Abstract
Autophagy regulates cellular homeostasis and response to environmental stress. Within the retinal pigment epithelium (RPE) of the eye, the level of autophagy can change with both age and disease. The purpose of this study is to determine the relationship between reduced autophagy and age-related degeneration of the RPE. The gene encoding RB1CC1/FIP200 (RB1-inducible coiled-coil 1), a protein essential for induction of autophagy, was selectively knocked out in the RPE by crossing Best1-Cre mice with mice in which the Rb1cc1 gene was flanked with Lox-P sites (Rb1cc1flox/flox). Ex vivo and in vivo analyses, including western blot, immunohistochemistry, transmission electron microscopy, fundus photography, optical coherence tomography, fluorescein angiography, and electroretinography were performed to assess the structure and function of the retina as a function of age. Deletion of Rb1cc1 resulted in multiple autophagy defects within the RPE including decreased conversion of LC3-I to LC3-II, accumulation of autophagy-targeted precursors, and increased numbers of mitochondria. Age-dependent degeneration of the RPE occurred, with formation of atrophic patches, subretinal migration of activated microglial cells, subRPE deposition of inflammatory and oxidatively damaged proteins, subretinal drusenoid deposits, and occasional foci of choroidal neovascularization. There was secondary loss of photoreceptors overlying the degenerated RPE and reduction in the electroretinogram. These observations are consistent with a critical role of autophagy in the maintenance of normal homeostasis in the aging RPE, and indicate that disruption of autophagy leads to retinal phenotypes associated with age-related degeneration.
Keywords: age-related macular degeneration, FIP200, photoreceptor, retina, retinal pigment epithelium
Abbreviations: AMD, age-related macular degeneration; ANOVA, analysis of variance; Bru, Bruch's membrane; CKO, conditional knockout; CTSD, cathepsin D; ERG, electroretinogram; GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segment; LAP, LC3-associated phagocytosis; MTOR, mechanistic target of rapamycin; OCT, optical coherence tomography; ONL, outer nuclear layer; OS, outer segment; PBS, phosphate-buffered saline; POS, photoreceptor outer segments; RB1CC1, RB1-inducible coiled-coil 1; RPE, retinal pigment epithelium; siRNA, small interfering ribonucleic acid.
Introduction
Macroautophagy (from this point referred to as autophagy) is an important intracellular pathway critical for maintaining cellular homeostasis and regulating its response to environmental stressors. Activation of this pathway results in the engulfment of cytoplasmic material within double-membrane vesicles, known as autophagosomes, that then fuse with lysosomes, resulting in degradation of the delivered material.1-3 Autophagy is classically described as being activated in conditions of nutrient deprivation.4 Increasing evidence, however, shows that autophagy is important in maintaining a stable intracellular environment by removing toxic aggregates, misfolded proteins, and damaged organelles.5,6 Dysregulation of autophagy has been implicated in many pathological conditions, including cancer, infectious disease, and neurodegeneration.7,8
The retinal pigment epithelium is a monolayer of cells located immediately adjacent to the photoreceptor layer of the neural retina. The retinal circulation provides nutrients to the inner retinal cell layers, whereas the photoreceptors, which are located on the outer surface of the retina, obtain metabolic support from the RPE.9,10 In addition to providing photoreceptor cells with nutrition, the RPE carries out receptor-mediated phagocytosis of photoreceptor outer segments shed into the subretinal space.11-15 Impairment of normal RPE function can lead to death of these cells, followed by a secondary degeneration of the overlying photoreceptors.
Previous studies have shown that changes in RPE autophagy correlate with both progressive age and outer retinal degeneration.16-21 For example, markers of autophagy and exosomes have been found in the RPE of aged mice as well as in drusen from cadaveric eyes from people with age-related macular degeneration (AMD),16 and there has been increasing evidence for dysregulation of autophagy in the RPE leading to accumulation of lipofuscin and reduced ability to clear intracellular debris. 22–25 However, the causal relationship between autophagy dysfunction and RPE degeneration has not been directly demonstrated in vivo. One method for assessing the role of a process in a specific cell type is to reduce or eliminate that function and assess the effect on the phenotype. Accordingly, in this study we suppress the activation of autophagy specifically within the RPE in an attempt to determine whether this would affect the survival of this cell in vivo. RB1CC1 is a component of the ULK1-ATG13-RB1CC1/FIP200 complex, which is inhibited by MTOR and is an essential upstream inducer of autophagy.26-29 Tissue-specific deletion of the Rb1cc1 gene has been studied in various cell types30–33 by crossing mice with the Rb1cc1 gene flanked by 2 loxP sites (Rb1cc1flox/flox) with mice having Cre-recombinase expression driving by a cell-specific promoter. Recently, Dunaief and coworkers described a mouse strain in which the Best1 promoter was used to provide RPE-specific expression of the Cre-recombinase and RPE-selective loss of a loxP-flanked gene.34
Kim and coworkers recently demonstrated the importance of autophagy components in LC3-associated phagocytosis (LAP).35 This study aims to assess the role of canonical autophagy, presumably downstream of LAP, in RPE homeostasis. We generated RPE-specific Rb1cc1 conditional knockout mice (Rb1cc1-CKO) by crossing Best1-Cre mice with Rb1cc1flox/flox mice. Our results show that selective reduction in the induction of autophagy in the RPE causes age-dependent degeneration of the RPE, and secondary degeneration of the overlying photoreceptors. Our data provide evidence suggesting a critical function for autophagy in the maintenance of normal homeostasis in the aging RPE, and we propose this novel animal model for the study of the molecular mechanisms by which loss of autophagy contributes to outer retinal degeneration.
Results
Knockout of RB1CC1 in ARPE-19 cells results in loss of POS-induced autophagy
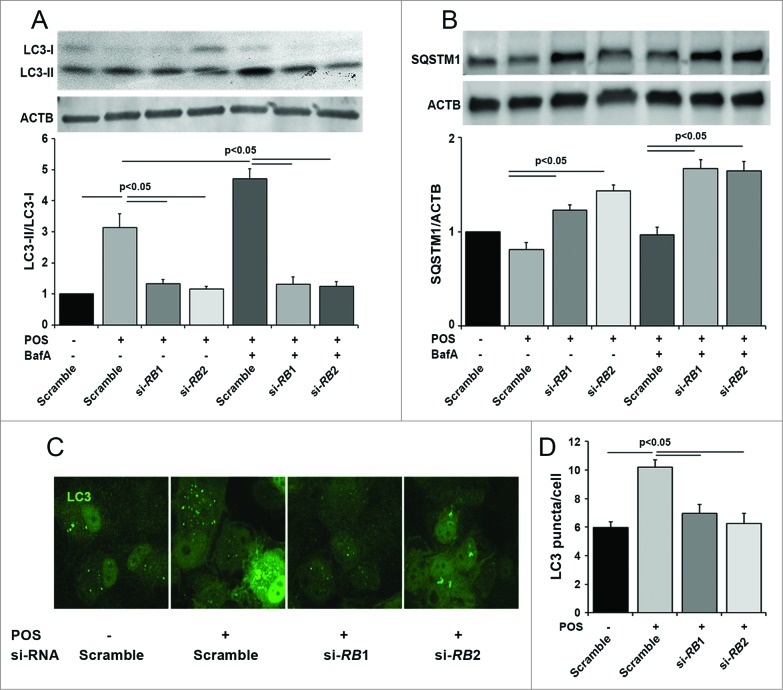
Autophagy in the retina pigment epithelium occurs, at least in part, in response to ingestion of photoreceptor outer segments (POS).36 Prior to testing the Rb1cc1 deletion in the RPE in vivo, we tested whether knockdown of RB1CC1 will result in decreased POS-induced autophagy in vitro. We reduced autophagy activation by siRNA knockdown of the RB1CC1 gene transcript in ARPE-19 cells. Western blot confirmed the loss of the RB1CC1 protein (data not shown), decreased lipidation of LC3-I (Fig. 1A), increased accumulation of SQSTM1/p62 (Fig. 1B), and decreased autophagosome formation (Fig. 1C and D). Inhibiting autophagosome-lysosome fusion with bafilomycin A1 showed an increase in LC3-II and SQSTM1 in the control cells, which rules out the possibility that the accumulation is due to lysosomal disruption in the absence of bafilomycin A1. Furthermore, immunostaining for LC3 confirmed that the number of GFP-LC3 puncta, presumably corresponding to autophagosomes, was reduced following RB1CC1 knockdown.
Figure 1.
Knockout of RB1CC1 results in loss of photoreceptor outer segment (POS)-induced autophagy in ARPE-19 cells. (A) Quantitative western blot analysis shows that siRNA against the RB1CC1 transcript (si-RB1 and si-RB2) significantly decreased the levels of LC3-II/LC3-I ratio, with or without the presence of bafilomycin A1 (BafA), after POS feeding. As a control, cells were also treated with a nonspecific siRNA sequence (Scramble). (B) Cells treated with RB1CC1-siRNA showed increased levels of SQSTM1, with or without the presence of BafA after POS feeding. SQSTM1 levels normalized to the ACTB/actin loading control. (C) Immunofluorescence analysis showed RB1CC1 siRNA-treated ARPE-19 cells have a significantly reduced punctate pattern of LC3 staining (green), consistent with the formation of fewer autophagosomes. Treatment of cells with si-RB1 or si-RB2 alone did not induce puncta formation. (D) Quantification of the number of autophagosomes per cell.
Conditional knockout of Rb1cc1 results in reduced autophagy activation in mouse RPE
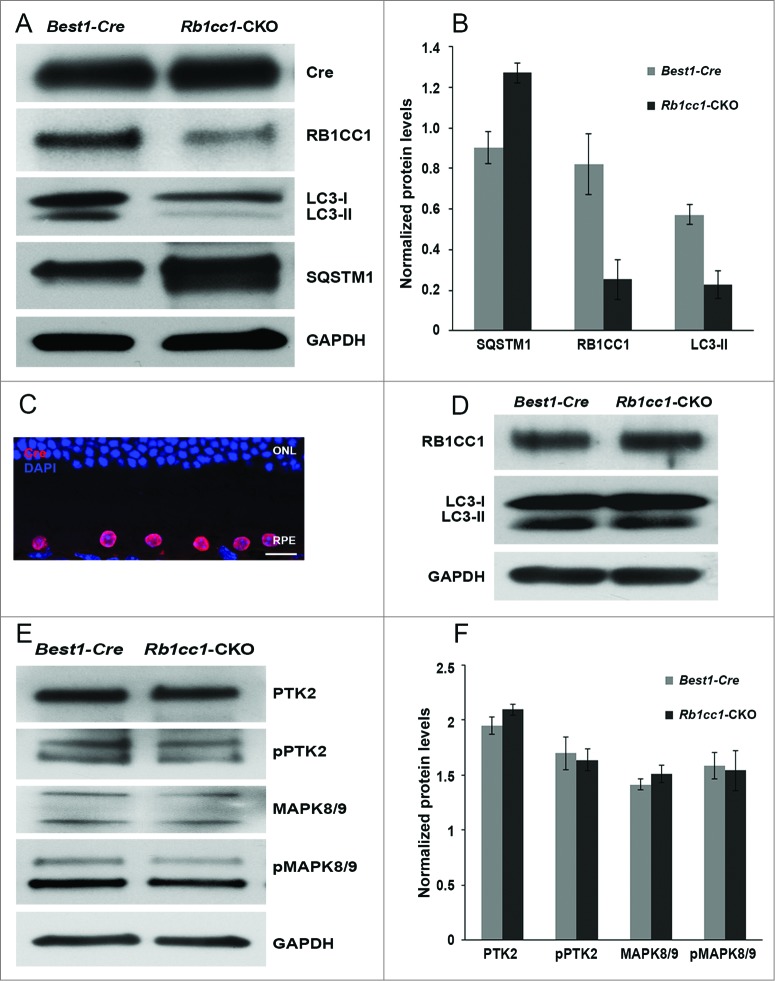
To investigate whether disrupting autophagy would have any effect on the RPE and overlying retina, we crossed the Rb1cc1flox/flox mouse with the Best1-Cre mouse, the latter strain providing the RPE-specific expression of the Cre-recombinase. In this study, the RPE-specific Rb1cc1-CKO (Rb1cc1flox/flox; Best1-Cre [+]) served as the experimental group, and the Rb1cc1wt; Best1-Cre(+) littermates served as the control group, as Cre alone can cause phenotypic changes in the RPE.37 In both groups, we detected robust expression of Cre within the RPE, and marked reduction of RB1CC1 expression only in the Rb1cc1-CKO mice (Fig. 2A and B). Immunohistochemical analysis confirms that the Cre expression is only within the RPE (Fig. 2C). There was also a markedly reduced level of LC3-II and a significant accumulation of SQSTM1, both consistent with defective autophagy in the RPE (Fig. 2A and B). Interestingly, the SQSTM1 appears as a doublet band. This is the typical appearance of this band on RPE samples. In the overlying retina there were normal levels of RB1CC1 and equal conversion of LC3-I to LC3-II, consistent with the cell-specific nature of the autophagy gene deletion (Fig. 2D).
Figure 2.
Conditional knockout of Rb1cc1 in the RPE results in the selective loss of autophagy activation in these cells. (A) Western blots, and (B) quantification of protein levels of RPE/choroid lysates from 2-mo-old Rb1cc1-CKO mice showed markedly reduced level of LC3-II and a significant accumulation of SQSTM1 as compared to littermate controls. Differences were significant at the P < 0.05 level. (C) Immunostaining confirmed the RPE-specific expression of Cre-recombinase (red). (D) Western blot of retina protein lysates showed normal levels of RB1CC1 and equal conversion of LC3-I to LC3-II. (E) Western blot, and (F) quantification of protein levels showed comparable amounts of phosphorylated-PTK2 (pPTK2), phosphorylated-MAPK8/JNK1-MAPK9/JNK2 (pMAPK8/9), and total PTK2 and MAPK, in the RPE from both Rb1cc1-CKO and control mice at the age of 2 mo. ONL, outer nuclear layer; RPE, retinal pigment epithelium.
Aside from the essential function of RB1CC1 in regulating autophagy in mammalian cells, it is also important in regulating a number of intracellular signaling pathways such as cell survival, differentiation, migration, proliferation, and protein synthesis.38 Those other functions are mediated by the kinases PTK2/FAK and MAPK/JNK. We found comparable levels of phosphorylated-PTK2 (pPTK2) and phosphorylated-MAPK8/JNK1-MAPK9/JNK2 (pMAPK8/9), as well as the levels of total PTK2 and MAPK8/9, in the RPE from both experimental and littermate control mice (Fig. 2D and F). These results suggested that knocking down RB1CC1 expression in the RPE did not significantly affect the levels of nonautophagy-related RB1CC1 targets, and were consistent with the Rb1cc1-CKO having primarily a defect on autophagy and not on the PTK2 and MAPK pathways.
Selective deletion of Rb1cc1 results in an age-dependent degeneration of the RPE
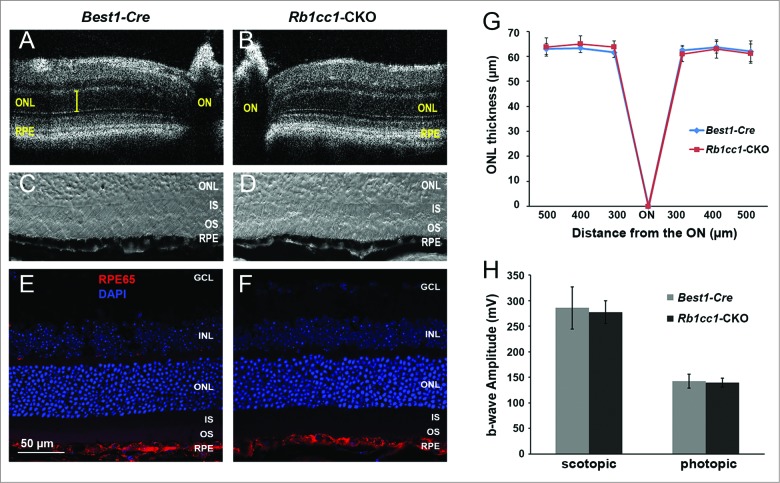
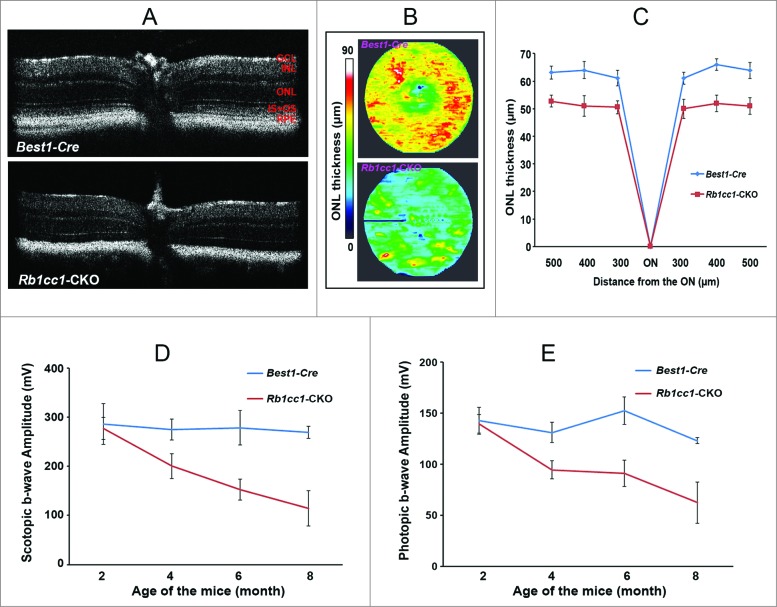
To assess the phenotype resulting from the RPE-specific deletion of Rb1cc1, we performed in vivo and ex vivo analyses. Optical coherence tomography (OCT), an in vivo assessment of retinal structure and thickness (Fig. 3A and B), as well as histological analysis (Fig. 3C–F), confirm that at the age of 2 mo the outer retinas of the Rb1cc1-CKO and littermate controls were similar in appearance. Quantitative analysis of total retinal and outer nuclear layer thickness was similar between the 2 groups of animals (Fig. 3G). Visual function, as evaluated by rod (scotopic) and cone (photopic) electroretinogram (ERG) showed no significant difference between Rb1cc1-CKO and control mice (Fig. 3H). The RPE of the Rb1cc1-CKO mice were identical to the littermate controls in terms of the RPE appearance and integrity of RPE65 staining (a RPE-specific marker indicative of normal RPE function; Fig. 3E and F).
Figure 3.
Retinal and RPE morphology are normal in Rb1cc1-CKO mice at 2 mo of age. (A–D) At the age of 2 mo, the outer retinas of the Rb1cc1-CKO and littermate controls were similar in appearance, as shown by OCT and DIC images of retina sections. (E, F) Immunostaining of RPE65 in Rb1cc1-CKO retina was identical to controls. (G) Quantification of outer nuclear layer thickness (yellow bar in [A]) of Rb1cc1-CKO and control mice measured with OCT. (H) Quantification of rod (scotopic) and cone (photopic) ERG showed no significant difference between Rb1cc1-CKO and control mice. GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
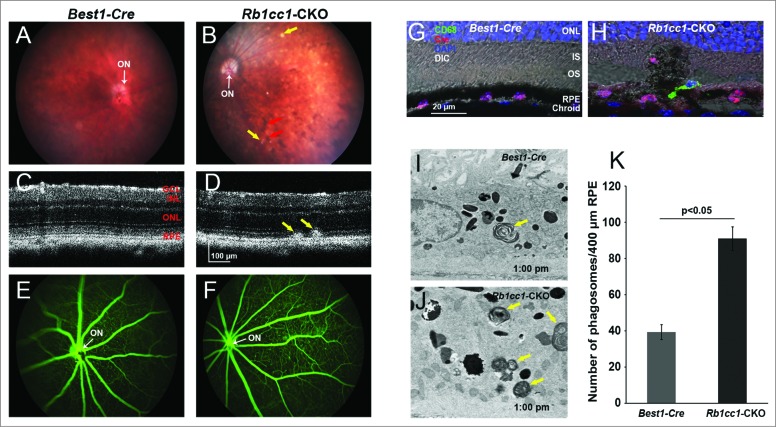
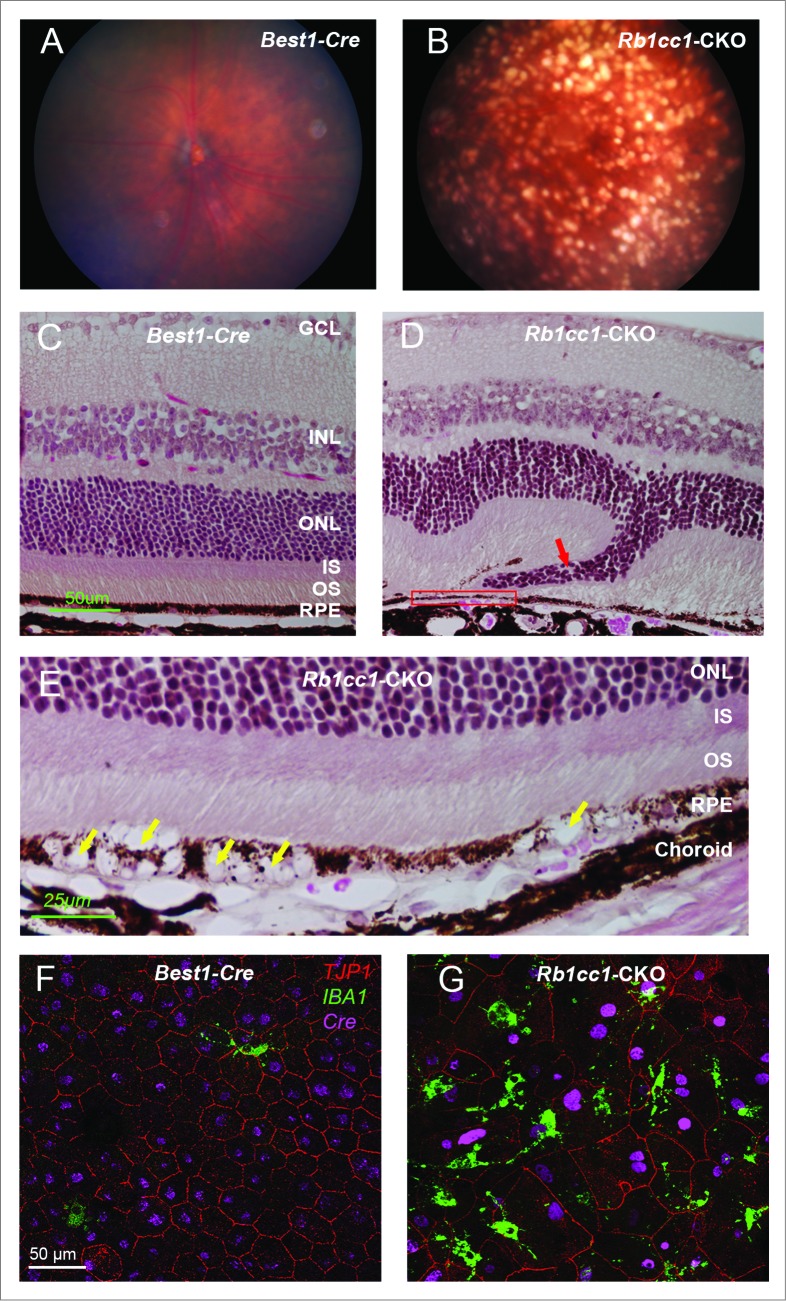
At 4 mo of age we detected significant differences in the outer retinal appearance of the Rb1cc1-CKO versus control mice. Examination of the Rb1cc1-CKO retinas revealed small, white-yellowish structures and areas of patchy RPE atrophy (Fig. 4A and B). We also observed scattered foci of RPE hyperpigmentation, usually at the border of RPE atrophic patches. Subretinal hyper-reflective spots that vary in shape and thickness were detected on OCT (Fig. 4C and D). These foci, as pointed out by the arrows, were located above the RPE reflectance layer, and appeared to correlate with the location of RPE pigment clumps seen in fundus photographs. Fluorescein angiography, a technique for visualizing the integrity of the retinal vasculature, showed that this was normal in both groups (Fig. 4E and F). Immunohistochemistry on Rb1cc1-CKO RPE confirmed that the areas of abnormalities correlated with the areas of Cre-recombinase expression (Fig. 4G and H). There were also CD68-positive staining cells in these foci, consistent with macrophage infiltration (Fig 4H). Finally, transmission electron microscopy showed increased accumulation of single-membrane phagosome-like structures in the Rb1cc1-CKO cells (Fig. 4I–K).
Figure 4.
RPE degeneration present by 4 mo of age. (A, B) Fundus photograph shows the significant RPE atrophy present in the Rb1cc1-CKO mice, as compared to littermate controls. ON signifies the optic nerve. Yellow and red arrows point to representative foci of RPE atrophy and clumping, respectively. (C, D) OCT imaging shows the increased granularity of the RPE reflectance and the presence of hyperreflective foci located above the RPE (yellow arrows). (E, F) Fluorescein angiography showed a normal retinal vasculature in both groups of mice. (G, H) Immunostaining for the Cre-recombinase (red) showed that there was no RPE degeneration in the Best1-Cre control retinas, but only in the Rb1cc1-CKO retinas where there was Cre expression. Staining for CD68 showed infiltration of macrophages into the areas of degenerating RPE. (I–K) Transmission electron microscopy showed that there were significantly more unprocessed phagosomes (yellow arrows) present in the RPE of the Rb1cc1-CKO mouse, as compared to littermate controls. IS, inner segment; OCT, optical coherence tomography; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
The abnormal phenotype increased in severity with increasing age, and by 8 mo there was widespread disruption of the RPE cell morphology and the overlying retina (Fig. 5A and B). There were areas of RPE vacuolization as well as areas of RPE atrophy, with disorganization of the overlying retina (Fig. 5C–E). Flat mounts of the RPE stained with an antibody against the tight junction protein TJP1/ZO-1 showed a marked disorganization of the hexagonal pattern normally seen in the RPE (Fig. 5F). Retinal microglial cells are intraretinal immune surveillance cells that can proliferate in response to injury or stress.39-41 They often have a neuroprotective effect, mitigating the consequences of oxidative stress and abnormal accumulation of debris. Activation and migration of microglia to the area of injury is commonly seen in disease. Though this response can diminish in some models of aging,42 we detected an increased presence of IBA1-positive cells in the Rb1cc1-CKO mice even at 8 mo of age (Fig. 5G); IBA1 being a marker of activated microglial cells and consistent with the activation of an immunological response to the RPE degeneration.
Figure 5.
Inability to activate autophagy results in continued age-dependent degeneration of the RPE. (A, B) Fundus photograph of retina from 8-mo-old Rb1cc1-CKO mouse shows significant RPE degeneration. (C–E) Photomicrographs show the vacuolization of the RPE (yellow arrows), and areas of atrophy. Note the disruption of the outer nuclear layer of the retina (red arrow) where the RPE is absent (rectangle). (F, G) Flat mounts stained for the tight junction protein TJP1/ZO-1 show marked disruption of the normal hexagonal pattern of the RPE, and increased staining of IBA1-positive cells, consistent with increased infiltration of activated microglia. GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
Disruption of Bruch's membrane and subRPE inflammation
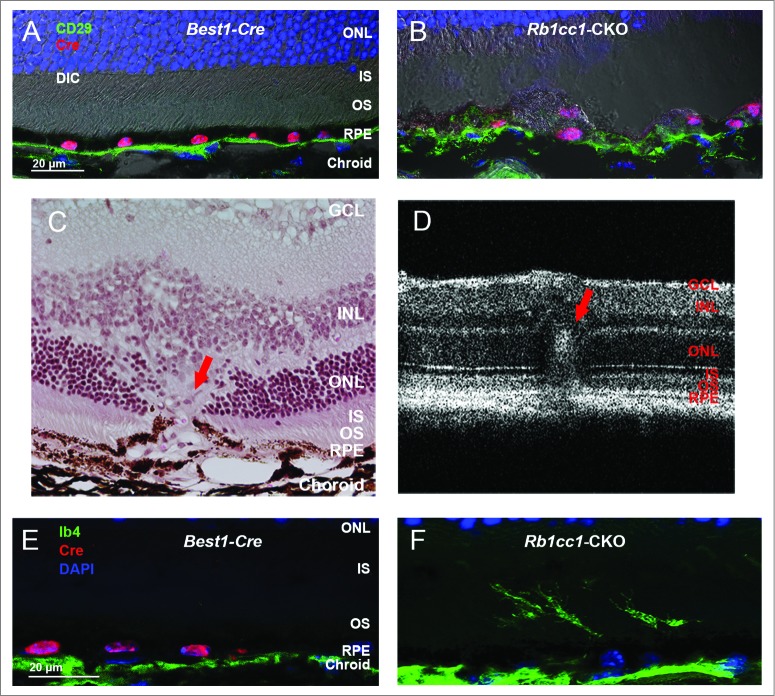
Bruch's membrane is a thick structure separating the choroid from the RPE, and is composed of the basement membranes of the choriocapillaris and RPE sandwiching collagenous and elastic fibers. Bruch's membrane is thought to be important in maintaining the structural integrity of the subRPE space and degeneration of Bruch's membrane is a core feature of age-related macular degeneration. Disruption of Bruch's membrane in the Rb1cc1-CKO cells was demonstrated by staining for CD29, a component of the ITGB1/integrin β-1 complex that localizes to the basal surface of the RPE and that serves as a surrogate marker for disruption of the Bruch's-RPE interface and integrity of this structure (Fig. 6A and B).43 We noted occasional ingrowth of vessels, as demonstrated with staining for the vascular marker IB4, from the choroid into the outer retina (red arrow), consistent with foci of neovascularization (Fig. 6C–F). These foci were infrequently seen, and only on histology. We did not detect any significant areas of hyperfluorescence on angiography, consistent with the self-limited nature of rodent choroidal neovascularization.44 While we cannot definitively state that the vessels are of choroidal origin, their morphology is consistent with what is seen on histological examination of other validated rodent models of choroidal neovascularization as compared to models of retinal neovascularization.44-46
Figure 6.
Loss of autophagy in the RPE results in disruption of the normal interface with Bruch's membrane. (A, B) Staining of retinal sections with an antibody against CD29, a component of the ITGB1/integrin β-1 complex and a marker of the basal surface of the RPE, shows disruption of the normally continuous linear structure. (C) Histological section of a retina from an 8-mo-old Rb1cc1-CKO mouse shows a focus of vascular ingrowth from the choroid through the RPE and into the retina (red arrow), consistent with a focus of choroidal neovascularization. (F) This finding was also seen on OCT imaging of the retina (red arrow). (E, F) Staining with the vascular marker, IB4, confirms that the ingrowth contained blood vessels. GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
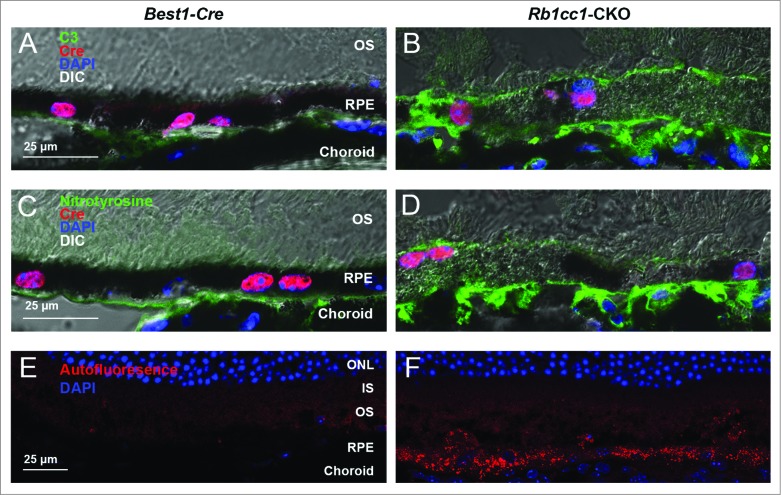
There was an accumulation of oxidatively damaged proteins on the basal surface of the RPE, with a resultant inflammatory response, as demonstrated by the presence of nitrotyrosine-positive staining and C3 (complement component 3) staining, respectively (Fig. 7A–D). We also observed markedly increased autofluorescence in the RPE of the Rb1cc1-CKO mice compared to the control animals, most likely due to accumulation of lipofuscin (Fig. 7E and F).
Figure 7.
Subretinal inflammation and increased autofluorescence was detected in Rb1cc1-CKO mice. (A, B) Loss of autophagy activation resulted in increased deposition of C3 (complement component 3) and (C, D) oxidatively-damaged proteins on the basal surface of the RPE. There was also increased accumulation of autofluorescent puncta within the RPE (E, F). Photomicrographs are of sections taken from 8-mo-old mice. In all cases, abnormalities were only seen in the Rb1cc1-CKO mouse, and not in the Best1-Cre mouse, even in areas of Cre expression. IS, inner segment; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
Selective deletion of Rb1cc1 results in accumulation of lipids and intracellular debris
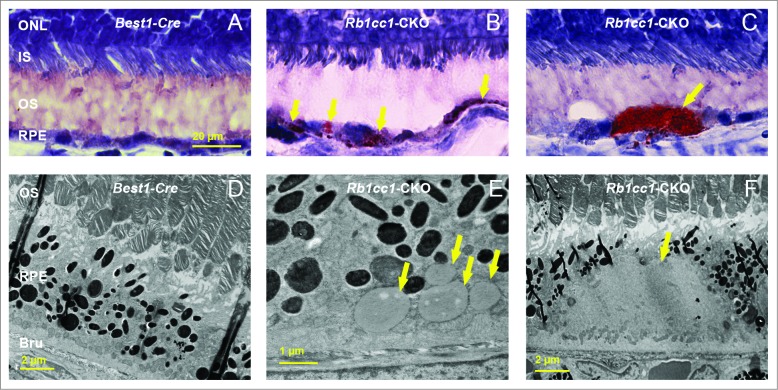
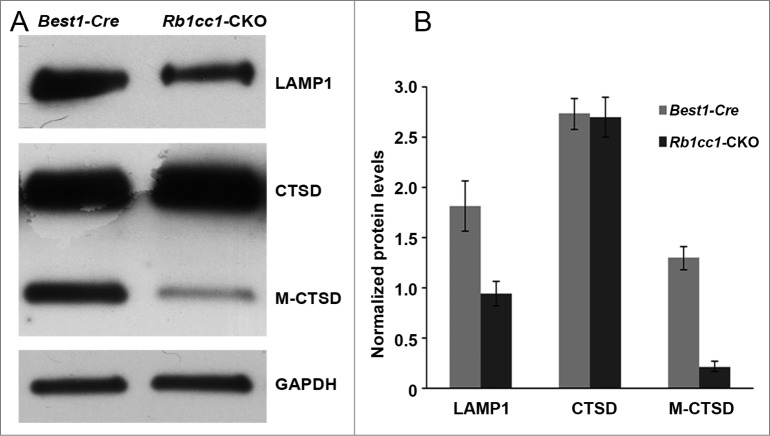
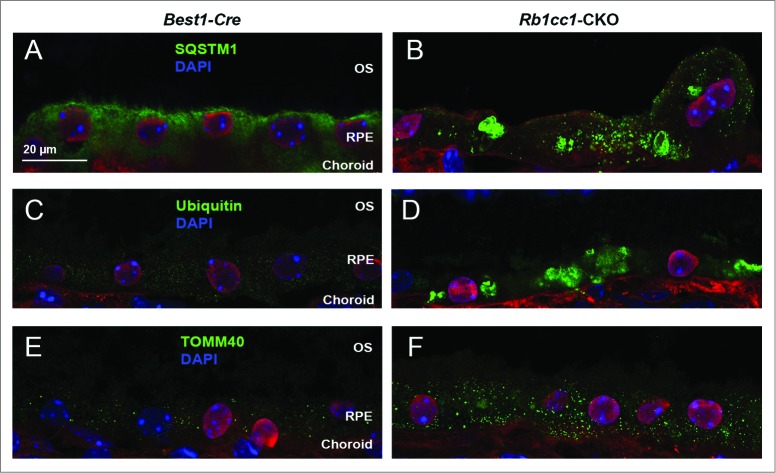
The accumulation of lipids in the RPE is another common phenotype seen in age-related macular degeneration. Staining with Oil Red O showed intracellular accumulation of neutral lipids (Fig. 8A–C). These accumulations localized primarily to the basal aspect of the RPE. The sizes of these accumulations varied, ranging from small round vesicles to large, oblong-shaped structures. Electron microscopy showed poorly processed phagosomes and accumulation of vacuoles, presumably lipid-filled and some large enough to force an apical displacement of the melanosomes. The larger vacuole-like structures seen on electron microscopy appeared to correspond to the large, oblong-shaped lipid accumulations seen with Oil Red O staining (Fig. 8D–F). There were decreased levels of lysosome markers, such as mature CTSD/cathepsin D and LAMP1 (Fig. 9), consistent with defective processing of intracellular debris. When staining cells for material targeted for degradation by autophagy47,48 using antibodies against SQSTM1 or ubiquitin, we saw distinct patterns of accumulation of this material in the Rb1cc1-CKO cells that did not overlap with the lipid-filled vacuoles (Fig. 10). Rather, the SQSTM1-positive material appeared to localize on the apical surface of the vesicles. The Rb1cc1-CKO mice also had an accumulation of mitochondria, as confirmed with TOMM40 staining.
Figure 8.
There was increased accumulation of lipid-filled vacuoles within the RPE of the Rb1cc1-CKO mice as compared to littermate controls. (A–C) Sections from eyes of 8-mo-old mice stained with Oil Red O show the disruption of the normal RPE architecture the presence of large, lipid-filled structures (yellow arrows). (D–F) Electron microscopy shows that the lipid is contained within variably sized vacuoles, some of which are large enough to displace the normal RPE melanin content. BRU, Bruch's membrane; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; RPE, retinal pigment epithelium.
Figure 9.
There were decreased levels of lysosome markers consistent with defective processing of intracellular debris. Western blots (A), and quantification (B) showing decreased levels of the lysosome proteins LAMP1 and mature CTSD/cathepsin D (M-CTSD) in the RPE of the Rb1cc1-CKO mice as compared to littermate controls.
Figure 10.
Loss of autophagy in the RPE results in accumulation of intracellular debris. The Rb1cc1-CKO mice had a significant accumulation of SQSTM1 (A, B) and ubiquitin-tagged debris (C, D) within the RPE, as well as increased accumulation of mitochondria (E, F), as compared to littermate controls. These findings are consistent with decreased autophagy flux in these cells. OS, outer segment; RPE, retinal pigment epithelium.
Reduction of autophagy in the RPE resulted in a secondary loss of photoreceptors and reduced retinal function
Given the intimate anatomical and functional interdependence between the RPE and photoreceptor cells, we wanted to determine whether autophagy deficiency in the former had an effect on the latter. The Rb1cc1-CKO had a selective deficiency of autophagy activation in the RPE, but the photoreceptors had normal levels of autophagy markers (as demonstrated in Fig. 2C). Despite this, however, the RPE degeneration still resulted in a secondary loss of photoreceptors. Outer nuclear layer thickness, as measured by OCT, was decreased in the Rb1cc1-CKO, and there was a progressive age-dependent reduction in both photopic and scotopic ERG as compared to littermate controls (Fig. 11).
Figure 11.
RPE degeneration leads to retinal degeneration. Loss of autophagy function in the RPE resulted in a secondary loss of photoreceptors. (A) The retinal thinning was due to reduction in the ONL as seen on cross section OCT imaging of 8-moold mice, and (B) the automated ONL thickness measurement shown as a heat map. (C) Manual measurement of the ONL thickness vs. total retinal thickness confirmed the automated measurements. (C, D) There was decreased scotopic and photopic electroretinography responses, consistent with the loss of photoreceptors.
Discussion
Age-related degeneration of the RPE contributes to a number of significant, blinding ophthalmic conditions, most notably age-related macular degeneration. A major obstacle to the study of these diseases is the fact that the etiology of the degeneration appears to be multifactorial, with genetic and environmental factors, along with age, influencing onset and progression of the pathology. This combination of genetic and environmental stressors is a feature shared with several other neurodegenerative diseases, including Alzheimer and Huntington diseases.49-51 Along with AMD, these diseases are characterized by the accumulation of deposits, for example amyloid deposits in Alzheimer disease, and drusen (chemically complex deposits that form under the RPE) in AMD. In neurodegenerations, autophagy deficiency has been implicated in the inadequate cellular response to the various disease-causing factors. In the present work, we specifically deleted a key factor necessary for induction of autophagy in the RPE, and demonstrated a resultant age-dependent degeneration that shares significant phenotypic similarity to human age-related degenerations of the RPE and photoreceptors, including accumulation of lipid-filled deposits and oxidatively damaged proteins, as well as local activation of the complement cascade.
The RPE performs many critical functions in support of the retina, and the photoreceptor cells in particular. One key function is the ingestion of shed photoreceptor outer segments.15 This shedding occurs in a circadian rhythm, and the daily burden of shed material represents approximately 10%, by weight, of total photoreceptor outer segments.11,52 The composition of the ingested material includes lipids, proteins, small molecules, and vitamin A. Normally, these constituents undergo orderly degradation, with specific sorting of various components to different locations. For example, there is selective sorting of lipids and cholesterol within the RPE, some for recycling and some for clearance and elimination.53 In addition, vitamin A re-enters the visual cycle for re-isomerization and transport back to the photoreceptors for regeneration of the opsin visual pigments.54
Our data confirm that in our model we successfully deleted Rb1cc1 from the RPE, and in this manner we were able to selectively prevent induction of canonical autophagy in these cells. There appeared to be normal phagocytic ingestion of the shed photoreceptor outer segments, suggesting that LAP was not affected. Rather, the deletion of Rb1cc1 resulted in inadequate processing of the ingested material. The kinetics of the degeneration suggests that at early ages there is sufficient breakdown of the phagocytized outer segments, as there is normal morphology and visual function. However, the absence of autophagy induction appears to result in cumulative damage that does not manifest as a detectable phenotype until later ages. It was recently reported by Kim and coworkers,35 that RPE-specific deletion of the gene encoding another autophagy-related protein, ATG5, does not result in a significant RPE degeneration, but rather in aberrant recycling of chromophore. They suggested that noncanonical autophagy, but not conventional autophagy, was active in the RPE and served to reintroduce vitamin A into the visual cycle. In our animals, we deleted a more upstream inducer of autophagy, Rb1cc1, and this might have allowed for a more complete reduction in autophagy than achieved by deletion of Atg5. Furthermore, Kim and coworkers focused their functional studies on mice 20 wk of age or less, and perhaps observation at older ages as in our study would have allowed for sufficient accumulation of autophagy-targeted content to result in RPE degeneration. We recently reported that autophagy in the RPE is activated in a circadian rhythm,36 with a phase-shift of several hours relative to the peak of disc shedding, consistent with the time required for normal phagocytic processing of the outer segments. While our data do not address the role of LC3-associated phagocytosis in the processing of shed outer segments by the RPE, they do strongly support the critical role of conventional macroautophagy in maintaining RPE homeostasis and thus preventing an age-related degeneration.
In human disease, AMD can manifest with varying phenotypes, with one or more defining phenotypic characteristics such as pigment abnormalities, drusen deposits, and variably sized areas of RPE atrophy.55 Some patients also have foci of neovascularization, usually extending from the choroid into the subRPE and subretinal spaces. Drusen, a hallmark feature of AMD, are not a uniform entity, but rather consist of deposits that can actually be found either under the retina (subretinal drusenoid deposits) or under the RPE (cuticular drusen and soft drusen).56 The derivation and composition of drusen is thought to originate from the RPE and contain a complex mixture of proteins and lipids. In addition, the presence of the AMD phenotype has been correlated with reduced autophagic function in the RPE.57 The model presented here demonstrates that blocking the induction of autophagy in the RPE results in many of the salient features of AMD, particularly RPE atrophy, the accumulation of lipid-filled inclusions, drusenoid deposits, abnormal accumulation of reactive oxygen species, complement activation, and occasional foci of neovascularization.
The RPE-specific defect in autophagy and resultant RPE degeneration also causes a loss of photoreceptor cells. Since autophagy in the photoreceptors is normal in this model, presumably their death is secondary to the loss of the RPE. This is similar to what is seen in other models of RPE-specific cell death, such as the sodium iodate toxicity model or genetic models that result in RPE degeneration.58,59 This effect is not altogether unexpected, as the photoreceptors are highly dependent on the RPE for metabolic support. Photoreceptor degeneration is seen in other conditions where the normal interface with the RPE is disrupted, such as in retinal detachment. The secondary loss of photoreceptors suggests that there is a potential for neuroprotective intervention to preserve these critical neurosensory cells and retain visual function even in the face of an underlying RPE degeneration. This is an area of active investigation.
While we cannot conclude that decreased autophagy is the primary cause of AMD, our studies support the conclusion that dysregulation of this homeostatic-maintenance pathway plays a critical role in development of the phenotypes characteristic of this disease. We propose that macroautophagy serves to mitigate the negative effects of genetic and environmental stressors associated with the development of AMD. For example, it has recently been proposed that a major contributor to the formation of atrophic AMD is the accumulation of RNA Alu repeats, and the subsequent activation of the inflammasome.60,61 Autophagy has been implicated in reducing the toxicity of these repeats as well as in reducing the disease burden caused by inflammasome activation.62-64 Another pathogenic pathway affected by autophagy is the processing of lipids, a well-known contributor to the AMD phenotype.65-67 Autophagy plays a critical role in lipid metabolism and reduces lipid toxicity in a number of animal models of disease.68,69 Going forward, the RB1CC1 model will provide a novel platform for studying the pathways by which these potential contributors to AMD exert their effects, and for evaluating modulators of these mechanisms that may have therapeutic potential.
Materials and Methods
Animals
Rb1cc1flox/flox and Best1-Cre mice have been described previously.33,34 These 2 strains were bred to generate RPE-specific Rb1cc1-CKO (Rb1cc1flox/flox; Best1-Cre [+]) and control (Rb1cc1wt; Best1-Cre[+]) littermates. Mice were genotyped by PCR analysis of genomic DNA isolated from tail tips using specific primers as described for Cre recombinase (F: 5′-ATG CCC AAG AAG AAG AAG AAG GTG TCC A-3′ and R: 5′-TGG CCC AAA TGT TGC TGG ATA GTT TTT A-3′) and for the wild-type and floxed Rb1cc1 allele (F: 5′-CAA AGA ACA ACG AGT GGC AGT AG-3′and R: 5′-CAT CAG ATA CAC TAG AGC TGG-3′).
Mice were bred and housed under standard 12-h light/12-h dark conditions in the University of Michigan Kellogg Eye Center animal facility. All animal experiments were conducted in accordance with the ARVO statement for the Use of Animals in Ophthalmic and Vision Research and approved by the University Committee on the Use and Care of Animals of the University of Michigan. All mice were genotyped by established methods and only those negative for mutation in the Crb1/Rd8 gene were used.70
Tissue collection
Retinal tissue and RPE-choroid complexes were carefully isolated at various ages as previously described.36 Due to the presence of circadian changes in autophagy levels in the RPE,36 all collections were performed at the same time of day, 1:00 p.m. Briefly, mice were euthanized and eyes were immediately enucleated and placed in a dish. The connective tissue, muscle, and optic nerve were removed from the back of the eye, and the cornea and lens were removed to form an eye cup. The retina was dissected off of the RPE, cut from the connection to the optic nerve head, and pulled away from the rest of the eye. The RPE/choroid was carefully scraped from the sclera with a pair of flat-top forceps.
ARPE-19 cell transfection and outer segment feeding
ARPE-19 cells were cultured in Ham's F10 (Corning Cellgro, 10–070-CV) medium supplemented with 10% fetal bovine serum (Sigma Aldrich, 12107C), and antibiotics (Corning Cellgro, 30–002-CI) at 37°C in 5% CO2.21 Cells were passaged and then cultured for 72 h in 12-well plates or 8-well chamber slides. When cells were 80–90% confluent, they were transfected with 2 different siRNAs against the human RB1CC1 transcript (Qiagen, Hs_RBA1CC1_1, Hs_RBA1CC1_3) or with scrambled siRNA (Ambion®, AM4611), followed by incubation with photoreceptor outer segments isolated from bovine eyes71 at 40 POS per cell at 37°C. Cells were harvested and prepared as previously described for immunohistochemistry and western blot analysis.72
Western blot analysis
RPE/choroid and retina samples were homogenized. The proteins were separated by 4–15% SDS-PAGE (Tris-HCl Ready Gels; Bio-Rad Laboratories) and blotted on polyvinylidene fluoride membranes (Bio-Rad, 162–0177). The membranes were incubated overnight with primary antibodies. A full list of antibodies is shown in Table 1. Secondary polyclonal goat Anti-Rabbit and Goat Anti-Mouse immunoglobulin/HRP antibodies were from Dako (P0488 or P0447). Detection was by SuperSignal West Dura Substrate (Thermo Scientific, 34075) according to the manufacturer's protocols. Quantitative densitometry of the immunoblots was performed using ImageJ software (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html), and expressed as the mean density (±SD ) from replicate experimental groups. All experiments were performed a minimum of 3 times.
Table 1.
List of antibodies used in this study
| Antibody | Host | Immunoblot | Immunostaining | Supplier |
|---|---|---|---|---|
| LC3 | rabbit | 1:1000 | 1:200 | Novus Biologicals, 4108 |
| ATG5 | rabbit | 1:1000 | — | Novus Biologicals, NB100–53818S |
| ATG12 | rabbit | 1:1000 | — | Cell Signaling Technology, 4180 |
| SQSTM1/p62 | rabbit | 1:1000 | — | Novus Biologicals, NBP1–48320S |
| C3 | mouse | 1:500 | 1:100 | Santa Cruz Biotechnology, cs-28294 |
| ITGB1/CD29 | mouse | 1:100 | Biolegend, 303001 | |
| CD68 | rat | — | 1:200 | AbD Serotec, MCA1957T |
| CTSD/cathepsin D | goat | 1:200 | — | Santa Cruz Biotechnology, sc-6487 |
| Cre-recombinase | mouse | 1:1000 | 1:500 | Millipore, MAB3120 |
| Cre-recombinase | rabbit | — | 1:1000 | Novagen, 69050–3 |
| RB1CC1/FIP200 | rabbit | 1:1000 | — | Proteintech, 10041–2-AP |
| Isolectin IB4 488 Conjugate | Griffoniasimplicifolia | — | 1:50 | Invitrogen, 121411 |
| IBA1 | goat | — | 1:500 | Novus Biologicals, NB100–1028 |
| PTK2/FAK | rabbit | 1:1000 | — | Cell Signaling Technology, 3285 |
| Phospho-PTK2 | rabbit | 1:500 | — | Cell Signaling Technology, 3284 |
| MAPK8/JNK1-MAPK9/JNK2 | rabbit | 1:1000 | — | Cell Signaling Technology, 9258 |
| Phospho MAPK8/JNK1-MAPK9/JNK/2 | rabbit | 1:500 | — | Cell Signaling Technology, 9234 |
| LAMP1 | mouse | 1:2000 | — | EnCor Biotechnology, MAC-6E2 |
| Nitrotyrosine | mouse | — | 1:100 | Santa Cruz Biotechnology, sc-32731 |
| RPE65 | mouse | — | 1:200 | Courtesy of Dr. Thompson 58 |
| TOMM40 | rabbit | — | 1:200 | Santa Cruz Biotechnology, sc-11414 |
| Ubiquitin | rabbit | — | 1: 600 | Cell Signaling Technology, 3933 |
| TJP1/ZO-1 | rabbit | — | 1:100 | Invitrogen, 61–7300 |
| GAPDH | mouse | 1:60,000 | — | Ambion Applied Biosystems, AM4300 |
Ratios listed are the dilution used for each particular experiment.
Histology and immunohistochemistry
For retina sections, eyes were enucleated and fixed with 4% paraformaldehyde at room temperature for 3 h. As described above, all collections were performed at the same time of day, 1:00 p.m. The cornea and lens were removed and the eye cup rinsed 3 times in 1×PBS (Sigma Life Science, P4417), then transferred to 10% and then 20% sucrose (Sigma Life Science, S9378) in PBS for 2 h each, before embedding in OCT (Sakura, 4583) mixed in a ratio of 1:1 with 20% sucrose. A cryostat was used to obtain serial retina/RPE/choroid sections (10 μm). Sections were washed in PBS, blocked with 10% goat serum (Sigma Life Science, G9023) and 0.1% Triton X-100 (Sigma Life Science, 9002–93–1) for one h, and incubated with primary antibodies overnight at 4°C. After incubation for one h at room temperature with secondary antibodies, sections were counterstained with ProLong Gold with DAPI (Invitrogen, P36941) to reveal cell nuclei.
For RPE flat mounts, eyes were enucleated and fixed with 4% paraformaldehyde at room temperature for one h. The anterior segment and retina were carefully removed under a dissecting microscope to minimize damage of the underlying RPE. The remaining RPE-choroid-sclera complexes were incubated for one h with blocking solution (10% goat serum with 0.3% Triton X-100 in PBS) before they were incubated with the primary antibodies overnight at 4°C. Six radial incisions were made, and the RPE-choroid-sclera complex was flat mounted on a glass slide.
For RPE autofluorescence detection, freshly-cut frozen sections were air dried at room temperature for 2 h before being mounted with ProLong Gold with DAPI. Confocal images were taken using excitation 488 nm and emission spectra from 500 to 620 nm.73,74
Images were obtained using a confocal microscope (Leica SP5, Leica Corp., Germany). Images were taken at the comparable area of sections. All images in each individual experiment were acquired with a fixed detection gain.
For hematoxylin and eosin staining, slides from histologically comparable positions of paraffin-embedded eye samples that were sectioned at 6-µm thickness were stained with hematoxylin (Fisher Scientific, 3536–16) and eosin (Fisher Scientific, E511). Oil Red O (American MasterTech, KTORO) staining was performed on 10-µm fresh cryosections. Images were captured with a Leica DM6000 microscope.
Transmission electron microscopy
For transmission electron microscopy, the superior part of the eye cups were dissected in the fixative (2.5% glutaraldehyde in 0.1 M Sorensen's buffer, pH 7.4) immediately after the eyes were enucleated and were then fixed overnight at 4°C. After several buffer rinses, samples were post-fixed in 1% osmium tetroxide in the same buffer. Samples were then rinsed in double-distilled water to remove phosphate salt and then en bloc stained with aqueous 3% uranyl acetate for one h. After being dehydrated in ascending concentrations of ethanol, tissues were rinsed 2 times in propylene oxide, and then embedded in epoxy resin. The samples were ultra-thin sectioned 70 nm in thickness and stained again with uranyl acetate and lead citrate. The sections were examined using a Philips CM100 electron microscope at 60 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA).
Quantification of phagosomes in the RPE
The number of phagosomes present in pigment epithelial cells was determined by counting directly on the electron microscope screen, at a magnification of 10,500×. Three representative thin sections crossing the optic nerve were used from each eye. In each section, 2 independent areas of 400-µm length of the RPE were counted. A phagosome was scored as within the area between the basal infolding and the base of microvilli.
Spectral domain optical coherence tomography
Optical coherence tomography was performed with a spectral domain optical coherence tomography system (Bioptigen Inc.., Durham, NC). A volume analysis centered on the optic nerve head was performed, using 100 horizontal, raster, and consecutive B scan lines, each one composed of 1200 A-scans. The volume size was 1.6 × 1.6 mm. Outer nuclear layer (ONL) thickness was assessed at distances of 500 µm, 400 µm, and 300 µm from the optic nerve head (nasally and temporally) and at 200 µm above and 400 µm below the optic nerve head. Quantification of the ONL thickness and generation of the heat map of the ONL were performed using InVivoVue™ Diver 2.4 software (Bioptigen Inc.., Durham, NC).
Electroretinography analysis
ERGs were performed using the Espion e2 recording system (Diagnosys, Lowell, MA) as described previously.75 After overnight dark adaptation, mice were anesthetized with an intraperitoneal injection of ketamine (93 mg/kg) and xylazine (8 mg/kg). Body temperature was maintained at 37°C with a heating pad. Corneal ERGs were recorded from both eyes using gold wire loops after pupil dilation with topical phenylephrine (2.5%) and tropicamide (1.0%) and a drop of 2% methylcellulose for corneal hydration. A gold wire loop placed in the mouth was used as reference, and a ground electrode was on the tail. The dark-adapted ERG was recorded at −2.31 log cd.s/m2 (candle seconds per meter squared) to elicit rod-isolated responses. After 10 min of light adaptation to a white 32 cd/m2 rod-suppressing background, light adapted ERGs were recorded at 1.09 log cd.s/m2. Ten to 25 responses were recorded at 3 to 60 s depending upon the stimulus intensity intervals. B-wave amplitude of the averaged response was measured from baseline to the positive peak of the waveform.
Fundus photography and fluorescein angiography
General fundus photography and fluorescein angiography were performed with a commercial camera and imaging system (Micron III, Phoenix research labs, Pleasanton, CA). Mice were intraperitoneally injected with 40 µl of 2% fluorescein sodium (Akorn, NDC17478–253–10) per 10 g body weight.
Statistical analysis
Experimental results are expressed as mean ± SD. Values of ERG, SD-OCT and quantifications of phagosome and autophagosome were analyzed by 2-tailed Student ttest and one-way ANOVA with Bonferroni correction for multiple comparison. Nonparametric Kruskal-Wallis (K-W) test was used for western blotting analysis. Differences were considered significant at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The National Eye Institute R01-EY-020823 (DNZ), Beckman Initiative for Macular Research (DNZ), Foundation Fighting Blindness (DNZ, DAT), Research to Prevent Blindness, Inc. (DNZ; DAT is a Research to Prevent Blindness Senior Scientific Investigator), and the University of Michigan Core Center for Vision Research (NEI-EY007003) provided support for this work.
References
- 1.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124-31; PMID:20034776; http://dx.doi.org/ 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun 2004; 313:453-8; PMID:14684184; http://dx.doi.org/ 10.1016/j.bbrc.2003.07.023 [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/ 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ 2005; 12:1535-41; PMID:16247501; http://dx.doi.org/ 10.1038/sj.cdd.4401728 [DOI] [PubMed] [Google Scholar]
- 5.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032-6; PMID:15525940; http://dx.doi.org/ 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al.. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425-34; PMID:15866887; http://dx.doi.org/ 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, et al.. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25:1025-40; PMID:15657430; http://dx.doi.org/ 10.1128/MCB.25.3.1025-1040.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al.. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/ 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- 9.Linsenmeier RA, Padnick-Silver L. Metabolic dependence of photoreceptor on the choroid in the normal and detached retina. Invest Ophthalmol Vis Sci 2000; 41:3117-23; PMID:10967072 [PubMed] [Google Scholar]
- 10.Wangsa-Wirawan ND, Linsenmeier RA. Retinal oxygen: fundamental and clinical aspects. Arch Ophthalmol 2003; 121:547-57; PMID:12695252; http://dx.doi.org/ 10.1001/archopht.121.4.547 [DOI] [PubMed] [Google Scholar]
- 11.Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010; 25:8-15; PMID:20134024; http://dx.doi.org/ 10.1152/physiol.00038.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev 2005; 85:845-81; PMID:15987797; http://dx.doi.org/ 10.1152/physrev.00021.2004 [DOI] [PubMed] [Google Scholar]
- 13.Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol 1967; 33:61-72; PMID:6033942; http://dx.doi.org/ 10.1083/jcb.33.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young RW, Bok D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J Cell Biol 1969; 42:392-403; PMID:5792328; http://dx.doi.org/ 10.1083/jcb.42.2.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaVail MM. Legacy of the RCS rat: impact of a seminal study on retinal cell biology and retinal degenerative diseases. Prog Brain Res 2001; 131:617-27; PMID:11420975; http://dx.doi.org/ 10.1016/S0079-6123(01)31048-8 [DOI] [PubMed] [Google Scholar]
- 16.van Lookeren Campagne M, LeCouter J, Yaspan BL, Ye W. Mechanisms of age related macular degeneration and therapeutic opportunities. J Pathol 2014; 232:151-64; PMID:24105633; http://dx.doi.org/ 10.1002/path.4266 [DOI] [PubMed] [Google Scholar]
- 17.Reme CE, Autophagy in visual cells and pigment epithelium. Invest. Ophthalmol Vis Sci 1977; 16:807-14; PMID:302253 [PubMed] [Google Scholar]
- 18.Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One 2009; 4:e4160; PMID:19129916; http://dx.doi.org/ 10.1371/journal.pone.0004160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krohne TU, Stratmann NK, Kopitz J, Holz FG. Effects of lipid peroxidation products on lipofuscinogenesis and autophagy in human retinal pigment epithelial cells. Exp Eye Res 2010; 90:465-71; PMID:20059996; http://dx.doi.org/ 10.1016/j.exer.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 20.Viiri J, Hyttinen JM, Ryhanen T, Rilla K, Paimela T, Kuusisto E, Siitonen A, Urtti A, Salminen A, Kaarniranta K. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol Vis 2010; 16:1399-414; PMID:20680098 [PMC free article] [PubMed] [Google Scholar]
- 21.Mitter SK, Song C, Qi X, Mao H, Rao H, Akin DA, Lewin AS, Grant M, Dunn WA, Ding JD, et al.. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 2014; 10(11):1989-2005; PMID:25484094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitter SK, Rao HV, Qi X, Cai J, Sugrue A, Dunn WA Jr, Grant MB, Boulton ME. Autophagy in the retina: a potential role in age-related macular degeneration. Adv Exp Med Biol 2012; 723:83-90; PMID:22183319; http://dx.doi.org/ 10.1007/978-1-4614-0631-0_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valapala M, Wilson C, Hose S, Bhutto IA, Grebe R, Dong A, Greenbaum S, Gu L, Sengupta S, Can M, et al.. Lysosomal-mediated waste clearance in retinal pigment epithelial cells is regulated by CRYBA1/bA3/A1-crystallin via V-ATPas-MTORC1 signaling. Autophagy 2014; 10:480-96; PMID:24468901; http://dx.doi.org/ 10.4161/auto.27292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D'Agostino VG, et al.. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One 2013; 8:e69563; PMID:23922739; http://dx.doi.org/ 10.1371/journal.pone.0069563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Muela N, Koga H, García-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 2013; 12:478-88; http://dx.doi.org/ 10.1111/acel.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DJ. ULK-Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992-2003; PMID:19225151; http://dx.doi.org/ 10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and-independent autophagy. Nat Struct Mol Biol 2013; 20:144-9; PMID:23262492; http://dx.doi.org/ 10.1038/nsmb.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 2013; 14:284-91; PMID:23392225; http://dx.doi.org/ 10.1038/embor.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181:497-510; PMID:18443221; http://dx.doi.org/ 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 2013; 16:532-42; PMID:23542691; http://dx.doi.org/ 10.1038/nn.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 2011; 25:1510-27; PMID:21764854; http://dx.doi.org/ 10.1101/gad.2051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan J-L. Role of FIP200 in cardiac and liver development and its regulation of TNF and TSC-mTOR signaling pathways. J Cell Biol 2006; 175:121-33; PMID:17015619; http://dx.doi.org/ 10.1083/jcb.200604129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iacovelli J, Zhao C, Wolkow N, Veldman P, Gollomp K, Ojha P, Lukinova N, King A, Feiner L, Esumi N, et al.. Generation of Cre transgenic mice with postnatal RPE-specific ocular expression. Invest Ophthalmol Vis Sci 2011; 52:1378-83; PMID:21212186; http://dx.doi.org/ 10.1167/iovs.10-6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al.. Noncanonical autophagy promotes the visual cycle. Cell 2013; 154:365-76; PMID:23870125; http://dx.doi.org/ 10.1016/j.cell.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J, Jia L, Shelby SJ, Ganios AM, Feathers K, Thompson DA, Zacks DN. Circadian and noncircadian modulation of autophagy in photoreceptors and retinal pigment epithelium. Invest Ophthalmol Vis Sci 2014; 55:3237-46; PMID:24781939; http://dx.doi.org/ 10.1167/iovs.13-13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L, Marioutina M, Dunaief JL, Marneros AG. Age- and gene-dosage-dependent cre-induced abnormalities in the retinal pigment epithelium. Am J Pathol 2014; 184:1660-7; PMID:24854863; http://dx.doi.org/ 10.1016/j.ajpath.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal 2008; 20:787-94; PMID:18036779; http://dx.doi.org/ 10.1016/j.cellsig.2007.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Yang P, Kijlstra A. Distribution, markers and functions of retinal microglia. Ocul Immunol Inflamm 2002; 10:27-39; PMID:12461701; http://dx.doi.org/ 10.1076/ocii.10.1.27.10328 [DOI] [PubMed] [Google Scholar]
- 40.Langmann T. Microglia activation in retinal degeneration. J Leukoc Biol 2007; 81:1345-51; PMID:17405851; http://dx.doi.org/ 10.1189/jlb.0207114 [DOI] [PubMed] [Google Scholar]
- 41.Cuenca N, Fernandez-Sanchez L, Campello L, Maneu V, De al Villa P, Lax P, Pinilla I. Cellular responses following retinal injuries and therapeutic approaches for neurodegenerative diseases. Prog Retin Eye Res 2014; 43:17-75; PMID:25038518; http://dx.doi.org/ 10.1016/j.preteyeres.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 42.Karlstetter M, Langmann T. Microglia in the aging retina. Adv Exp Med Biol 2014; 801:207-12; PMID:24664700; http://dx.doi.org/ 10.1007/978-1-4614-3209-8_27 [DOI] [PubMed] [Google Scholar]
- 43.Rizzolo LJ. Basement membrane stimulates the polarized distribution of integrins but not the Na,K-ATPase in the retinal pigment epithelium. Cell Regul 1991; 2:939-49.; PMID:1667092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grossniklaus HE, Kang SJ, Berglin L. Animal models of choroidal and retinal neovascularization. Prog Retin Eye Res 2010; 29:500-19; PMID:20488255; http://dx.doi.org/ 10.1016/j.preteyeres.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zacks DN, Ezra E, Terada Y, Michaud N, Connolly E, Gragoudas ES, Miller JW. Verteporfin photodynamic therapy in the rat model of choroidal neovascularization: angiographic and histologic characterization. Invest Ophthalmol Vis Sci 2002; 43:2384-91; PMID:12091441 [PubMed] [Google Scholar]
- 46.Jiang A, Hu W, Meng H, Gao H, Qiao X. Loss of VLDL receptor activates retinal vascular endothelial cells and promotes angiogenesis. Invest Ophthalmol Vis Sci 2009; 50:844-50; PMID:18936153; http://dx.doi.org/ 10.1167/iovs.08-2447 [DOI] [PubMed] [Google Scholar]
- 47.Lippai M, Low P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int 2014; 2014:832704; PMID:25013806; http://dx.doi.org/ 10.1155/2014/832704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryhänen T, Hyttinen JM, Kopitz J, Rilla K, Kuusisto E, Mannermaa E, Viiri J, Holmberg CI, Immonen I, Meri S, et al.. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med 2009; 13:3616-31; http://dx.doi.org/ 10.1111/j.1582-4934.2008.00577.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer disease. Prog Retin Eye Res 2011; 30:217-38; PMID:21440663; http://dx.doi.org/ 10.1016/j.preteyeres.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, Bhattacharjee S, Jones BM, Hill JM, Clement C, Sambamurti K, Dua P, Lukiw WJ. Beta-amyloid precursor protein (bAAP) processing in Alzheimer disease (AD) and age-related macular degeneration (AMD). Mol Neurobiol 2014; Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyttinen JM, Amadio M, Viiri J, Pascale A, Salminen A, Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev 2014:18C:16-28; http://dx.doi.org/ 10.1016/j.arr.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 52.Mazzoni F, Safa H, Finnemann SC. Understanding photoreceptor outer segment phagocytosis: use and utility of RPE cells in culture. Exp Eye Res 2014; 126:51-60; PMID:24780752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng W, Reem RE, Omarova S, Huang S, DiPatre PL, Charvet CD, Curcio CA, Pikuleva IA. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One 2012; 7(5):e37926; PMID:22629470; http://dx.doi.org/ 10.1371/journal.pone.0037926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang PH, Kono M, Koutalos Y, Ablonczy Z, Crouch RK. New insights into retinoid metabolism and cycling within the retina. Prog Retin Eye Res 2013; 32:48-63; PMID:23063666; http://dx.doi.org/ 10.1016/j.preteyeres.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferris FL 3rd, Wilkinson CP, Bird A, Chakravarthy U, Chew E, Scaky K, Sadda SR, Beckman Initiative for Macular Research Classification Committee . Clinical classification of age-related macular degeneration. Ophthalmology 2013; 120:844-51; PMID:23332590; http://dx.doi.org/ 10.1016/j.ophtha.2012.10.036 [DOI] [PubMed] [Google Scholar]
- 56.Spaide RF, Curcio CA. Drusen characterization with multimodal imaging. Retina 2010; 30:1441-54; PMID:20924263; http://dx.doi.org/ 10.1097/IAE.0b013e3181ee5ce8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaarniranta K, Sinha D, Blasiak J, Kauppinen A, Vereb Z, Salminen A, Boulton ME, Petrovski G. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy 2013; 9:973-84; PMID:23590900; http://dx.doi.org/ 10.4161/auto.24546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Machalinska A, Lubinski W, Klos P, Kawa M, Baumer B, Penkala K, Grzegrzolka R, Karczewicz D, Wiszniewska B, Machalinski B. Sodium iodate selectively injures the posterior pole of the retina in a dose-dependent manner: morphological and electrophysiological study. Neurochem Res 2010; 35:1819-27; PMID:20725778; http://dx.doi.org/ 10.1007/s11064-010-0248-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramkumar HL, Zhang J, Chan CC. Retinal ultrastructure of murine models of dry age-related macular degeneration (AMD). Prog Retin Eye Res 2010; 29:169-90; PMID:20206286; http://dx.doi.org/ 10.1016/j.preteyeres.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al.. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011; 471:325-30; PMID:21297615; http://dx.doi.org/ 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarallo V, Hiran Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al.. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012:149; 847-59; PMID:22541070; http://dx.doi.org/ 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibbings D, Mostowy S, Voinnet O. Autophagy selectively regulates miRNA homeostasis. Autophagy 2013; 14:1314-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams-Bey Y, Boularan C, Vural A, Huang NN, Hwang IY, Shan-Shi C, Kehri JH. Omega-3 free fatty acids suppress macrophage inflammasome activation by inhibiting NF-kB activation and enhancing autophagy. PLoS One 2014; 9(6):e97957; PMID:24911523; http://dx.doi.org/ 10.1371/journal.pone.0097957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo W, Sun Y, Liu W, Wu X, Guo L, Cai P, Wu X, Wu X, Shen Y, Shu Y, Gu Y, Xu Q. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014; 10:972-85; PMID:24879148; http://dx.doi.org/ 10.4161/auto.28374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malek G, Johnson LV, Mace BE, Saloupis P, Schmechel DE, Rickman DW, Toth CA, Sullivan PM, Bowes Rickman C. Apolipoprotein E allele-dependent pathogenesis: a model for age-related macular degeneration. Proc Nat Acad Sci 2005; 102:11900-5; PMID:16079201; http://dx.doi.org/ 10.1073/pnas.0503015102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curcio CA, Johnson M, Rudolf M, Huang JD. The oil spill in ageing Bruch membrane. Br J Ophthalmol 2011; 95:1638-45; PMID:21890786; http://dx.doi.org/ 10.1136/bjophthalmol-2011-300344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pikuleva IA, Curcio CA. Cholesterol in the retina: the best is yet to come. Prog Retin Eye Res 2014; 41C:64-89; http://dx.doi.org/ 10.1016/j.preteyeres.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu K, Czaia MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 2013; 20:3-11; PMID:22595754; http://dx.doi.org/ 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Christian P, Sacco J, Adeli K. Autophagy: emerging roles in lipid homeostasis and metabolic control. Biochim Biophys Acta 2013; 1831:819-24; PMID:23274236; http://dx.doi.org/ 10.1016/j.bbalip.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 70.Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci 2012; 53:2921-7; PMID:22447858; http://dx.doi.org/ 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Papermaster DS. Preparation of retinal rod outer segments. Methods Enzymol 1982; 81:48-52; PMID:6212746; http://dx.doi.org/ 10.1016/S0076-6879(82)81010-0 [DOI] [PubMed] [Google Scholar]
- 72.Shelby SJ, Colwill K, Dhe-Paganon S, Pawson T, Thompson DA. MERTK interactions with SH2-domain proteins in the retinal pigment epithelium. PLoS One 2013; 8(2):e53964; PMID:23390493; http://dx.doi.org/ 10.1371/journal.pone.0053964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp Eye Res 2005; 80:595-606; PMID:15862166; http://dx.doi.org/ 10.1016/j.exer.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 74.Hadziahmetovic M1, Song Y, Ponnuru P, Iacovelli J, Hunter A, Haddad N, Beard J, Connor JR, Vaulont S, Dunaief JL. Age-dependent retinal iron accumulation and degeneration in hepcidin knockout mice. Invest Ophthalmol Vis Sci 2011; 52:109-18; PMID:20811044; http://dx.doi.org/ 10.1167/iovs.10-6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson DA, Khan NW, Othman MI, Chang B, Jia L, Grahek G, Wu Z, Hiriyanna S, Nellissery J, Li T, et al.. Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS ONE 2012; 7(5):e35865; PMID:22563472; http://dx.doi.org/ 10.1371/journal.pone.0035865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemati N, Feathers KL, Chrispell JD, Reed DM, Carlson TJ, Thompson DA. RPE65 surface epitopes, protein interactions, and expression in rod- and cone-dominant species. Mol Vis 2005; 11:1151-65; PMID:16379027 [PubMed] [Google Scholar]