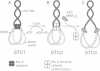Abstract
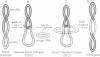
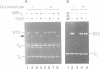
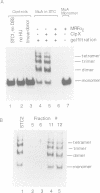
During transposition bacteriophage Mu transposase (MuA) catalyzes the transfer of a DNA strand at each Mu end to target DNA and then remains tightly bound to the Mu ends. Initiation of Mu DNA replication on the resulting strand transfer complex (STC1) requires specific host replication proteins and host factors from two partially purified enzyme fractions designated Mu replication factors alpha and beta (MRFalpha and beta). Escherichia coli ClpX protein, a molecular chaperone, is a component required for MRFalpha activity, which removes MuA from DNA for the establishment of a Mu replication fork. ClpX protein alters the conformation of DNA-bound MuA and converts STC1 to a less stable form (STC2). One or more additional components of MRFalpha (MRFalpha2) displace MuA from STC2 to form a nucleoprotein complex (STC3), that requires the specific replication proteins and MRFbeta for Mu DNA synthesis. MuA present in STC2 is essential for its conversion to STC3. If MuA is removed from STC2, Mu DNA synthesis no longer requires MRFalpha2, MRFbeta and the specific replication proteins. These results indicate that ClpX protein activates MuA in STC1 so that it can recruit crucial host factors needed to initiate Mu DNA synthesis by specific replication enzymes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of Mu B protein. Cell. 1988 Apr 22;53(2):257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- Alfano C., McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J Biol Chem. 1989 Jun 25;264(18):10709–10718. [PubMed] [Google Scholar]
- Alfano C., McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem. 1989 Jun 25;264(18):10699–10708. [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Dodson M., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda. Protein association and disassociation reactions responsible for localized initiation of replication. J Biol Chem. 1989 Jun 25;264(18):10719–10725. [PubMed] [Google Scholar]
- Geuskens V., Mhammedi-Alaoui A., Desmet L., Toussaint A. Virulence in bacteriophage Mu: a case of trans-dominant proteolysis by the Escherichia coli Clp serine protease. EMBO J. 1992 Dec;11(13):5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., de Crecy-Lagard V., Maurizi M. R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993 Oct 25;268(30):22618–22626. [PubMed] [Google Scholar]
- Hwang B. J., Park W. J., Chung C. H., Goldberg A. L. Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5550–5554. doi: 10.1073/pnas.84.16.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. J., Woo K. M., Goldberg A. L., Chung C. H. Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988 Jun 25;263(18):8727–8734. [PubMed] [Google Scholar]
- Katayama-Fujimura Y., Gottesman S., Maurizi M. R. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem. 1987 Apr 5;262(10):4477–4485. [PubMed] [Google Scholar]
- Kiehm D. J., Ji T. H. Photochemical cross-linking of cell membranes. A test for natural and random collisional cross-links by millisecond cross-linking. J Biol Chem. 1977 Dec 10;252(23):8524–8531. [PubMed] [Google Scholar]
- Kruklitis R., Nakai H. Participation of the bacteriophage Mu A protein and host factors in the initiation of Mu DNA synthesis in vitro. J Biol Chem. 1994 Jun 10;269(23):16469–16477. [PubMed] [Google Scholar]
- Kuo C. F., Zou A. H., Jayaram M., Getzoff E., Harshey R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 1991 Jun;10(6):1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J Biol Chem. 1990 Jan 25;265(3):1623–1627. [PubMed] [Google Scholar]
- Lavoie B. D., Chan B. S., Allison R. G., Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991 Oct;10(10):3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I., Luo L., Baker T. A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995 Oct 1;9(19):2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- Maurizi M. R., Clark W. P., Katayama Y., Rudikoff S., Pumphrey J., Bowers B., Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990 Jul 25;265(21):12536–12545. [PubMed] [Google Scholar]
- Mhammedi-Alaoui A., Pato M., Gama M. J., Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994 Mar;11(6):1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker T. A., Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992 Jul 24;70(2):303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell. 1989 Jul 28;58(2):399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- Nakai H. Amplification of bacteriophage Mu DNA by rolling circle DNA replication in vitro. J Biol Chem. 1993 Nov 15;268(32):23997–24004. [PubMed] [Google Scholar]
- Nakai H., Kruklitis R. Disassembly of the bacteriophage Mu transposase for the initiation of Mu DNA replication. J Biol Chem. 1995 Aug 18;270(33):19591–19598. doi: 10.1074/jbc.270.33.19591. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Reich C. Instability of transposase activity: evidence from bacteriophage mu DNA replication. Cell. 1982 May;29(1):219–225. doi: 10.1016/0092-8674(82)90106-4. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Reich C. Stoichiometric use of the transposase of bacteriophage Mu. Cell. 1984 Jan;36(1):197–202. doi: 10.1016/0092-8674(84)90089-8. [DOI] [PubMed] [Google Scholar]
- Resibois A., Pato M., Higgins P., Toussaint A. Replication of bacteriophage mu and its mini-mu derivatives. Adv Exp Med Biol. 1984;179:69–76. doi: 10.1007/978-1-4684-8730-5_7. [DOI] [PubMed] [Google Scholar]
- Ross W., Shore S. H., Howe M. M. Mutants of Escherichia coli defective for replicative transposition of bacteriophage Mu. J Bacteriol. 1986 Sep;167(3):905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hearst J. E. Molecular matchmakers. Science. 1993 Mar 5;259(5100):1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Tippe-Schindler R., Zahn G., Messer W. Control of the initiation of DNA replication in Escherichia coli. I. Negative control of initiation. Mol Gen Genet. 1979 Jan 10;168(2):185–195. doi: 10.1007/BF00431444. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M. The dependence of temperate phage Mu-1 upon replication functions of E. coli K12. Mol Gen Genet. 1974;131(3):209–214. doi: 10.1007/BF00267960. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A., Wojtkowiak D., Marszalek J., Banecki B., Jonsen M., Graves B., Georgopoulos C., Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995 May 1;14(9):1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M. R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowiak D., Georgopoulos C., Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993 Oct 25;268(30):22609–22617. [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Yoo S. J., Seol J. H., Kang M. S., Ha D. B., Chung C. H. clpX encoding an alternative ATP-binding subunit of protease Ti (Clp) can be expressed independently from clpP in Escherichia coli. Biochem Biophys Res Commun. 1994 Sep 15;203(2):798–804. doi: 10.1006/bbrc.1994.2253. [DOI] [PubMed] [Google Scholar]
- Zylicz M., Ang D., Liberek K., Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989 May;8(5):1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]