reagent (Invitrogen, 11668–019). After 24 h with or without rapamycin incubation (50 nM), immunostaining was performed. The number of puncta was quantified under confocal microscopy.
Kidney proximal tubule-specific atg7-KO mice
The mouse model of kidney proximal tubule-specific atg7-KO was generated in our recent work.50 Kidneys were harvested after sacrifice of the mice for immunostaining. Animal experiments were conducted according to the guidelines approved by the Institutional Animal Care and Use Committee of the Charlie Norwood VA Medical Center and Medical College of Georgia.
Proteasome activity assay
Five micrograms of cell lysates were used for the in vitro 20S proteasome activity assay according to the manufacturer's instructions (Millipore, APT280) using a TECAN GENios fluorescence microplate reader (Research Triangle Park, NC 27709). HK2 and Atg5+/+ and atg5−/− cells were cultured in the presence of 3-MA and/or MG132 for 2 h. Proteins were extracted with 1× assay buffer containing 0.05% NP-40 (USBiological, N3500) and 0.001% SDS (Fisher, S529). Relative fluorescent units (RFU) were measured after 1-h incubation at 60°C with the substrate (Suc-LLVY-AMC; Millipore, APT280) using a 380/460 nm filter set.
Statistics
Values were expressed as mean ± standard deviation (SD). The Student t test was used for comparison. Statistical significance for the Pearson correlation coefficient was calculated based on the 4 pairs of parameters. P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The study was supported in part by grants from National Natural Science Foundation of China [81430017], National Basic Research Program of China 973 Program No. 2012CB517601, the National Institutes of Health and Veterans Health Administration (VA) of USA.
References
- 1.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 2007; 179:321–30; PMID:17954613; http://dx.doi.org/ 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 1998; 141:993–1008; PMID:9585417; http://dx.doi.org/ 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Lab Invest 2005; 85:452–63; PMID:15723088; http://dx.doi.org/ 10.1038/labinvest.3700253 [DOI] [PubMed] [Google Scholar]
- 4.Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse IFT complex B. Cell Motil Cytoskeleton 2009; 66:457–68; PMID:19253336; http://dx.doi.org/ 10.1002/cm.20346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 2011; 12:222–34; PMID:21427764; http://dx.doi.org/ 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- 6.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 1999; 401:386–9; PMID:1051763811832437 [DOI] [PubMed] [Google Scholar]
- 7.Yoder BK, Tousson A, Millican L, Wu JH, Bugg CE Jr., Schafer JA, Balkovetz DF. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am J Physiol Renal Physiol 2002; 282:F541–52; PMID:11832437; http://dx.doi.org/ 10.1152/ajprenal.00273.2001 [DOI] [PubMed] [Google Scholar]
- 8.Abou Alaiwi WA, Lo ST, Nauli SM. Primary cilia: highly sophisticated biological sensors. Sensors 2009; 9:7003–20; PMID:22423203; http://dx.doi.org/ 10.3390/s90907003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet 2008; 40:69–77; PMID:18066062; http://dx.doi.org/ 10.1038/ng.2007.54 [DOI] [PubMed] [Google Scholar]
- 10.Yoder BK. Role of primary cilia in the pathogenesis of polycystic kidney disease. J Am Soc Nephrol 2007; 18:1381–8; PMID:17429051; http://dx.doi.org/ 10.1681/ASN.2006111215 [DOI] [PubMed] [Google Scholar]
- 11.Baldari CT, Rosenbaum J. Intraflagellar transport: it's not just for cilia anymore. Curr Opin Cell Biol 2010; 22:75–80; PMID:19962875; http://dx.doi.org/ 10.1016/j.ceb.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ., et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 2003; 33:129–37; PMID:12514735; http://dx.doi.org/ 10.1038/ng1076 [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Dong Z. Primary cilia and kidney injury: current research status and future perspectives. Am J Physiol Renal Physiol 2013; 305:F1085–98; PMID:23904226; http://dx.doi.org/ 10.1152/ajprenal.00399.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Reeuwijk J, Arts HH, Roepman R. Scrutinizing ciliopathies by unraveling ciliary interaction networks. Hum Mol Genet 2011; 20:R149–57; PMID:21862450; http://dx.doi.org/ 10.1093/hmg/ddr354 [DOI] [PubMed] [Google Scholar]
- 15.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 2006; 7:125–48; PMID:16722803; http://dx.doi.org/ 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N. Physiological functions of autophagy. Curr Top Microbiol Immunol 2009; 335:71–84; PMID:19802560 [DOI] [PubMed] [Google Scholar]
- 17.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol 2012; 22:R29–34; PMID:22240478; http://dx.doi.org/ 10.1016/j.cub.2011.11.034 [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Feng LQ, Zheng XX. Microtubule and kinesin/dynein-dependent, bi-directional transport of autolysosomes in neurites of PC12 cells. Int J Biochem Cell Biol 2011; 43:1147–56; PMID:21530677; http://dx.doi.org/ 10.1016/j.biocel.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445–544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belibi F, Zafar I, Ravichandran K, Segvic AB, Jani A, Ljubanovic DG, Edelstein CL. Hypoxia-inducible factor-1alpha (HIF-1alpha) and autophagy in polycystic kidney disease (PKD). Am J Physiol Renal Physiol 2011; 300:F1235–43; PMID:21270095; http://dx.doi.org/ 10.1152/ajprenal.00348.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 2013; 502:254-7; PMID:24089205; http://dx.doi.org/ 10.1038/nature12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature 2013; 502:194-200; PMID:24089209; http://dx.doi.org/ 10.1038/nature12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Wei Q, Dong G, Dong Z. ERK-mediated suppression of cilia in cisplatin-induced tubular cell apoptosis and acute kidney injury. Biochim Biophys Acta 2013; 1832:1582–90; PMID:23727409; http://dx.doi.org/ 10.1016/j.bbadis.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Periyasamy-Thandavan S, Jiang M, Schoenlein P, Dong Z. Autophagy: molecular machinery, regulation, and implications for renal pathophysiology. Am J Physiol Renal Physiol 2009; 297:F244–56; PMID:19279132; http://dx.doi.org/ 10.1152/ajprenal.00033.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J Biol Chem 2007; 282:5641–52; PMID:17182613; http://dx.doi.org/ 10.1074/jbc.M609532200 [DOI] [PubMed] [Google Scholar]
- 26.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D., et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201–6; PMID:23364696; http://dx.doi.org/ 10.1038/nature11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 2008; 4:141–50; PMID:17986870; http://dx.doi.org/ 10.4161/auto.5190 [DOI] [PubMed] [Google Scholar]
- 28.Huber TB, Edelstein CL, Hartleben B, Inoki K, Dong Z, Koya D, Kume S, Lieberthal W, Pallet N, Quiroga A., et al. Emerging role of autophagy in kidney function, diseases and aging. Autophagy 2012; 8:1009–31; PMID:22692002; http://dx.doi.org/ 10.4161/auto.19821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez-Sanchez M, Menzies FM, Chang YY, Simecek N, Neufeld TP, Rubinsztein DC. The Hedgehog signalling pathway regulates autophagy. Nat Commun 2012; 3:1200; PMID:23149744; http://dx.doi.org/ 10.1038/ncomms2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol 2008; 183:377–84; PMID:18981227; http://dx.doi.org/ 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell PD, Fitzgibbon W, Sas K, Stenbit AE, Amria M, Houston A, Reichert R, Gilley S, Siegal GP, Bissler J., et al. Loss of primary cilia upregulates renal hypertrophic signaling and promotes cystogenesis. J Am Soc Nephrol 2011; 22:839–48; PMID:21493775; http://dx.doi.org/ 10.1681/ASN.2010050526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X., et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012; 21:374–87; PMID:22439934; http://dx.doi.org/ 10.1016/j.ccr.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr Top Dev Biol 2008; 85:225–60; PMID:19147008; http://dx.doi.org/ 10.1016/S0070-2153(08)00809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007; 317:372–6; PMID:17641202; http://dx.doi.org/ 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- 35.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet 2006; 38:303–11; PMID:16493421; http://dx.doi.org/ 10.1038/ng1753 [DOI] [PubMed] [Google Scholar]
- 36.Shillingford JM, Murcia NS, Larson CH, Low SH, Hedgepeth R, Brown N, Flask CA, Novick AC, Goldfarb DA, Kramer-Zucker A., et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A 2006; 103:5466–71; PMID:16567633; http://dx.doi.org/ 10.1073/pnas.0509694103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer DC, Jacoby U, Pape L, Ward CJ, Kuwertz-Broeking E, Renken C, Nizze H, Querfeld U, Rudolph B, Mueller-Wiefel DE., et al. Activation of the AKT/mTOR pathway in autosomal recessive polycystic kidney disease (ARPKD). Nephrol Dial Transplant 2009; 24:1819–27; PMID:19176689; http://dx.doi.org/ 10.1093/ndt/gfn744 [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Wu M, Yao G, Zhang J, Zhou J. The cytoplasmic tail of FPC antagonizes the full-length protein in the regulation of mTOR pathway. PLoS One 2014; 9:e95630; PMID:24851866; http://dx.doi.org/ 10.1371/journal.pone.0095630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao Y, Kim J, Schrier RW, Edelstein CL. Rapamycin markedly slows disease progression in a rat model of polycystic kidney disease. J Am Soc Nephrol 2005; 16:46–51; PMID:15563559; http://dx.doi.org/ 10.1681/ASN.2004080660 [DOI] [PubMed] [Google Scholar]
- 40.Harris PC, Torres VE. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J Clin Invest 2014; 124:2315–24; PMID:24892705; http://dx.doi.org/ 10.1172/JCI72272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravichandran K, Edelstein CL. Polycystic kidney disease: a case of suppressed autophagy? Semin Nephrol 2014; 34:27–33; PMID:24485027; http://dx.doi.org/ 10.1016/j.semnephrol.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 42.Huang K, Diener DR, Rosenbaum JL. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J Cell Biol 2009; 186:601–13; PMID:19704024; http://dx.doi.org/ 10.1083/jcb.200903066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Praetorius HA, Spring KR. A physiological view of the primary cilium. Annu Rev Physiol 2005; 67:515–29; PMID:15709968; http://dx.doi.org/ 10.1146/annurev.physiol.67.040403.101353 [DOI] [PubMed] [Google Scholar]
- 44.Praetorius HA, Frokiaer J, Nielsen S, Spring KR. Bending the primary cilium opens Ca2+-sensitive intermediate-conductance K+ channels in MDCK cells. J Membr Biol 2003; 191:193–200; PMID:12571753; http://dx.doi.org/ 10.1007/s00232-002-1055-z [DOI] [PubMed] [Google Scholar]
- 45.Nauli SM, Jin X, AbouAlaiwi WA, El-Jouni W, Su X, Zhou J. Non-motile primary cilia as fluid shear stress mechanosensors. Methods Enzymol 2013; 525:1–20; PMID:23522462; http://dx.doi.org/ 10.1016/B978-0-12-397944-5.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 2006; 17:3781–92; PMID:16775004; http://dx.doi.org/ 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 2004; 101:13368–73; PMID:15326289; http://dx.doi.org/ 10.1073/pnas.0403453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol 2013; 9:437–43; PMID:23666116; http://dx.doi.org/ 10.1038/nchembio.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pabla N, Dong G, Jiang M, Huang S, Kumar MV, Messing RO, Dong Z. Inhibition of PKCdelta reduces cisplatin-induced nephrotoxicity without blocking chemotherapeutic efficacy in mouse models of cancer. J Clin Invest 2011; 121:2709–22; PMID:21633170; http://dx.doi.org/ 10.1172/JCI45586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang M, Wei Q, Dong G, Komatsu M, Su Y, Dong Z. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int 2012; 82:1271–83; PMID:22854643; http://dx.doi.org/ 10.1038/ki.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]