Abstract
Recent evidence that excessive lipid accumulation can decrease cellular levels of autophagy and that autophagy regulates immune responsiveness suggested that impaired macrophage autophagy may promote the increased innate immune activation that underlies obesity. Primary bone marrow-derived macrophages (BMDM) and peritoneal macrophages from high-fat diet (HFD)-fed mice had decreased levels of autophagic flux indicating a generalized impairment of macrophage autophagy in obese mice. To assess the effects of decreased macrophage autophagy on inflammation, mice with a Lyz2-Cre-mediated knockout of Atg5 in macrophages were fed a HFD and treated with low-dose lipopolysaccharide (LPS). Knockout mice developed systemic and hepatic inflammation with HFD feeding and LPS. This effect was liver specific as knockout mice did not have increased adipose tissue inflammation. The mechanism by which the loss of autophagy promoted inflammation was through the regulation of macrophage polarization. BMDM and Kupffer cells from knockout mice exhibited abnormalities in polarization with both increased proinflammatory M1 and decreased anti-inflammatory M2 polarization as determined by measures of genes and proteins. The heightened hepatic inflammatory response in HFD-fed, LPS-treated knockout mice led to liver injury without affecting steatosis. These findings demonstrate that autophagy has a critical regulatory function in macrophage polarization that downregulates inflammation. Defects in macrophage autophagy may underlie inflammatory disease states such as the decrease in macrophage autophagy with obesity that leads to hepatic inflammation and the progression to liver injury.
Keywords: autophagy, innate immunity, Kupffer cells, lipopolysaccharide, macrophage, obesity, polarization, steatohepatitis
Abbreviations: ARG1, arginase 1; BMDM, bone marrow-derived macrophages; CCL, chemokine (C-C motif) ligand; CHIL3/CHI3L3, chitinase-like 3; CD, chow diet; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPT, glutamic pyruvic transaminase, soluble; GFP, green fluorescent protein; HFD, high-fat diet; IFNG, interferon gamma; IL, interleukin; MAP1LC3/LC3B, microtubule-associated protein 1 light chain 3 β; LPS, lipopolysaccharide; MGL2, macrophage galactose N-acetyl-galactosamine specific lectin 2; MAPK, mitogen-activated protein kinase; NOS2, nitric oxide synthase 2, inducible; PBS, phosphate-buffered saline; PTGS2, prostaglandin-endoperoxide synthase 2; qRT-PCR, quantitative real-time PCR; RETNLA, resistin like α;; STAT, signal transducer and activator of transcription; TNF, tumor necrosis factor; TUNEL, terminal deoxynucleotide transferase-mediated deoxyuridine triphosphate nick end-labeling; WAT, white adipose tissue
Introduction
Recent studies have begun to elucidate important functions for macroautophagy in the control of immune responses and inflammation.1,2 Investigations focused initially on the importance of autophagy in the direct elimination of infectious organisms. The sequestration of pathogens into autophagosomes and delivery to lysosomes for degradation is an important mechanism to limit both the harmful effects of an infection and the resultant immune reaction. More recent studies have shown that autophagy has more far-reaching effects on immunity through the ability to directly regulate immune cell responsiveness to a broad array of stimuli. Prominent among these effects is the ability of autophagy to downregulate the proinflammatory response of macrophages. The increasing realization that many human diseases result from an overactive innate immune response attests to the potential importance of immune regulation by autophagy in the pathogenesis of these diseases.
The extent of tissue inflammation that develops as the consequence of an innate immune response is determined in large part by the balance between macrophage polarization into proinflammatory M1 and anti-inflammatory M2 macrophages. In response to an activating stimulus M1 polarization predominates, leading initially to release of proinflammatory cytokines, reactive oxygen species, and nitric oxide, whereas increased M2 polarization subsequently follows to mediate resolution of the inflammation.3 Autophagy has been shown to have an anti-inflammatory effect through downregulation of the inflammasome, an intracellular structure that promotes the generation of active proinflammatory cytokines interleukin IL1B and IL18.4-7 These studies failed to find an effect of autophagy on macrophage M1 polarization based on the failure of an inhibition of autophagy to alter levels of inflammasome-independent, M1-generated proinflammatory cytokines such as TNF (tumor necrosis factor) and IL6.5,6 Little is known about the ability of autophagy to affect M2 polarization except for recent reports of the nonspecific lysosomal inhibitors chloroquine8 and bafilomycin A19 impairing M2 polarization in a tumor microenvironment.
An increasingly prevalent condition that leads to human disease in association with overactivation of innate immunity is obesity. Obesity is associated with chronic, low-grade inflammation that generates proinflammatory cytokines which promote obesity-associated diseases such as nonalcoholic fatty liver disease.10,11 Endotoxemia occurs with high-fat diet (HFD) feeding in mice, and gut-derived lipopolysaccharide (LPS) triggers innate immune activation in the setting of obesity.12 Adipose stores in both obese rodents13 and humans14 become infiltrated with M1 polarized macrophages in contrast to the predominant M2 phenotype found in macrophages in lean adipose tissue.15 Similar events occur in the liver where resident hepatic macrophages or Kupffer cells become overactivated and promote the development of nonalcoholic fatty liver disease.16,17 Several factors have been implicated recently in mediating M1 and M2 polarization;18-20 however, additional as yet unidentified cellular pathways altered with obesity likely regulate macrophage polarization.
Recent studies have linked the function and regulation of autophagy with cellular lipid metabolism.21,22 We and others have demonstrated that diet-induced or genetic obesity decreases autophagic function in the liver.23,24 In contrast, increased autophagy has been reported in adipocytes from obese mice,25 suggesting that the effects of obesity on autophagy may be cell-type specific. Autophagic function in macrophages in obesity and the role of autophagy in the associated innate immune response have not been examined.
The emerging recognition that autophagy regulates immune responses, together with the ability of the obese state to alter autophagy, suggested that obesity-induced changes in macrophage autophagy could be a mechanism underlying the excessive inflammation that marks this condition. Here we report a new mechanism by which autophagy downregulates the innate immune response. We demonstrate that HFD-induced obesity is associated with decreased macrophage autophagy. Loss of autophagic function in macrophages promotes inflammation as a knockout of macrophage Atg5 in HFD-fed, LPS-stimulated mice led to a proinflammatory state. The mechanism of this effect is that autophagy is required to both suppress M1 and promote M2 polarization in macrophages. Inflammation in the liver generated by loss of macrophage autophagy is sufficient to trigger liver injury. These findings demonstrate a significant role for autophagy in macrophage polarization that regulates tissue innate immune responses such as the inflammatory state associated with obesity.
Results
Diet-induced obesity impairs macrophage autophagy
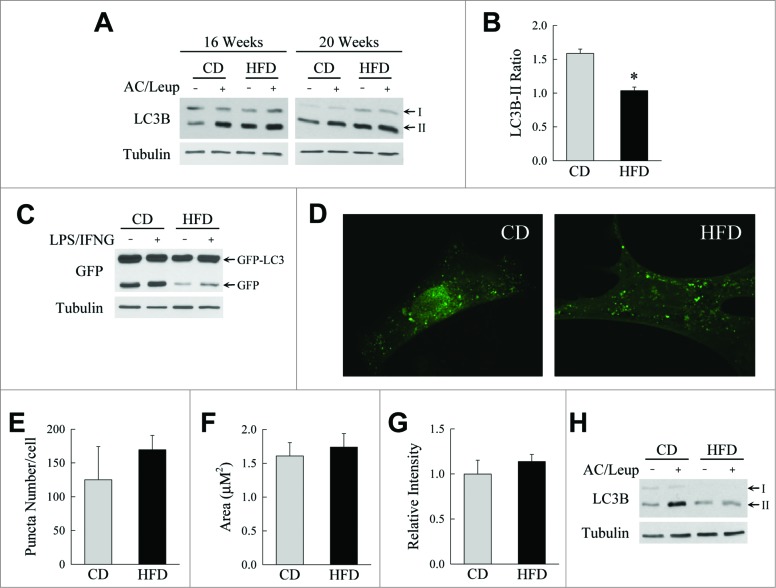
To begin to examine the potential importance of altered macrophage autophagy as a mechanism for the overactive inflammatory response in obesity, the effect of HFD-induced obesity on macrophage autophagic function was determined. Bone marrow cells were isolated from chow diet (CD)- and HFD-fed mice, differentiated into macrophages in vitro, and examined for levels of autophagic function by measure of autophagic flux.26 Bone marrow-derived macrophages (BMDM) from mice fed a HFD for 16 to 20 wk had elevated steady-state microtubule associated protein 1 light chain 3B (LC3B)-II protein levels by immunoblotting (Fig. 1A). Levels of LC3B-II increased in cells from CD-fed mice but were unchanged in BMDM from HFD-fed mice with the lysosomal inhibitors ammonium chloride and leupeptin, indicating a decrease in macrophage autophagic flux in obesity (Fig. 1A). Quantification of the levels of autophagic flux in mice fed 20 wk of HFD by densitometric scanning of the LC3B-II band intensities normalized to the intensity of the corresponding tubulin signal, and calculating the ratio the normalized LC3B-II signal in ammonium chloride and leupeptin-treated BMDM to that in untreated BMDM, confirmed that autophagic function was significantly impaired in macrophages from HFD-fed mice (Fig. 1B). This defect in BMDM autophagy was also demonstrated by studies of cleavage of an adenovirally expressed green fluorescent protein (GFP), in a GFP-LC3 fusion construct. Levels of free GFP, which reflect levels of autophagy, were markedly decreased in BMDM from HFD-fed mice unstimulated or treated with LPS and IFNG (interferon gamma) (Fig. 1C). No significant increase in free GFP occurred with LPS and IFNG treatment, which is consistent with previous findings that LPS does not alter autophagic function in BMDM,6 although other reports have shown LPS to induce autophagy in these cells.27
Figure 1.
HFD-induced obesity inhibits macrophage autophagy. (A) Immunoblots of total protein from BMDM from wild-type C57 BL/6 mice fed CD or HFD for 16 or 20 wk. Some cells were treated with ammonium chloride and leupeptin (AC/Leup) for 2 h. Proteins were probed for LC3B and tubulin, and LC3B-I and LC3B-II bands are indicated by arrows. (B) Quantification of the ratio of LC3B-II in ammonium chloride- and leupeptin-treated cells to untreated cells from CD- and HFD-fed BMDM from 20-wk-old animals by densitometric scanning of immunoblots (n = 4). Data presented as the mean ± SEM. *P < 0.003 as compared to control. (C) Western blots of total protein from BMDM from CD- or HFD-fed mice infected with GFP-LC3 adenovirus that were untreated or treated with LPS and IFNG for 8 h probed for GFP and tubulin. The GFP-LC3 and free GFP bands are highlighted with arrows. (D) Confocal images of GFP-LC3 puncta in BMDM from CD- and HFD-fed mice. (E to G) Quantification of GFP-LC3 puncta number, area and intensity in the BMDM from these mice (n = 9 to 13). (H) Western blots of total protein from peritoneal macrophages isolated from CD- and HFD-fed mice untreated or ammonium chloride and leupeptin-treated for 2 h.
Decreased macrophage autophagy in obesity could result from a defect in autophagosome formation or from problems in subsequent steps of the autophagic pathway. To determine whether autophagosome formation was altered by HFD feeding, BMDM were infected with GFP-LC3 adenovirus, stimulated with rapamycin to induce autophagy, and LC3 puncta representing autophagosomes examined by confocal microscopy. In response to rapamycin, high numbers of LC3 puncta were present in BMDM from mice fed either CD or HFD (Fig. 1D). Rapamycin induced equivalent numbers of autophagosomes in BMDM from CD- and HFD-fed mice by measures of LC3 puncta number, area, and fluorescent intensity (Fig. 1E to G). The finding that the cellular defect in macrophage autophagy occurs after the stage of autophagosome formation is consistent with previous studies in other cell types in which excessive cellular lipid accumulation impaired autophagic function downstream of autophagosome formation.28,29
To further establish that autophagy is impaired in macrophages in obese mice, autophagic flux was assessed in a second, primary macrophage type. Peritoneal macrophages were isolated from mice after thioglycollate injection. In contrast to peritoneal macrophages from CD-fed mice in which LC3B-II levels increased markedly with lysosomal inhibition, LC3B-II levels in cells from HFD-fed mice were unchanged by ammonium chloride and leupeptin treatment (Fig. 1H), confirming a defect in macrophage autophagy in these animals. Thus, HFD-induced obesity leads to impaired macrophage autophagy.
Inhibition of autophagy skews macrophage polarization to an M1 phenotype
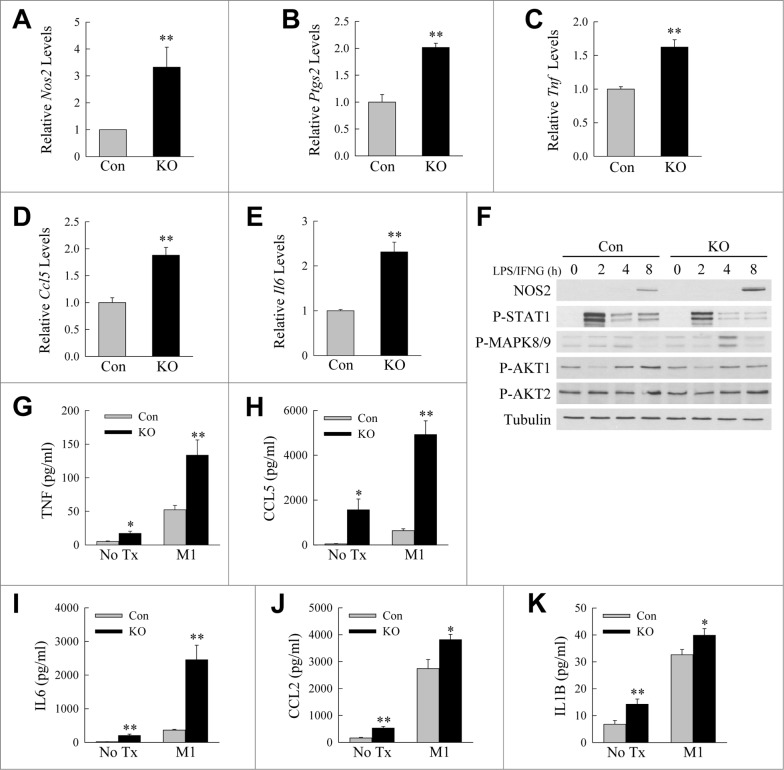
Findings of impaired autophagy in macrophages from obese mice, together with the fact that the heightened immune response in obesity is associated with an imbalance in M1 and M2 macrophages, suggested that defective autophagy may be a mechanism of altered macrophage polarization in obesity. We therefore examined the effects of the loss of autophagy on BMDM polarization in vitro. As a source of primary macrophages with an inhibition of macrophage autophagy, mice were generated with a macrophage-specific knockout of the autophagy gene Atg5. Atg5f/f mice with floxed alleles for the autophagy gene Atg530 were crossed with Lyz2-Cre mice31 expressing a myeloid cell-specific Cre to generate atg5myeΔ mice which have been previously demonstrated to have decreased macrophage autophagy.32 BMDM were isolated from littermate control and atg5myeΔ mice, M1 polarized by treatment with LPS and IFNG, and levels of M1 biomarker genes measured by quantitative real-time PCR (qRT-PCR). M1 biomarker genes increased dramatically with LPS and IFNG treatment (data not shown). BMDM from atg5myeΔ mice had a significantly increased induction of M1 markers by qRT-PCR as compared to cells from littermate control mice. Levels of M1 genes such as the markers Nos2 (nitric oxide synthase 2, inducible) and Ptgs2/Cox2 (prostaglandin-endoperoxide synthase 2/cyclooxygenase 2), and the cytokines Tnf, Ccl5 (chemokine [C-C motif] ligand 5) and Il6, were increased in knockout BMDM 1.5-3.6 fold at 12 h (Fig. 2A to E) and 1.4 to 13.4 fold at 24 h after LPS and IFNG treatment (Fig. S1). Increased induction of the M1 marker NOS2 in knockout macrophages was confirmed by immunoblotting (Fig. 2F). Knockout cells secreted significantly greater amounts of proinflammatory cytokines including TNF, CCL5, IL6, CCL2, and IL1B unstimulated and with LPS and IFNG treatment (Fig. 2G to K). Levels of secreted TNF, CCL5, and IL6 in particular were increased 2.5- to 6.8-fold with LPS and IFNG treatment (Fig. 2G to I). Analysis of cell signaling pathways associated with M1 polarization revealed that knockout cells had decreased phosphorylation and therefore reduced activation of STAT1 (signal transducer and activator of transcription 1) and increased activation of JUN/c-Jun N-terminal kinase or MAPK8/9 (mitogen-activated protein kinase 8/9), effects that would promote M1 polarization,33,34 but no change in AKT1 and AKT2 phosphorylation (Fig. 2F). Inhibition of macrophage autophagy therefore promotes M1 polarization resulting in increased proinflammatory cytokine production.
Figure 2.
Loss of ATG5-dependent macrophage autophagy increases M1 polarization. ((A)to E) Relative mRNA levels of Nos2, Ptgs2, Tnf, Ccl5, and Il6 in BMDM from littermate control (Con) and atg5myeΔ knockout (KO) mice 12 h after treatment with LPS and IFNG (n = 4). (F) Immunoblots of total protein isolates from control and knockout BMDM treated with LPS and IFNG for the indicated number of hours and probed for the total and phosphorylated (P-) proteins shown. (G to K) Levels of TNF, CCL5, IL6, CCL2, and IL1B secreted into the culture medium by control and knockout BMDM with no treatment (No Tx) or stimulated with LPS and IFNG (M1) for 24 h (n = 6 to 8). All data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01 versus control cells with the same treatment.
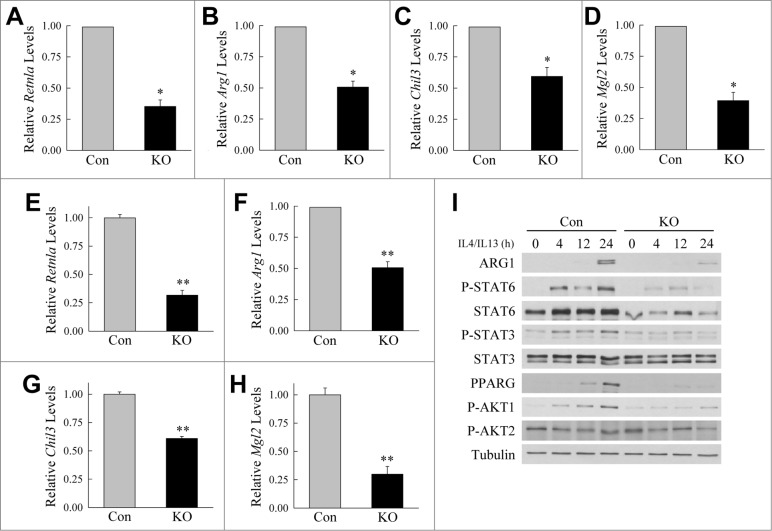
To determine whether autophagy also regulated M2 polarization, BMDM were polarized into M2 cells with IL4 and IL13. M2 gene expression was virtually undetectable in differentiated BMDM but increased markedly with IL4 and IL13 (data not shown). A comparison between control and Atg5 null BMDM revealed a significant reduction in the levels of M2 marker genes in knockout cells at 12 h (Fig. 3A to D) and 24 h (Fig. 3E to H). Levels of mRNA induction for the M2 genes Retnla (resistin like α), Arg1 (arginase 1), Chil3/Chi3 l3 (chitinase-like 3) and Mgl2 (macrophage galactose N-acetyl-galactosamine specific lectin 2) were significantly decreased by 40% to 69% in macrophages lacking autophagy. Knockout cells also had decreased levels of ARG1 protein (Fig. 3I). In response to IL4 and IL13, knockout cells had marked defects in signaling pathways that promote M2 polarization including P-STAT6, P-STAT3, PPARG (peroxisome proliferator-activated receptor gamma), and P-AKT1 with no change in P-AKT2 (Fig. 3I). Thus, defective autophagy promoted a proinflammatory macrophage phenotype by 2 mechanisms—promotion of M1 and inhibition of M2 macrophage polarization.
Figure 3.
Atg5-deficient macrophages undergo decreased M2 polarization. (A to D) Relative mRNA levels of Retnla, Arg1, Chil3, and Mgl2 in BMDM from littermate control (Con) and atg5myeΔ knockout (KO) mice 12 h after treatment with IL4 and IL13 (n = 4). (E to H) Relative mRNA levels of Retnla, Arg1, Chil3, and Mgl2 in control and knockout BMDM 24 h after treatment with IL4 and IL13 (n = 6 to 12). All data are presented as the mean ± SEM. *P < 0.01 and **P < 0.00001 vs. control BMDM. (I) Immunoblots of total protein isolates from control and knockout BMDM treated with IL4 and IL13 for the number of hours shown and probed for the indicated total and phosphorylated (P-) proteins.
BMDM from HFD-fed mice have altered M1 and M2 polarization
If autophagic function in macrophages is significantly inhibited by HFD feeding, and autophagy regulates macrophage polarization, then macrophages from HFD-fed mice should have altered polarization similar to atg5 null macrophages. We therefore compared differentiated BMDM from CD- and HFD-fed mice for their responses to M1 and M2 stimulation. BMDM from HFD-fed mice had increased M1 polarization in comparison to cells from CD-fed mice as determined by induction of M1 marker genes in response to LPS and IFNG by qRT-PCR (Fig. S2A to C). With IL4 and IL13 treatment to induce M2 polarization, levels of M2 marker genes were decreased in BMDM from HFD-fed mice (Fig. S2D to F). HFD feeding therefore mimicked the effects of a genetic inhibition of autophagy by promoting a proinflammatory macrophage phenotype from increased M1 and decreased M2 polarization.
Inhibition of myeloid cell autophagy has minor effects on body mass and metabolism in obese mice with low-grade endotoxemia
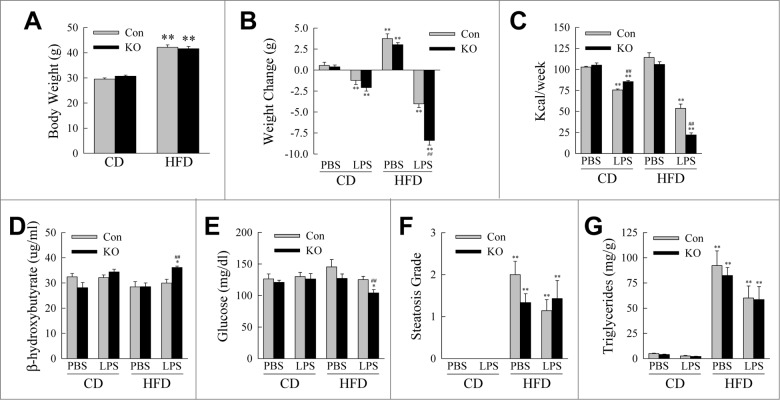
Findings that obesity affected macrophage autophagy and that autophagy regulated the inflammatory phenotype of macrophages suggested that decreased autophagy may play a role in the dysregulation of the innate immune response in obesity. To examine this possibility we determined the effects of a knockout of macrophage Atg5 on obesity-associated inflammation in vivo using a combined HFD feeding and low-dose LPS mouse model.35 Littermate control Atg5f/f and atg5myeΔ mice were fed CD or HFD for 12 wk after weaning and then injected daily with phosphate-buffered saline (PBS) or low dose LPS (0.25 mg/kg) for 2 wk. Prior to LPS treatment, Atg5f/f and atg5myeΔ mice on CD or HFD had equivalent body weights (Fig. 4A). All LPS-treated mice had significant weight loss in comparison to CD-fed, PBS-injected control mice, but only with HFD feeding was the decrease in weight significantly greater in knockout mice (Fig. 4B). Weight loss from LPS treatment resulted in part from decreased food intake with the greatest reduction occurring in HFD-fed knockout mice (Fig. 4C). HFD-fed knockout mice also had slightly increased rates of β-oxidation as indicated by elevated serum levels of β-hydroxybutyrate (Fig. 4D). HFD-fed, LPS-treated mice also had a modest but significant decrease in serum glucose levels (Fig. 4E). These metabolic effects were not sufficient to affect hepatic lipid accumulation as the histological degree of steatosis and liver triglyceride content were equivalent in control and knockout mice (Fig. 4F and G). Therefore decreased macrophage autophagy had only minor effects on metabolic function in obese mice.
Figure 4.
Metabolic effects of the knockout of Atg5 in macrophages with HFD feeding and LPS. (A) Body weights of littermate control (Con) and atg5myeΔ knockout (KO) mice after 12 wk of CD or HFD feeding (n = 21 to 26). (B) Change in body weight after 2 wk of phosphate-buffered saline (PBS) or LPS injections (n = 9 to 13). (C) Caloric intake in the different groups of mice (n = 4 or 5). (D) Serum β-hydroxybutyrate levels (n = 3 to 7). (E) Serum glucose levels (n = 9 to 13). (F) Histological grades of steatosis (n = 5 to 8). (G) Liver triglyceride content (n = 9 to 13). All data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01 versus CD-fed, PBS-treated littermate control mice.##P < 0.01 vs. littermate control mice with the same diet and treatment.
Lack of macrophage autophagy promotes a proinflammatory state
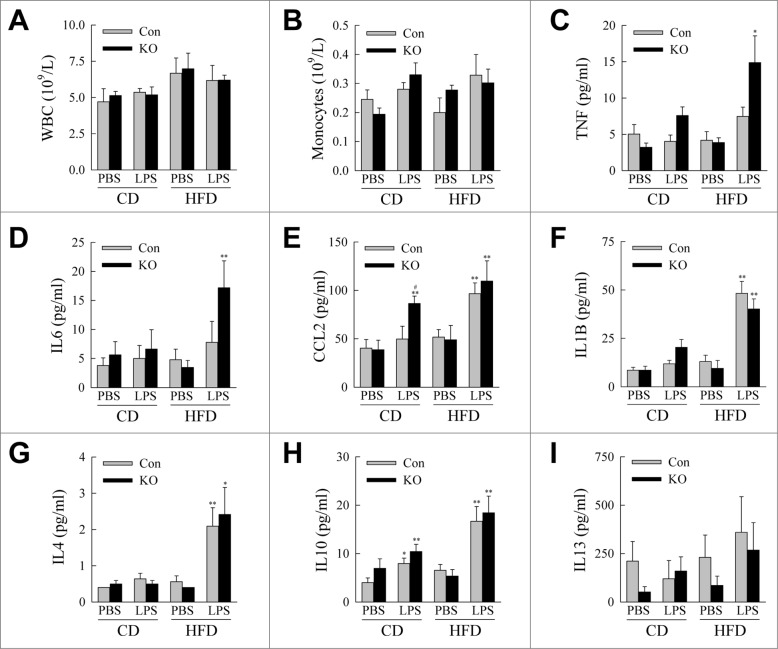
We then examined whether decreased macrophage autophagy alters the development of obesity-associated inflammation. As expected serum total white blood cell and monocyte counts were unaffected in knockout mice (Fig. 5A and B), consistent with the previously reported phenotype of atg5 macrophage knockout mice,36 and with the fact that serum white cell counts are unaffected by obesity. Serum levels of the proinflammatory cytokines TNF and IL6 were significantly increased in HFD-fed, LPS-treated knockout mice (Fig. 5C and D), although other inflammatory cytokines such as CCL2 and IL1B were increased by the HFD and LPS but equivalent in control and knockout mice (Fig. 5E and F). Serum levels of the anti-inflammatory cytokines IL4, IL10, and IL13 were unaltered by the loss of autophagy irrespective of whether they were increased by HFD or LPS (Fig. 5G to I). These findings indicate that with decreased macrophage autophagy a selective increase in proinflammatory cytokines occurs in response to HFD feeding and endotoxemia.
Figure 5.
atg5-knockout mice exhibit evidence of systemic inflammation. (A) Serum white blood cell (WBC) counts (n = 4 to 6). (B) Serum monocyte counts (n = 4 to 6). (C to I) Serum levels of TNF, IL6, CCL2, IL1B, IL4, IL10 and IL13 (n = 6 to 10). All data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01 versus CD-fed, PBS-treated littermate control mice.#P < 0.05 vs. littermate control mice with the same diet and treatment.
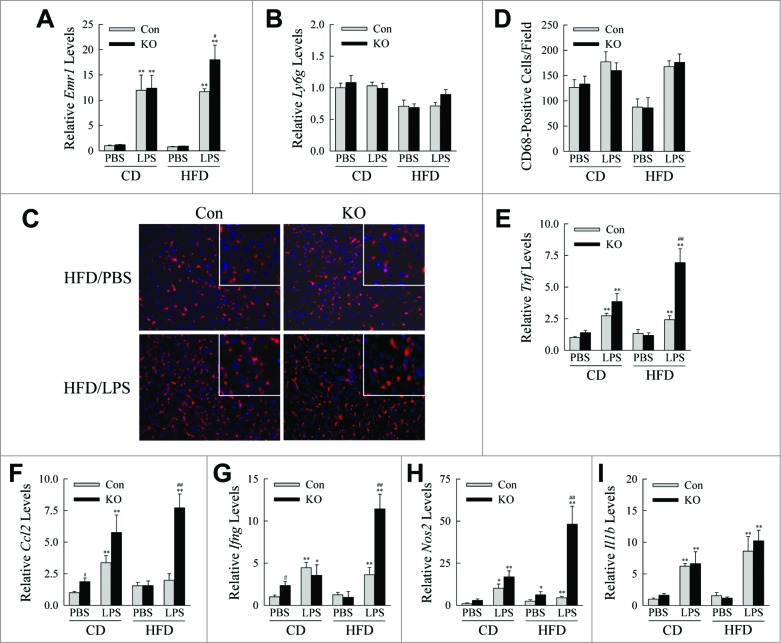
In light of systemic evidence of a proinflammatory state, the effects of the loss of macrophage autophagy on tissue-specific inflammation were determined. In the liver, a critical target organ for the heightened inflammation in obesity, hepatic macrophage but not neutrophil infiltration occurred in both control and knockout livers in response to LPS as indicated by markedly increased mRNA levels by qRT-PCR for Emr1/F4/80 with no change in Ly6 g (Fig. 6A and B). Immunofluorescence staining for CD68+ macrophages revealed a trend to increased numbers of cells in the livers of all LPS-injected mice, but no difference between control and knockout animals (Fig. 6C and D). However, HFD-fed, LPS-treated atg5myeΔ mice had markedly increased hepatic mRNA levels for the proinflammatory cytokines and M1 markers Tnf, Ccl2, Ifng and Nos2 (Fig. 6E to H). Although Il1b mRNA content was markedly induced by LPS, the levels were unaffected by the lack of autophagy (Fig. 6I). These findings demonstrate that loss of macrophage autophagy promoted a proinflammatory macrophage phenotype in the liver without altering macrophage number.
Figure 6.
HFD-fed, LPS-injected knockout mice have increased hepatic inflammation. (A, B) Relative levels of Emr1 and Ly6 g mRNA in the livers of CD- and HFD-fed mice treated with PBS or LPS (n = 4 to 6). (C) Immunofluorescence staining for CD68 in livers (200 × magnification). Enlarged images are shown in the inserts. (D) Numbers of CD68+ cells per field (n = 3). (E to I) Relative hepatic mRNA levels for Tnf, Ccl2, Ifng, Nos2, and Il1b (n = 4 to 6). All data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01 versus CD-fed, PBS-treated littermate control mice. #P < 0.05 and ##P < 0.01 vs. littermate control mice with the same diet and treatment.
Another critical site for obesity-associated inflammation is white adipose tissue (WAT).10 Levels of the macrophage marker Emr1 were increased in epididymal WAT with either LPS treatment or HFD feeding, but to a lesser extent in HFD-fed, LPS-treated knockout mice (Fig. S3A). In addition, mRNA levels of the proinflammatory cytokines and M1 markers Tnf, Ccl2, Ifng and Nos2, although markedly increased by HFD feeding and/or LPS treatment, were unaffected by loss of macrophage autophagy in contrast to the findings in liver (Fig. S3B to E). The loss of macrophage autophagy had no effect on the degree of WAT inflammation as indicated by equivalent histological grades of inflammation in control and atg5myeΔ mice (Fig. S3F and S4). Thus, despite the global nature of the atg5 macrophage knockout, a heightened proinflammatory immune response developed in the liver but not in WAT.
Hepatic macrophages from knockout mice hyperpolarize to an M1 phenotype
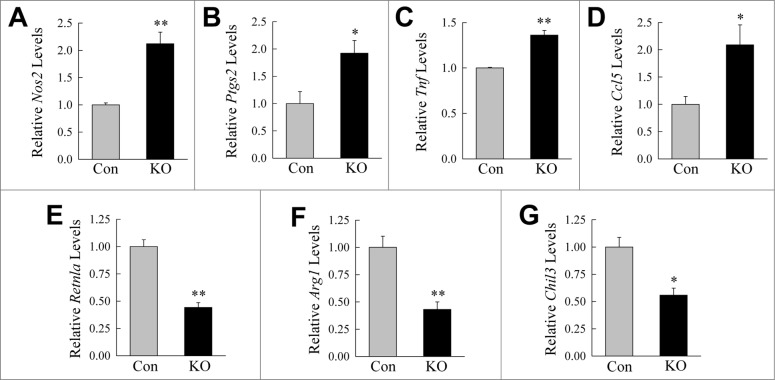
The combined findings that autophagy regulated macrophage polarization, and that macrophage autophagy-deficient mice developed increased hepatic inflammation, suggested that with obesity, defective autophagy leads to hepatic macrophage proinflammatory polarization. To examine this possibility the effect of the loss of ATG5-dependent autophagy on macrophage polarization in primary hepatic resident macrophages or Kupffer cells was determined. Macrophages were isolated from the livers of HFD-fed, LPS-treated control and atg5myeΔ mice and induced to undergo M1 or M2 polarization. Hepatic macrophages isolated from knockout mice exhibited increased M1 polarization in response to LPS and IFNG as determined by levels of M1 markers Nos2, Ptgs2, Tnf, and Ccl5 (Fig. 7A to D). M2 polarization from IL4 and IL13 was decreased in knockout Kupffer cells as reflected in reduced mRNA levels of Arg1, Retnla, and Chil3 (Fig. 7E to G). Thus, consistent with findings in BMDM, autophagy-defective Kupffer cells hyperpolarize to an M1 phenotype. These findings, together with the data demonstrating that there was no increase in macrophage number in the livers of HFD-fed, LPS-treated atg5myeΔ mice, demonstrate that liver inflammation developed in these mice due to increased proinflammatory hepatic macrophage polarization.
Figure 7.
Autophagy regulates Kupffer cell M1 and M2 polarization. (A to D) Relative mRNA levels in Kupffer cells from control (Con) and knockout (KO) mice for Nos2, Ptgs2, Tnf and Ccl5 in response to LPS and IFNG treatment for 12 h. (E to G) mRNA levels 12 h after IL4 and IL13-induced M2 polarization for Retnla, Arg1, and Chil3 (n = 3). All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01 versus control mice.
Overactivation of the hepatic immune response leads to steatohepatitis
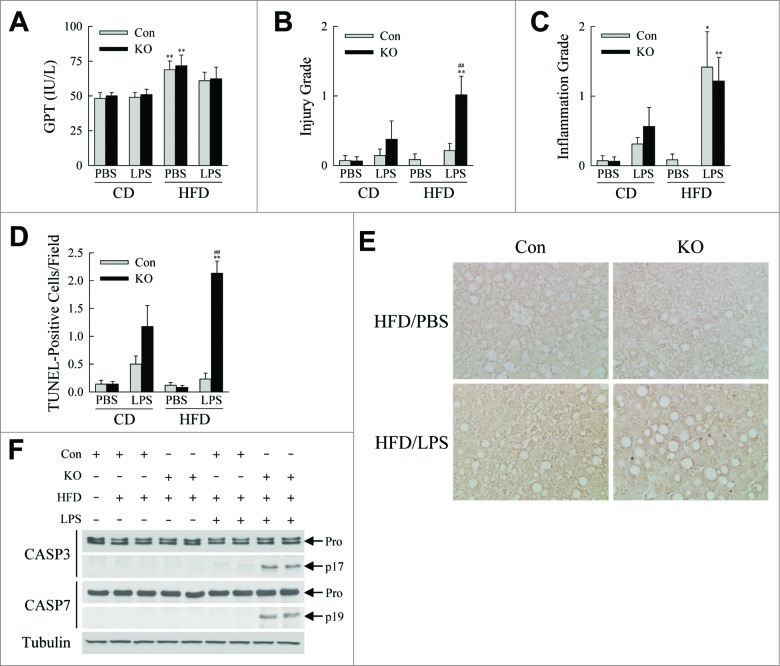
The proinflammatory state associated with obesity is thought to mediate organ-specific complications such as nonalcoholic fatty liver disease. Autophagy-deficient mice were therefore examined for whether their heightened state of hepatic inflammation promoted liver injury. Serum levels of GPT (glutamic-pyruvic transaminase, soluble), a marker of liver injury, were not significantly different between control and knockout mice (Fig. 8A). However, blinded grading of the degree of histological liver injury revealed that the livers of HFD-fed, LPS-treated knockout mice had significantly increased hepatocyte injury and cell death (Fig. 8B and S5). In contrast, consistent with the qRT-PCR and immunofluorescence findings, hepatic inflammation was induced by LPS treatment, but the histological extent of inflammatory cell infiltration was unaffected by the loss of autophagy (Fig. 8C). Increased hepatocyte death was confirmed by the finding of significantly increased numbers of terminal deoxynucleotide transferase-mediated deoxyuridine triphosphate nick end-labeling (TUNEL)-positive or apoptotic cells only in the livers of HFD-fed, LPS-treated knockout mice (Fig. 8D and E). This increase in apoptotic hepatocyte death was further supported by evidence of cleaved CASP3/caspase 3 and CASP7 only in the knockout livers with positive TUNEL staining (Fig. 8F). The LPS-induced overactivation of the innate immune response that occurred in the livers of obese mice from an inhibition of macrophage autophagy therefore triggered hepatocyte injury and cell death.
Figure 8.
HFD-fed, LPS-treated knockout mice develop liver injury. (A) Serum GPTs in CD- and HFD-fed mice treated with PBS or LPS. (B) Histological grades of liver injury in the same mice. (C) Histological grades of hepatic inflammation. (D) Numbers of TUNEL-positive cells per field (400× magnification) (n = 6 to 8). All data are presented as the mean ± SEM. *P < 0.05 and **P < 0.01 vs. CD-fed, PBS-treated littermate control mice.##P < 0.01 versus littermate control mice with the same diet and treatment. (E) Representative TUNEL staining (400× magnification). (F) Immunoblots of total hepatic protein from CD- and HFD-fed control and knockout mice that were PBS- (-LPS) or LPS (+LPS) treated. Immunoblots were probed with antibodies for CASP3/caspase 3, CASP7/caspase 7 and tubulin. Arrows indicate the procaspase (Pro), the cleaved CASP3 (p17) and the cleaved CASP7 (p19) forms. The images with the cleaved forms are longer exposures of the procaspase immunoblots.
Discussion
Considerable interest exists in understanding whether defects in autophagy underlie the pathogenesis of human diseases. One disease-associated state marked by alterations in autophagic function is obesity in which opposing effects on autophagy have been reported depending on the cell type studied. Autophagy is decreased in hepatocytes,23,24 but increased in pancreatic β cells37 in obese rodents, and increased in adipocytes from both obese rodents and humans.25,38 The present study examined the effects of obesity on macrophage autophagy for the first time and demonstrates that autophagy is decreased in both primary BMDM and peritoneal macrophages from mice with HFD-induced obesity. The finding that bone marrow cells from HFD-fed mice retain decreased autophagic function even after considerable time in culture undergoing differentiation suggests that epigenetic changes have been induced in these cells by the obese state.
The impairment in macrophage autophagic function with obesity, together with the known involvement of autophagy in immune responsiveness, suggested that decreased macrophage autophagy may contribute to the dysregulated inflammatory response that characterizes obesity. The current study demonstrates that impaired macrophage autophagy leads to the development of obesity-associated inflammatory disease based on findings that: (1) macrophages isolated from bone and liver of atg5 knockout mice have increased M1 polarization and decreased M2 polarization in vitro; (2) macrophages from obese mice with defective autophagy have an identical proinflammatory phenotype with elevated M1 and decreased M2 polarization; (3) HFD-induced weight gain and LPS treatment, which by themselves were insufficient to trigger inflammation, led to systemic and hepatic inflammation in mice with a knockout of macrophage Atg5; and (4) HFD-fed, LPS-treated mice lacking ATG5-dependent macrophage autophagy develop a classic inflammatory disease of obesity, steatohepatitis.
The current findings identify a new mechanism by which autophagy regulates innate immunity through effects on M1 and M2 polarization. Other investigations have demonstrated that autophagy regulates immune responsiveness, but studies to date have largely implicated autophagy in the sequestration and removal of infectious organisms.2,39,40 Autophagy has been suggested to play a role in macrophage production by regulating haematopoietic stem cell fate, monocyte recruitment, and macrophage differentiation.41 However, circulating monocyte numbers and hepatic macrophage recruitment were unaffected in our model. Anti-inflammatory functions of autophagy have been demonstrated in the intestine of a mouse model of colitis,6 and polymorphisms of autophagy genes are associated with the human inflammatory intestinal condition of Crohn disease.42 Autophagy inhibits inflammation through the downregulation of inflammasome formation and IL1B cleavage to its active form.5,6 Effects of autophagy on the inflammasome cannot explain the increased inflammatory response in obese mice as serum IL1B and hepatic Il1b mRNA levels were unchanged in knockout mice. Rather the mechanism by which autophagy regulates the hepatic immune response in obesity is through an effect on macrophage polarization. In contrast to the lack of change in IL1B, serum protein and liver mRNA for other proinflammatory, inflammasome-independent cytokines such as TNF and IL6 were increased in knockout mice. M1-stimulated knockout BMDM and Kupffer cells also had more marked increases in these cytokines than in IL1B. Other studies have found autophagy to only affect inflammasome-mediated proinflammatory cytokine production.5,6 This discrepancy may result from differences in experimental design as other studies relied strictly on thioglycollate-activated peritoneal macrophages and inflammasome-specific stimuli such as LPS and ATP, or the failure to employ fully differentiated macrophages as loss of autophagy blocks macrophage differentiation.43
Mice with a knockout of myeloid cell Atg5 were phenotypically normal after HFD feeding. When HFD feeding was combined with low-dose LPS to mimic the endotoxemia of obesity, knockout mice developed systemic inflammation as indicated by increased serum levels of the proinflammatory cytokines TNF and IL6. Knockout mice had minor decreases in body weight and serum glucose as compared to control mice, effects that would only reduce rather than promote a proinflammatory state. Therefore inflammation was not secondary to a metabolic effect induced by the knockout of macrophage Atg5. Knockout mice also had a markedly increased proinflammatory immune response in the liver, a critical site of inflammation in obesity. Although myeloid cell-specific Lyz2-Cre is expressed in neutrophils as well as macrophages, the lack of effect on neutrophil number and the macrophage-specific profile of cytokine production indicated that the hepatic inflammation was macrophage mediated. Autophagy therefore functions in macrophages to dampen the innate immune response to obesity. Effects of the loss of autophagy were only apparent in HFD-fed, obese mice as in lean mice the absence of macrophage autophagy had little impact on the inflammatory response to LPS. Thus, in obesity, which leads to increased immune cell stimulation from factors other than LPS such as free fatty acids, macrophage autophagy is critical to downregulate the inflammatory response.
Despite generalized immune activation from HFD feeding and LPS, loss of macrophage autophagy specifically affected the liver as the amount of inflammation in WAT was unchanged. The confinement of the proinflammatory effect of the loss of ATG5-dependent autophagy to the liver explains the specific rise in the systemic circulation of the cytokines TNF and IL6, which are produced in large quantities in the liver. The restricted liver effect could represent tissue-specific differences in macrophage populations that are mediated by autophagy. Alternatively, the results could reflect unique aspects of liver anatomy and immune responsiveness. Due to the vascular supply, endogenous, gut-derived LPS travels first to the liver where the LPS is removed preventing its systemic circulation. Although the present study employed exogenous LPS, the mode of experimental LPS delivery mimicked normal endogenous LPS exposure as intraperitoneal absorption of the injected LPS also carried it directly to the liver. In addition to its unique LPS exposure, the liver contains the vast majority (80% to 90%) of the resident macrophages in the body, making this organ particularly susceptible to damage from a dysregulated immune response.44 The unique extent of LPS exposure and extensive macrophage population make the liver a major determinant of systemic responses to LPS,45 and may dictate additional mechanisms of hepatic macrophage plasticity not required in other tissues including a function for autophagy. Consistent with the concept that the effects specific to the liver were secondary to organ environment is the finding of similar effects of the loss of autophagy in both BMDM and Kupffer cells.
The livers of HFD-fed, LPS-treated knockout mice had a heightened proinflammatory response in the absence of an increase in macrophage number indicating that the macrophages in the liver had been skewed to a more inflammatory phenotype. Consistent with the in vivo findings, in both BMDM and Kupffer cells loss of autophagy increased M1 and decreased M2 polarization. The similar findings in BMDM as well as Kupffer cells indicate that there is no inherent difference in hepatic macrophages that led to the liver-specific increase in inflammation. Rather, factors in the hepatic environment must contribute to this process such as high LPS exposure or free fatty acids as discussed previously. A variety of cell signaling pathways have been implicated in the regulation of macrophage polarization.18-20 Effects were seen on several factors that regulate M1 and M2 polarization. Knockout cells had decreased STAT1 and increased MAPK8/9 activation in response to LPS and IFNG, suggesting a multifactorial mechanism for M1 promotion. Similarly decreased activation of multiple promoters of M2 polarization occurred in knockout macrophages with IL4 and IL13 stimulation also indicating a complex mode of inhibition of M2 polarization.
An unrestrained M1 macrophage response has been previously implicated in prolonging tissue injury,46 but the present findings demonstrate that an M1-skewed immune response is sufficient in itself to initiate injury as knockout livers had increased hepatic injury and cell death despite equivalent steatosis. In nonalcoholic fatty liver disease the mechanisms by which simple fatty livers progress to the more serious liver disease of steatohepatitis remain unknown. The present findings suggest that defects in macrophage autophagy may be a factor mediating the development of hepatic inflammation and injury that characterizes steatohepatitis. Similarly a study of HFD-fed aged mice who suffer from impaired autophagy,47 reveals that aging has no effect on WAT inflammation or hepatic steatosis, but increases liver inflammation and injury, as well as hepatocyte apoptosis.48 The loss of macrophage autophagic function with aging may explain in part the increased prevalence of steatohepatitis,49 as well as of the metabolic syndrome,50 in aged humans. In the liver, heightened levels of inflammation may also promote later complications such as hepatocellular carcinoma, making autophagic function in macrophages an important therapeutic target in steatohepatitis and other obesity-related diseases.
Materials and Methods
Mouse models
Male mice were maintained under 12 h light/dark cycles with unlimited access to food and water. C57 BL/6 mice were the source of wild-type macrophages. Atg5f/f mice30 with floxed alleles for the autophagy gene Atg5 were crossed with Lyz2-Cre mice31 with the mouse lysozyme M promoter-driven Cre recombinase to generate Lyz2-Cre-Atg5f/f or atg5myeΔ mice. Both strains of transgenic mice were on a C57 BL/6 background. Control mice for all experiments were littermate Atg5f/f mice lacking the Cre transgene. CD (5% fat; LabDiet, PicoLab Rodent Diet 20, 5053) or HFD (60% fat; Research Diets, D12492) feeding was started after weaning. After 12 wk of CD or HFD, mice received daily intraperitoneal injections of 0.25 mg/kg of LPS (E. coli 0111:B4; Sigma, L4391), a concentration of LPS commonly used to mimic the low-grade endotoxemia of the metabolic syndrome,12,35 or an equivalent volume of PBS (Mediatech, MT21031CV) for 2 wk. All animal studies were approved by the Animal Care and Use Committee of the Albert Einstein College of Medicine and followed the National Institutes of Health guidelines for animal care.
Macrophage isolation, culture, and polarization
Bone marrow flushed from mouse femurs and tibias was plated in Dulbecco's modified Eagle medium (Mediatech, 10–017-CV) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, S11550 H), 2% 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (Fisher, BP-299–100), 1% nonessential amino acids (Sigma, M7145) and 1% antibiotic-antimycotic solution (Life Technologies, 15240–096). To obtain differentiated macrophages, 5 ng/ml of macrophage colony-stimulating factor (R&D Systems, 416-ML) were added every 2 d for 6 d. BMDM were stimulated with 10 ng/ml LPS (Sigma, L4391) and 100 g/ml IFNG (R&D Systems, 485-MI) to promote M1 polarization, or 10 ng/ml IL4 (R&D Systems, 404-ML) and 20 ng/ml IL13 (R&D Systems, 413-ML) to induce M2 polarization. To measure autophagic flux cells were treated with 20 mM ammonium chloride (Sigma, A0171) and 100 μM leupeptin (Fisher, 50824371) for 2 h. To induce autophagy, cells were treated for 24 h with 100 nM rapamycin (EMD Millipore, 53123-88-9).
Peritoneal macrophages were isolated from thioglycollate-injected mice. Mice were injected intraperitoneally with 1 ml of 3% thioglycollate (Sigma, B2551) and their ascites collected 4 d later and centrifuged to obtain the cell pellet. Pellets were pooled from 4 or 5 mice per group, resuspended in red blood cell lysis buffer, and then the cells seeded in the same medium as for BMDM. After 3 h, unattached cells were removed and the adherent cells washed twice with medium alone before the addition of fresh culture medium. Cells were treated the next morning with ammonium chloride and leupeptin as above.
Protein isolation and western blotting
Total macrophage and liver protein fractions were isolated as previously described.51,52 Protein concentrations were determined by the Bio-Rad protein assay, and western blotting performed as previously described.51 Membranes were exposed to antibodies that recognized LC3B (Cell Signaling Technology, 2775), GFP (Cell Signaling Technology, 2555), tubulin (Cell Signaling Technology, 2148), P-STAT1 (Cell Signaling Technology, 9177), P-MAPK8/9 (Cell Signaling Technology, 9251), P-AKT1 (Cell Signaling Technology, 9018), P-AKT2 (Cell Signaling Technology, 8599), NOS2 (Cell Signaling Technology, 2977), P-STAT6 (Cell Signaling Technology, 9361), STAT6 (Cell Signaling Technology, 9362), P-STAT3 (Cell Signaling, 9131), STAT3 (Cell Signaling Technology, 9132), PPARG (Cell Signaling Technology, 2443), CASP3/caspase 3 (Cell Signaling Technology, 9665), CASP7/caspase 7 (Cell Signaling Technology, 9492) and ARG1 (Santa Cruz Biotechnology, 20150). Western blot signals were quantitated by a FluorChem densitometer (Alpha Innotech, San Leandro, CA).
Adenovirus preparation and infection
Autophagic function was also determined by GFP-LC3 cleavage using the adenovirus AdGFP-LC3 (provided by Xiao-Ming Yin, Indiana University School of Medicine) which expresses a cleavable GFP-LC3 fusion protein.53 Viruses were amplified in 293 cells, purified by banding twice on cesium chloride gradients, as previously described,54 and titered by plaque assay. BMDM were infected at an MOI of 50, as previously described,55 and 18h later untreated or LPS and IFNG-treated cells were harvested for protein isolation. Levels of free GFP cleaved from adenoviral GFP-LC3 were determined by immunoblotting.
Confocal microscopy for LC3 puncta
To evaluate the formation of LC3 puncta, images were acquired of AdGFP-LC3-infected BMDM with a Leica SP5 confocal microscope (Buffalo Grove, IL). An argon 488 nm wavelength laser was used as the excitation source with an emission of 500 to 600 nm. Oil immersion objective HCX PL APO 63.0 × 1.40 was used to scan a 1024 × 1024 frame size. All images used 1 Airy unit pinhole size 95.5 μm. Images were analyzed using Imaris software to segment the puncta structure. Using the intensity-based thresholding method, the number, size and intensity of each puncta were compiled.
Real-time polymerase chain reaction
RNA was isolated from cells or mouse tissues using the commercial kit RNeasy Plus (QIAGEN, 74106). Reverse transcription was carried out with 1 μg of RNA in an Eppendorf Mastercycler (Hauppauge, NY), using a high capacity cDNA reverse transcription kit (ABI, 4368813). Annealing of primers was done at 25°C for 10 min, followed by elongation at 37°C for 2 h and inactivation of the enzyme at 85°C for 5 min. PCR was performed in triplicate in a 7500 Fast Real-Time PCR System (ABI, Grand Island, NY). The primer sequences are shown in Supplemental Table S1 and were purchased from Integrated DNA Technologies. PCR was carried out using Power SYBR Green Master Mix (ABI, 4367659). Taq polymerase was activated at 95°C for 10 min. The cycling parameters were denaturation at 95°C for 30 sec and extension at 60°C for 1 min (for 40 cycles). Data analysis was performed using the 2−ΔΔCT method for relative quantification. All samples were normalized to Gapdh (glyceraldehyde-3-phosphate dehydrogenase) RNA content.
Cytokine assays
Cytokines in serum or cell culture medium 24 h after M1 treatment were measured with a multiplex kit from Millipore (MCYTOMAG-70 K) and Luminex® xMAP® Technology.
Blood and serum assays
Blood was drawn into ethylene glycol tetraacetic acid-coated capillary blood collection tubes (BD Biosciences, 365973) and the complete blood count determined by blood analyzer (Forcyte Hematology Analyzer, Oxford Sciences, Oxford, CT). Serum glucose was assayed with an Ascensia Contour glucose meter (Bayer HealthCare, Whippany, NJ). The indicated commercial kits were used for the assay of serum levels of GPT (TECO Diagnostics, A526–120) and β-hydroxybutyrate (Sigma, MAK041).
Histology
Mouse livers were fixed in 10% neutral formalin, stained with hematoxylin and eosin and graded in a blinded fashion by a single pathologist for the degree of liver injury and inflammation. The percentage of hepatic parenchyma with apoptosis and/or necrosis, or inflammation was semi-quantitatively graded on a sliding scale of: 0, absent; 0.5, minimal; 1, mild; 1.5, mild to moderate; 2, moderate; 2.5, moderate to marked; and 3, marked. Epididymal WAT was similarly fixed and stained and inflammation graded on a scale of 0, absent; 1, minimal; 2, mild; 3, moderate; and 4, severe.
Liver triglyceride assay
Liver triglyceride content was determined using a kit (Sigma, TR0100–1KT) according to the manufacturer's instructions and normalized to liver weight.
Immunofluorescence microscopy
Livers were frozen in 2-methylbutane, sectioned, blocked for 1 h in 2% donkey serum (Sigma, D9663), 1% bovine serum albumin (Sigma, A9647) and 0.05% Tween 20 (Fisher, BP337) and incubated overnight with anti-CD68 antibody (Abd Serotec, MCA1957 GA) at 4°C. The tissues were then washed with PBS and incubated with Cy3-conjugated secondary antibody (Jackson ImmunoResearch, 111–165–152) for 1 h at room temperature. After washing with PBS, tissues were mounted in anti-fading medium containing 4′,6-diamidino-2-phenylindole (Life Technologies, P-36931) overnight before visualization by fluorescence microscopy.
Kupffer cell isolation and culture
Mouse liver nonparenchymal cells were isolated by Liberase (Roche Applied Science, 05–401–119–001) perfusion and a series of centrifugations at 50 g to remove hepatocytes. Kupffer cells were isolated from the nonparenchymal cell population by magnetic antibody sorting (Miltenyi Biotech, anti-biotin microbeads 130–090–485, separation columns 130–042–9) with a mouse biotin-labeled anti-EMR1 antibody (eBioscience, 13–4801–85). Cells were cultured in the same medium as for the BMDM, which was replaced 1 h after plating to remove all unattached cells. M1 and M2 treatments were started after an overnight incubation.
TUNEL assay
TUNEL-positive cells in liver sections were detected by commercial kit (Promega, G7130). Tissue sections were deparaffinized in xylenes, gradually rehydrated in decreasing concentrations of ethanol, and the assay performed according to the manufacturer's instructions. Under light microscopy, the numbers of TUNEL-positive cells in 10 randomly selected high power fields (400 × magnification) were counted per liver sample.
Statistical analysis
Numerical results are reported as means ± SEM and derived from at least 3 independent experiments unless otherwise indicated. The unpaired Student t test was used to assess significance between control and treated groups with P < 0.05 considered significant.
Acknowledgment
The GFP-LC3 adenovirus was the kind gift of Xiao-Ming Yin.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by NIH grants R01DK061498, R01DK044234 (MJC) and F32DK096791 (KL) and an American Liver Foundation Postdoctoral Research Fellowship Award (KL).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13:722-37; PMID:24064518; http://dx.doi.org/ 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/ 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958-69; PMID:19029990; http://dx.doi.org/ 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 2011; 30:4701-11; PMID:22068051; http://dx.doi.org/ 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12:222-30; PMID:21151103; http://dx.doi.org/ 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 2008; 456:264-8; PMID:18849965; http://dx.doi.org/ 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- 7. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011; 469:221-5; PMID:21124315; http://dx.doi.org/ 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- 8. Yang M, Liu J, Shao J, Qin Y, Ji Q, Zhang X, Du J. Cathepsin S-mediated autophagic flux in tumor-associated macrophages accelerate tumor development by promoting M2 polarization. Mol Cancer 2014; 13:43; PMID:24580730; http://dx.doi.org/ 10.1186/1476-4598-13-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang CP, Su YC, Hu CW, Lei HY. TLR2-dependent selective autophagy regulates NF-kB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell Death Differ 2013; 20:515-23; PMID:23175187; http://dx.doi.org/ 10.1038/cdd.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol 2011; 29:415-45; PMID:21219177; http://dx.doi.org/ 10.1146/annurev-immunol-031210-101322 [DOI] [PubMed] [Google Scholar]
- 11. Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 2010; 72:219-46; PMID:20148674; http://dx.doi.org/ 10.1146/annurev-physiol-021909-135846 [DOI] [PubMed] [Google Scholar]
- 12. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007; 56:1761-72; PMID:17456850; http://dx.doi.org/ 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- 13. Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003; 112:1821-30; PMID:14679177; http://dx.doi.org/ 10.1172/JCI19451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cancello R, Henegar C, Viguerie N, Taleb S, Poitou C, Rouault C, Coupaye M, Pelloux V, Hugol D, Bouillot JL, et al. Reduction of macrophage infiltration and chemoattractant gene expression changes in white adipose tissue of morbidly obese subjects after surgery-induced weight loss. Diabetes 2005; 54:2277-86; PMID:16046292 [DOI] [PubMed] [Google Scholar]
- 15. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 2007; 117:175-84; PMID:17200717; http://dx.doi.org/ 10.1172/JCI29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, Goforth MH, Subramanian V, Mukundan L, Ferrante AW, Chawla A. Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metabol 2008; 7:496-507; PMID:18522831; http://dx.doi.org/ 10.1016/j.cmet.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang W, Metlakunta A, Dedousis N, Zhang P, Sipula I, Dube JJ, Scott DK, O'Doherty RM. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010; 59:347-57; PMID:19934001; http://dx.doi.org/ 10.2337/db09-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity 2010; 32:593-604; PMID:20510870; http://dx.doi.org/ 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 19. Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol 2011; 6:275-97; PMID:21034223; http://dx.doi.org/ 10.1146/annurev-pathol-011110-130138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012; 122:787-95; PMID:22378047; http://dx.doi.org/ 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 2013; 20:3-11; PMID:22595754; http://dx.doi.org/ 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab 2011; 22:234-40; PMID:21419642; http://dx.doi.org/ 10.1016/j.tem.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; PMID:19339967; http://dx.doi.org/ 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabol 2010; 11:467-78; PMID:20519119; http://dx.doi.org/ 10.1016/j.cmet.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jansen HJ, van Essen P, Koenen T, Joosten LA, Netea MG, Tack CJ, Stienstra R. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 2012; 153:5866-74; PMID:23117929; http://dx.doi.org/ 10.1210/en.2012-1625 [DOI] [PubMed] [Google Scholar]
- 26. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/ 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27:1110-21; PMID:18337753; http://dx.doi.org/ 10.1038/emboj.2008.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun 2011; 412:618-25; PMID:21856284; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 29. Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J 2010; 24:3052-65; PMID:20375270; http://dx.doi.org/ 10.1096/fj.09-144519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; PMID:16625204; http://dx.doi.org/ 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- 31. Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 1999; 8:265-77; PMID:10621974 [DOI] [PubMed] [Google Scholar]
- 32. Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008; 4:458-69; PMID:18996346; http://dx.doi.org/ 10.1016/j.chom.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han MS, Jung DY, Morel C, Lakhani SA, Kim JK, Flavell RA, Davis RJ. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 2013; 339:218-22; PMID:23223452; http://dx.doi.org/ 10.1126/science.1227568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity 2003; 19:793-802; PMID:14670297 [DOI] [PubMed] [Google Scholar]
- 35. Imajo K, Fujita K, Yoneda M, Nozaki Y, Ogawa Y, Shinohara Y, Kato S, Mawatari H, Shibata W, Kitani H, et al. Hyperresponsivity to low-dose endotoxin during progression to nonalcoholic steatohepatitis is regulated by leptin-mediated signaling. Cell Metabol 2012; 16:44-54; PMID:22768838; http://dx.doi.org/ 10.1016/j.cmet.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 36. Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest 2011; 121:2736-49; PMID:21670502; http://dx.doi.org/ 10.1172/JCI45444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metabol 2008; 8:325-32; PMID:18840363; http://dx.doi.org/ 10.1016/j.cmet.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 38. Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab 2011; 96:E268-77; PMID:21047928; http://dx.doi.org/ 10.1210/jc.2010-1681 [DOI] [PubMed] [Google Scholar]
- 39. Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A 2012; 109:E3168-76; PMID:23093667; http://dx.doi.org/ 10.1073/pnas.1210500109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol 2012; 24:21-31; PMID:22118953; http://dx.doi.org/ 10.1016/j.coi.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen P, Cescon M, Bonaldo P. Autophagy-mediated regulation of macrophages and its applications for cancer. Autophagy 2014; 10:192-200; PMID:24300480; http://dx.doi.org/ 10.4161/auto.26927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007; 39:207-11; PMID:17200669; http://dx.doi.org/ 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood 2012; 119:2895-905; PMID:22223827; http://dx.doi.org/ 10.1182/blood-2011-08-372383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laskin DL, Sunil VR, Gardner CR, Laskin JD. Macrophages and tissue injury: agents of defense or destruction? Annu Rev Pharmacol Toxicol 2011; 51:267-88; PMID:20887196; http://dx.doi.org/ 10.1146/annurev.pharmtox.010909.105812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iimuro Y, Yamamoto M, Kohno H, Itakura J, Fujii H, Matsumoto Y. Blockade of liver macrophages by gadolinium chloride reduces lethality in endotoxemic rats-analysis of mechanisms of lethality in endotoxemia. J Leukoc Biol 1994; 55:723-8; PMID:8195698 [DOI] [PubMed] [Google Scholar]
- 46. Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H, Hainzl A, Schatz S, Qi Y, Schlecht A, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest 2011; 121:985-97; PMID:21317534; http://dx.doi.org/ 10.1172/JCI44490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell 2011; 146:682-95; PMID:21884931; http://dx.doi.org/ 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- 48. Fontana L, Zhao E, Amir M, Dong H, Tanaka K, Czaja MJ. Aging promotes the development of diet-induced murine steatohepatitis but not steatosis. Hepatology 2013; 57:995-1004; PMID:23081825; http://dx.doi.org/ 10.1002/hep.26099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Noureddin M, Yates KP, Vaughn IA, Neuschwander-Tetri BA, Sanyal AJ, McCullough A, Merriman R, Hameed B, Doo E, Kleiner DE, et al. Clinical and histological determinants of nonalcoholic steatohepatitis and advanced fibrosis in elderly patients. Hepatology 2013; 58:1644-54; PMID:23686698; http://dx.doi.org/ 10.1002/hep.26465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA 2002; 287:356-9; PMID:11790215; http://dx.doi.org/jbr10281 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y, Singh R, Lefkowitch JH, Rigoli RM, Czaja MJ. Tumor necrosis factor-induced toxic liver injury results from JNK2-dependent activation of caspase-8 and the mitochondrial death pathway. J Biol Chem 2006; 281:15258-67; PMID:16571730; http://dx.doi.org/ 10.1074/jbc.M512953200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y, Schattenberg JM, Rigoli RM, Storz P, Czaja MJ. Hepatocyte resistance to oxidative stress is dependent on protein kinase C-mediated down-regulation of c-Jun/AP-1. J Biol Chem 2004; 279:31089-97; PMID:15145937; http://dx.doi.org/ 10.1074/jbc.M404170200 [DOI] [PubMed] [Google Scholar]
- 53. Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 2010; 139:1740-52; PMID:20659474; http://dx.doi.org/ 10.1053/j.gastro.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu Y, Bialik S, Jones BE, Iimuro Y, Kitsis RN, Srinivasan A, Brenner DA, Czaja MJ. NF-kB inactivation converts a hepatocyte cell line TNF-α response from proliferation to apoptosis. Am J Physiol 1998; 275:C1058-66; PMID:9755059 [DOI] [PubMed] [Google Scholar]
- 55. Liu H, Lo CR, Jones BE, Pradhan Z, Srinivasan A, Valentino KL, Stockert RJ, Czaja MJ. Inhibition of c-Myc expression sensitizes hepatocytes to tumor necrosis factor-induced apoptosis and necrosis. J Biol Chem 2000; 275:40155-62; PMID:11016920; http://dx.doi.org/ 10.1074/jbc.M001565200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.