Abstract
To examine the interaction between molecular, electrical, and behavioral circadian rhythms, we combined optogenetic manipulation of suprachiasmatic nucleus (SCN) firing rate with bioluminescence imaging and locomotor activity monitoring. Manipulating firing rate reset circadian rhythms both ex vivo and in vivo and this resetting required spikes and network communication. This suggests that SCN firing rate is fundamental to circadian pacemaking as both an input onto and output of the molecular clockworks.
The brain's biological clock – the suprachiasmatic nucleus (SCN) – provides a unique model for studying the interaction between gene networks and behavior. The individual cellular oscillators that comprise the SCN network exhibit endogenous molecular and electrical rhythms. Additionally, a collection of intrinsic currents allows these neurons to fire action potentials in the absence of synaptic drive and, importantly, fire at elevated frequency (up to 8–12 Hz) during the day while being nearly silent at night (typically <1 Hz)1,2. Network communication by the neuropeptides vasoactive intestinal peptide (VIP), arginine vasopressin (AVP), and the neurotransmitter GABA allow these oscillators to form a tissue-level clock, orchestrating daily changes in physiology and behavior3-6. Thus interlocking molecular and electrical loops in the SCN interact to drive behavior; however, the precise interplay of these molecular, electrical, and behavioral components of the brain's biological clock remains unknown7-12.
The inability to precisely manipulate firing rate in SCN neurons without confounding ionic or pharmacological stimuli has hindered the examination of these relationships. To address this problem, we used SCN-directed expression of the optogenetic constructs channelrhodopsin (ChR2) and halorhodopsin (NpHR) to drive or inhibit SCN neuron firing rate, respectively, both ex vivo and in vivo. Here we show that optogenetic induction or suppression of firing rate within SCN neurons is sufficient to reset the phase and alter the period of the molecular clockworks, that this resetting requires action potentials and VIPergic network communication, and that in vivo optogenetic stimulation of the SCN synchronizes behavioral rhythms. We therefore conclude that SCN firing rate is a key component in circadian rhythmicity and entrainment, rather than solely an output of the molecular clock.
To manipulate firing rate in the SCN, we generated mouse lines that expressed either ChR2 or NpHR under an SCN-directed Cre driver (dopamine receptor D1a; ‘Drd1a-ChR2’ or ‘Drd1a-NpHR’ mice) and confirmed transgene expression in the SCN using immunohistochemistry (Methods). Optogenetic constructs were highly expressed throughout the SCN in ~90% of AVP+ and VIP+ neurons, as well as a high proportion of AVP−/VIP− SCN neurons (Supplementary Figs. 1 and 2). We were able to increase or decrease SCN neuron firing rate in vitro for an hour or more with appropriate light input (8 Hz 470 nm for Drd1a-ChR2 and continuous 590 nm for Drd1a-NpHR SCN neurons, respectively). Optogenetic stimulation of ex vivo SCN slices from Drd1a-ChR2 mice at the typical daytime peak of spontaneous firing rate in SCN neurons (470 nm, 8 Hz, 1 hour) resulted in widespread cellular activation in the SCN, with ~90% of ChR2-expressing cells exhibiting gene activation as assayed by cFos immunohistochemistry (Supplementary Fig. 3).
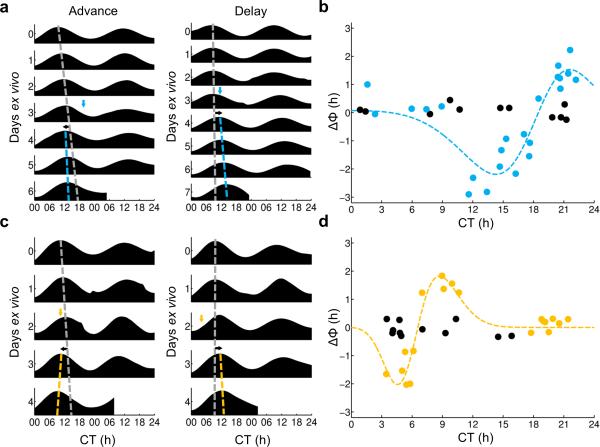
By crossing Drd1a-ChR2 or Drd1a-NpHR mice with a PER2::LUC reporter line in which the clock protein PERIOD2 (PER2) is fused to luciferase, we were able to assay the effect of optogenetic manipulation on the molecular clockworks. Ex vivo Drd1a-ChR2 × PER2::LUC SCN slices or Drd1a-NpHR × PER2::LUC SCN slices were optogenetically stimulated or inhibited, respectively, at varying times relative to the peak of the PER2::LUC rhythm (defined as circadian time 12 [CT 12]). In Drd1a-ChR2 × PER2::LUC SCN slices, optogenetic stimulation elicited delaying resets of the molecular clockworks from CT 12–18, advancing resets from CT 18–2, and no or minimal shifts from CT 2–12 (Figs. 1a,b). There were also small but significant changes in the period of the PER2 rhythm, with period lengthening resulting from delaying stimuli (CT 12–18), period shortening from advancing stimuli (CT 18–24), and no change in period from stimuli that did not result in phase shifts (CT 0–12; Supplementary Fig. 4). Identical optogenetic stimuli delivered to PER2::LUC SCN slices lacking ChR2 did not result in phase shifts or period changes. In Drd1a-NpHR × PER2::LUC SCN slices, optogenetic inhibition resulted in a different pattern of resets, with delay resets from CT 0–6, advances from CT 6–12, and no resets from CT 12–24 (Figs. 1c,d). As with ChR2 stimulation, NpHR inhibition resulted in changes in period, with period lengthening from delay-inducing treatments (CT 3–6), period shortening from advancing treatments (CT 9–12), and no change in period if shifts were not induced (CT 12–24; Supplementary Fig. 4). Identical optogenetic stimuli delivered to PER2::LUC SCN slices lacking NpHR did not result in phase shifts or period changes. In both Drd1a-ChR2 × PER2::LUC and Drd1a-NpHR × PER2::LUC SCN slices, effects on period and phase were found to persist for 5–6 days (n = 3 slices per genotype, data not shown).
Figure 1. Optogenetic manipulation of SCN neurons ex vivo produces changes in phase.
(a) Representative PER2::LUC actograms demonstrating the effects of ChR2-mediated stimulation of Drd1a-ChR2 organotypic slices resulting in advances (left) and delays (right). In these traces, the dashed gray line depicts the peak times of the undisturbed pre-stimulation rhythm extrapolated past the time of manipulation; the dashed blue line depicts the peak times of the post-stimulation rhythm; the blue arrow depicts the time of stimulation (470 nm, 1 hour, 8 Hz); and the black arrow depicts the direction of the resulting phase shift. (b) Phase response curve depicting changes of phase of PER2::LUC bioluminescence in Drd1a-ChR2 organotypic slices in response to ChR2-mediated stimulation at varying circadian times; dashed line, Gaussian fit, r2 = 0.8510; blue dots, ChR2+, n = 23 slices, 20 animals; black dots, ChR2−, n = 11 slices, 11 animals. (c) Representative PER2 LUC actograms demonstrating the effects of NpHR-mediated inhibition of Drd1a-NpHR organotypic slices resulting in advances (left) and delays (right). In these traces, the dashed gray line depicts the peak times of the extrapolated pre-treatment rhythm; the dashed yellow line depicts the peak times of the post-treatment rhythm; the yellow arrow depicts the time of stimulation (590 nm, 1 hour); and the black arrow depicts the direction of the resulting phase shift. (d) Phase response curve depicting changes of phase of PER2::LUC bioluminescence in Drd1a-NpHR organotypic slices in response to NpHR-mediated inhibition at varying circadian times; dashed line, Gaussian fit, r2 = 0.8895; yellow dots, NpHR+, n = 19 slices, 14 animals; black dots, NpHR−, n = 11 slices, 9 animals.
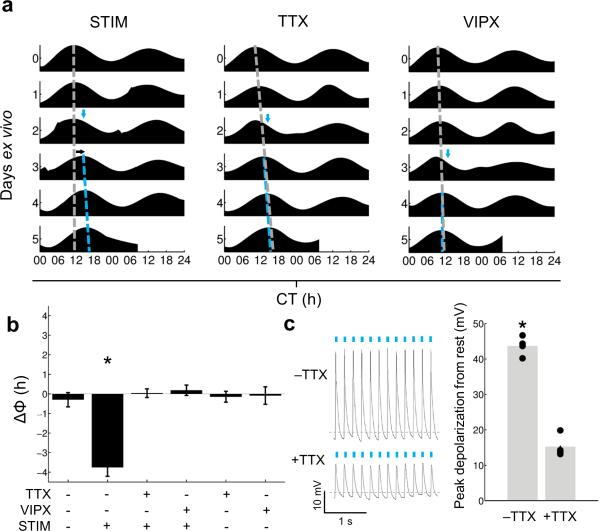
To investigate the roles of action potentials and intercellular communication in ChR2-mediated changes in phase and period of the molecular clockworks, we used ex vivo optogenetic stimulation of Drd1a-ChR2 × PER2::LUC SCN slices in the presence of the sodium channel blocker tetrodotoxin (TTX) or the VIP receptor blocker [D-p-Cl-Phe6,Leu17]-VIP (VIPX) . Stimulation during the delay zone (CT 12–18) in control media resulted in substantial phase delays and small but significant period lengthening, as before; however, these changes were inhibited when slices were stimulated in media containing TTX or VIPX (Figs. 2a,b; Supplementary Fig. 5). Importantly, transfer to TTX, VIPX, or control media had no effect on unstimulated Drd1a-ChR2 × PER2::LUC SCN slices (Fig. 2b; Supplementary Fig. 5). To determine the effects of TTX on ChR2-induced depolarization, we performed whole-cell current clamp recording on Drd1a-ChR2 neurons from acute SCN slices in response to ChR2 stimulation in the absence or presence of TTX (Fig. 2c). In the absence of TTX, the average peak depolarization amplitude in response to ChR2 stimulation was 43.71 ± 0.47 mV, which included the action potentials riding on top of the ChR2-induced depolarization. In the presence of TTX, however, depolarization by ChR2 stimulation persisted, but was severely reduced in amplitude to 15.25 ± 0.17 mV. In other studies, neural plasticity can be induced by ChR2 stimulation even in the presence of TTX13; however, our data suggest that in the SCN the subthreshold depolarization induced by ChR2 stimulation is insufficient to reset the circadian clock.
Figure 2. Pharmacological blockade of action potential generation or VIP signaling ablates phase changes induced by ChR2 stimulation.
(a) Representative PER2::LUC actograms of Drd1a-ChR2 organotypic slices undergoing a media change followed by ChR2 stimulation (left, ‘STIM’; n = 4 slices, 3 animals); a change to media containing 0.5 μM TTX followed by ChR2 stimulation (middle, ‘TTX’; n = 6 slices, 5 animals); or a change to media containing 1 μM [D-p-Cl-Phe6,Leu17]-VIP followed by ChR2 stimulation (right, ‘VIPX’; n = 5 slices, 4 animals); dashed gray line, peak times extrapolated past the time of manipulation in order to project the timing of the undisturbed rhythm; dashed blue line, peak times of post-manipulation rhythms; blue arrow, time of stimulation (470 nm, 1 hour, 8 Hz); black arrow, direction of resulting phase shift, if any. (b) Summary of changes in phase of PER2 LUC rhythms in response to combinations of ChR2 stimulation (470 nm, 1 hour, 8 Hz; ‘STIM’), TTX, and/or [D-p-Cl-Phe6,Leu17]-VIP (‘VIPX’); from left to right, n = 4 slices, 4 animals; 4 slices, 3 animals; 6 slices, 5 animals; 5 slices, 4 animals; 3 slices, 3 animals; and 3 slices, 3 animals; One-Way ANOVA with post-hoc HSD, F: 21.47, p<0.0001. (c) (Left) Representative voltage traces in response to ChR2 stimulation in the absence of or presence of TTX (−TTX or +TTX, respectively; dashed gray line in each trace indicates −50 mV); (Right) Peak depolarization amplitudes in response to ChR2 stimulation are significantly reduced in the presence of TTX; Mann-Whitney U, p = 0.0286; n = 4 cells. Data are presented as means ± SEM.
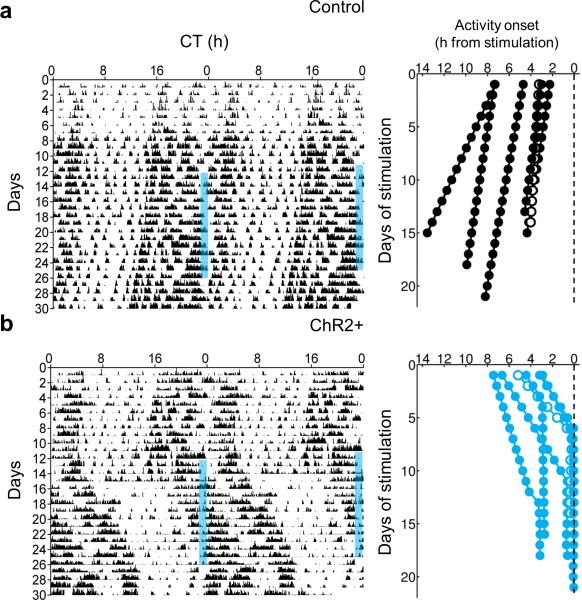
Finally, to investigate the effects of manipulating SCN neuron firing rate on circadian behavior, we optogenetically stimulated the SCN of Drd1a-ChR2 mice in vivo over multiple days at a frequency similar to that of the daytime firing rate of SCN neurons14. While ChR2 stimulation allows exact temporal control of firing rate phase locked to pulsed illumination, NpHR inhibition requires continuous illumination and does not allow for such precise control (Supplementary Fig. 2). Thus, we chose ChR2 excitation over NpHR inhibition to test the specific role of clock neuron firing rate in vivo. Importantly, SCN-targeted expression of ChR2 combined with a limited range of blue light penetration in the brain allowed for specific activation of the SCN (Supplementary Fig. 6). While control mice that lacked ChR2 expression showed no apparent response to repeated optogenetic stimulation of the SCN, Drd1a-ChR2 mice were entrained by the same stimulation, aligning the onset of their locomotor behavioral rhythms to the time of stimulation then free-running after cessation of the stimulus (Fig. 3). Control mice continued to free-run independently of the optogenetic stimulus, with their time of activity onset drifting away from the time of stimulation; conversely, Drd1a-ChR2 mice entrained to the stimulus, with activity onset progressively moving towards the time of stimulation until activity onset locked on to the time of stimulation or shortly after.
Figure 3. Repeated stimulation of ChR2 in the SCN in vivo results in entrainment.
(a) Left, representative double-plotted actogram of locomotor activity for a control ChR2– animal stimulated intracranially with blue light targeted to the SCN with a fiber optic cannula; blue bars represent time of stimulation (1 hour per day, 470 nm, 8 Hz); Right, activity onset time difference from time of stimulation (dashed line; n = 6 trials; black filled circles, n = 5 mice; black open circles, n = 1 mouse re-stimulated after free-running for multiple days). (b) Left, representative double-plotted actogram of locomotor activity for a Drd1a-ChR2 animal stimulated intracranially with blue light targeted to the SCN with a fiber optic cannula; blue bars represent time of stimulation (1 hour per day, 470 nm, 8 Hz); Right, activity onset time difference from time of stimulation (dashed line; n = 6 trials; blue filled circles, n = 5 mice; blue open circles, n = 1 mouse re-stimulated after free-running for multiple days).
The application of optogenetics to the SCN has allowed us to test the fundamental role of firing rate in influencing molecular and behavioral circadian rhythms. Artificial induction or suppression of firing rate across the SCN ex vivo has upstream effects on the phase and period of clock gene expression: the pattern of phase shifts elicited by ChR2 stimulation is essentially identical to that of light, which acts on the SCN through depolarizing glutamate release from retinal ganglion afferents15,16, while the pattern of phase shifts resulting from NpHR inhibition is similar to clock-resetting by dark pulses or other non-photic stimuli that are thought to act through inhibition of SCN neuron activity17,18. Induction of firing rate in vivo also has downstream effects on locomotor behavior consistent with its phase-shifting effects observed ex vivo, suggesting that increasing SCN firing rate per se is potentially behaviorally equivalent to light stimulation in its action on the circadian system. Additionally, our results show that pharmacological blockade of coupling or firing rate prevents phase shifts ex vivo, which suggests a stronger role for VIPergic network communication and for action potentials versus sub-threshold depolarizations in the resetting of the molecular clockworks. Together, these data indicate that manipulation of SCN firing rate is sufficient to produce lasting changes within the circadian clock. These results, and the stimulation system used to produce them, will serve as a foundation for future experiments in circadian neurobiology investigating the interaction between the molecular, electrical, and behavioral components of the brain's biological clock.
ONLINE METHODS
Animals
Drd1a-Cre (B6.FVB(Cg)-Tg(Drd1a-Cre)EY266Gsat/Mmucd, GENSAT; Supplementary Fig. 1) mice were crossed with Cre-dependent ChR2 (Ai27D, B6.Cg-Gt(ROSA)26Sortm27.1(CAG-COP4*H134R/tdTomato)Hze/J, Jackson Laboratories) or NpHR (Ai39, B6;129SGt(ROSA)26Sortm39(CAG-hop-/EYFP)Hze/J, Jackson Laboratories) mice to yield Drd1a-ChR2 or Drd1a-NpHR mice. A subset of Drd1a-ChR2 and Drd1a-NpHR mice were crossed with PER2::LUC (B6.129S6-Per2tm1Jt/J, Jackson Laboratories) mice to yield Drd1a-ChR2 × PER2::LUC mice or Drd1a-NpHR × PER2::LUC mice. All animals used (males and females; no obvious differences were observed between sexes) were on a C57BL/6J background, ~1–3 months of age, housed in a temperature- and humidity-controlled facility (~22°C, ~40% humidity) in single-sexed cages of no more than five animals from weaning until experimental use on a 12:12 light/dark (LD; light intensity ~300 lux) cycle, and were provided with food and water ad libitum. All animal care and experiments were conducted in concordance with Vanderbilt University's Institutional Animal Care and Use Committee guidelines.
Immunohistochemistry
Drd1a-ChR2 mice were administered a single injection of colchicine (2.5 mM; Sigma, St. Louis, MO) in the lateral ventricle. ~24 hours later, colchicine-injected mice were deeply anesthetized and transcardially perfused with 4% (w/v) paraformaldehyde (PFA; Sigma). Brains were removed and post-fixed with 4% PFA overnight, and cryoprotected in 20% sucrose in PBS. A cryostat was used to obtain 40 μm coronal slices containing the SCN. Slices were then labeled for AVP using rabbit polyclonal anti-vasopressin (1:5000, ab1565, Millipore, Billerica, MA) or VIP using rabbit polyclonal anti-VIP (1:2500, ab43841, Abcam, Cambridge, MA). For cFos experiments, membrane-attached organotypic SCN cultures were fixed for 1 hour in 4% PFA post-ChR2 stimulation and labeled for cFos using rabbit polyclonal anti-cFos (1:1000, ab7963, Abcam). For visualization, slices were incubated with Alexa Fluor 488 goat anti-rabbit IgG (1:500, Invitrogen). Slices were examined under a confocal microscope (LSM510; Zeiss; Thornwood, NY) at 488 nm for Alexa Fluor 488 and 543 nm for ChR2-tdTomato. Colocalization of Drd1a-ChR2 and AVP or VIP was determined manually using ImageJ and was defined as an AVP or VIP-positive (i.e., green fluorescent) neuron completely surrounded by red tdTomato membrane-bound fluorescence. ImageJ was also used to perform manual quantification of cFos-positive cell numbers.
Slice preparation and electrophysiological recording
Brains were removed and blocked in cold, oxygenated 95% O2–5% CO2 dissecting solution (in mM: 114.5 NaCl, 3.5 KCl, 1 NaH2PO4, 1.3 MgSO4, 2.5 CaCl2, 10 D-glucose, and 35.7 NaCHO3). SCN slices (200 μm) were cut on a vibroslicer (WPI, Sarasota, FL) at 4–10°C and transferred directly to an open recording chamber continually superfused with warmed (35 ± 0.5°C) extracellular solution (in mM: 124 NaCl, 3.5 KCl, 1 NaH2PO4, 1.3 MgSO4, 2.5 CaCl, 10 D-glucose, and 26 NaCHO3). Slices were allowed to recover for 1 hour before recording. SCN neurons were visualized using a Leica DMLFS microscope (Leica Microsystems, Buffalo Grove, IL) equipped with near-infrared (IR)-differential interference contrast and fluorescence optics. For cell-attached recordings, patch electrodes (4-6 MΩ) pulled from glass capillaries (WPI) on a multistage puller (DMZ; Zeitz, Martinsried, Germany) were filled with extracellular solution. For whole-cell current-clamp recordings, patch electrodes (8–10 MΩ) were filled with intracellular solution containing, in mM, 135 K-Gluconate, 10 KCl, 10 HEPES, 0.5 EGTA, and 2 mM MgCl2. Recordings were obtained with an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) and monitored online with pClamp 10.0 software (Molecular Devices). Slices were subjected to pulsed blue light (470 nm; 8 Hz; 10 ms duration) from a mounted high-power LED (Thorlabs, Newton, NJ) controlled by a Grass SD-9 stimulus generator (Grass Technologies, Warwick, RI) or continuous yellow light (590 nm) from a mounted high-power LED (Thorlabs). In some experiments, superfused extracellular solution was switched to extracellular solution containing 0.5 μM tetrodotoxin (TTX; Sigma) after 1–2 minutes of whole-cell current clamp recording.
Ex vivo culture and PER2::LUC imaging
Brains from mice killed without anesthesia by cervical dislocation were removed and blocked in cold HBSS supplemented with 100 U/ml penicillin/streptomycin, 10 mM HEPES, and 4.5 mM sodium bicarbonate. Hypothalamic coronal slices (200 μm) containing the SCN were cut on a vibroslicer (WPI) at 4–10°C, trimmed to ~1.5 × 1.5 mm squares, and transferred directly to culture membranes (Millipore) in vacuum grease-sealed 35 mm culture dishes with recording media containing 1.0 ml of DMEM (D-2902; Sigma) supplemented with 3.5 g/L D-glucose, 10 mM HEPES, 25 U/ml penicillin/streptomycin, 2% B27, and 0.1 mM beetle luciferin (Promega, Madison, WI). Slice cultures containing the SCN were maintained in an incubator at 36.8°C. Bioluminescence was monitored in real time with a LumiCycle (Actimetrics). After a minimum of two cycles of bioluminescence recording, slice cultures were removed from the LumiCycle and, while still in the incubator, placed under a custom-built high-power LED array consisting of blue Cree XP-E or yellow Luxeon Rebel LEDs (LEDSupply, Randolph, VT) soldered to an 0.25” aluminum sheet heatsink and driven at 1000 mA using a LEDD1B high-powered LED driver (Thorlabs) and subjected to either pulsed blue light (470 nm; 8 Hz; 10 ms duration) controlled by a Grass SD-9 stimulus generator (Grass Technologies, Warwick, RI) or continuous yellow light (590 nm) for 1 hour. Light intensity inside the 35 mm culture dish was determined to be 20.1 mW ± 0.5 mW (470 nm LEDs) or 16.27 mW ± 0.3 mW (590 nm LEDs) when driven at 1000 mA using a PM100D Optical Power Meter (Thorlabs). Control unstimulated slices were instead removed from the LumiCycle and kept in the incubator for an equivalent time without stimulation. Slice temperature before and after stimulation was measured with an infrared thermometer (Fluke, Norwich, UK) and was determined to not change (± 0.1°C) after stimulation (data not shown). Slice cultures were then returned to the LumiCycle and bioluminescence was recorded for at least two additional cycles; data were excluded if bioluminescence did not persist for at least five total cycles.
Pharmacological manipulation of PER2::LUC cultures
For some experiments, slices were cultured and recorded as above, but when they were removed from the incubator, slice cultures were either transferred to pre-warmed fresh recording media, fresh recording media containing 0.5 μM TTX, or fresh recording media containing 1 μM [D-p-Cl-Phe6,Leu17]-VIP (Tocris cat. no. 3045; Bristol, UK). TTX concentration was selected from Brancaccio et al. 2013 and Kudo et al. 20133,4; [D-p-Cl-Phe6,Leu17]-VIP concentration was based on values from Evans et al. 2013 and Atkins et al. 201019,20. The original recording media was sealed in its culture dish and kept warm inside the incubator. Slice cultures were then kept in the incubator without stimulation for 1 hour, or stimulated as above; after which all slice cultures were rinsed, transferred to their original recording media, resealed with vacuum grease, and returned to the LumiCycle for further bioluminescence recording.
In vivo optogenetic stimulation and locomotor activity monitoring
Anesthetized mice (100 mg/kg ketamine, 10 mg/kg xylazine) were placed into a stereotaxic device and implanted with a fiber optic cannula (5 mm in length, 400 μm diameter core, 0.39 NA; Thorlabs) sheathed to prevent light leak. The cannula was targeted to the SCN using the coordinates of +0.0 mm anterior and +0.0 mm lateral to bregma. After at least three days of recovery, mice were placed individually into litter-filled cages equipped with an IR motion detector (Spy2, Visonic, Tel Aviv, Israel) inside light-tight boxes, and food and water was provided ad libitum. Animals were chronically tethered to a fiber optic cable (400 μm diameter core, 0.39 NA; Thorlabs) attached to the implanted cannula and connected to a high-powered blue (470 nm) LED (Thorlabs) under the control of a LED driver (DC4100; Thorlabs). Mice were kept in constant darkness (DD) and allowed to free-run for at least four days before stimulation. Locomotor activity was monitored in 5 minute bins using ClockLab software (Actimetrics, Evanston, IL). Light pulses (470 nm; 8 Hz; 10 ms duration; 1 hour) were generated at various circadian times (CTs), where CT 12 is defined as the start of locomotor activity, by a Grass SD-9 stimulus generator attached to the LED driver under the control of a light timer, and repeated daily at the same clock time. Light intensity at the cannula tip was determined to be 10.9 mW ± 0.1 mW when driven at 1000 mA using a PM100D Optical Power Meter (Thorlabs). Calculated irradiances21, along with the estimated ChR2 activation threshold22 are shown in Supplementary Fig. 3. Animals were excluded from analysis if they did not successfully free-run in DD during the four days after surgery but before stimulation.
PER2::LUC data analysis
Sample sizes were chosen so as to be sufficient for statistical analysis based upon previous publications detailing similar measurements both in vivo and in vitro23,24,25. Data analysis was performed blind to genotype, while there were no methods to randomize mice to experimental groups or to blind investigators to genotype for the duration of the experiment. Baseline subtracted bioluminescence data were obtained using a 24 hour running average from the raw data using LumiCycle data analysis software (Actimetrics). Subtracted bioluminescence data were then loaded into Matlab (Mathworks, Natick, MA) for further analysis. Data were smoothed using a Loess local regression filter and a damped sine wave was fit from the beginning of the smoothed data until the time of manipulation. The damped sine wave was then extrapolated past the time of manipulation in order to project the timing of the undisturbed rhythm. Phase shifts were determined by calculating the mean difference in the post-manipulation peaks and troughs of the actual rhythms and the extrapolated unstimulated rhythms on the first and second cycles after stimulation. Period changes were calculated in ClockLab analysis software (Actimetrics) by measuring the best fit line through the calculated acrophases before and after stimulation. For data visualization, smoothed baseline-subtracted bioluminescence rhythms were depicted as double-plotted actograms created using Matlab. CT 12 was defined as the peak of PER2::LUC bioluminescence.
In vivo data analysis
Sample sizes were chosen so as to be sufficient for statistical analysis based upon previous publications detailing similar measurements both in vivo and in vitro23,24,25. Data analysis was performed blind to genotype, while there were no methods to randomize mice to experimental groups or to blind investigators to genotype for the duration of the experiment. A best fit line was drawn between the times of activity onset until the time at which the change in time of activity onset was less than ± 0.1 hours a day (i.e., when the animal “locked on” to the stimulus) or until cessation of the stimulus (such as in the case of control actograms). If the animal “locked on” to the stimulus as defined above, a second best fit line was drawn between these times of activity onset. The clock time of the best fit line(s) was then subtracted from the clock time of the daily optogenetic stimuli and the absolute value (i.e., activity onset in hours from time of stimulation) was plotted against days of stimulation for both ChR2+ and control animals.
Statistical analysis
All statistical analyses (Student's t-test, One-Way ANOVA, Tukey's HSD, Mann-Whitney U) were performed in Matlab, with α defined as 0.05. A Bartlett's multiple-sample test and a Kolmogorov-Smirnov test were used to confirm equal variance and normality. Curves were fit using Matlab's Curve Fitting Toolbox (Mathworks). Data are presented as means ± SEM.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Iwamoto for assistance with the whole-cell recording experiments and T. Page, C. Johnson, and L. Funkhouser-Jones for helpful discussion and manuscript comments. This work was supported by US National Institutes of Health grants R01 EY015815 (D.G.M.), T32 MH064931 (J.R.J.) and F31 NS082213 (J.R.J.), NSF Graduate Research Fellowship 0909667 (M.C.T.), and a Vanderbilt University Discovery Grant (D.G.M.).
Footnotes
Contributions
J.R.J., M.C.T., and D.G.M. prepared the paper. J.R.J. analyzed data. J.R.J. performed all in vitro and ex vivo experiments. J.R.J. and M.C.T. performed all in vivo experiments. J.R.J., M.C.T., and D.G.M. designed experiments.
Competing financial interests
The authors declare no competing financial interests.
A Supplementary Methods Checklist is available.
REFERENCES
- 1.Colwell CS. Nat. Rev. Neurosci. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhlman SJ, McMahon DG. J. Biol. Rhythms. 2006;21:470–481. doi: 10.1177/0748730406294316. [DOI] [PubMed] [Google Scholar]
- 3.Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. Neuron. 2013;78:714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kudo T, et al. J. Neurophysiol. 2013;110:1097–1106. doi: 10.1152/jn.00114.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman GM, Krock RM, Aton SJ, Thaben P, Herzog ED. Neuron. 2013;78:799–806. doi: 10.1016/j.neuron.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh DK, Takahashi JS, Kay SA. Annu. Rev. Phsyiol. 2009;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herzog ED, Takahashi JS, Block GD. Nat. Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 8.Kuhlman SJ, Silver R, Le Sauter J, Bult-Ito A, McMahon DG. J. Neurosci. 2003;23:1441–1450. doi: 10.1523/JNEUROSCI.23-04-01441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitabach MN, Blau J, Holmes TC. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi S, et al. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 11.Welsh DK, Logothetis DE, Meister M, Reppert SM. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz WJ, Gross RA, Morton MT. Proc. Natl. Acad. Sci. USA. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goold CP, Nicoll RA. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai K. Neuroscience. 2014;260:249–64. doi: 10.1016/j.neuroscience.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 15.Ding JM, et al. Science. 1994;266:1713–1717. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 16.Daan S, Pittendrigh CS. J. Comp. Physiol. 1976;106:253–266. [Google Scholar]
- 17.Quintero JE, McMahon DG. J. Neurophysiol. 1999;82:533–539. doi: 10.1152/jn.1999.82.2.533. [DOI] [PubMed] [Google Scholar]
- 18.Morin LP, Allen CN. Brain Res. Rev. 2006;51:1–60. doi: 10.1016/j.brainresrev.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkins N, et al. PLoS ONE. 2010;5:e12612. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Aravanis AM, et al. J. Neural Eng. 2007;4:S143–156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 23.Gamble KL, Allen GC, Zhou T, McMahon DG. J. Neurosci. 2007;27:12078–12087. doi: 10.1523/JNEUROSCI.1109-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruan GX, Allen GC, Yamazaki S, McMahon DG. PLoS Biol. 2008;6:2248–2265. doi: 10.1371/journal.pbio.0060249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan GX, Gamble KL, Risner ML, Young LA, McMahon DG. PLoS ONE. 2012;7:e38985. doi: 10.1371/journal.pone.0038985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.