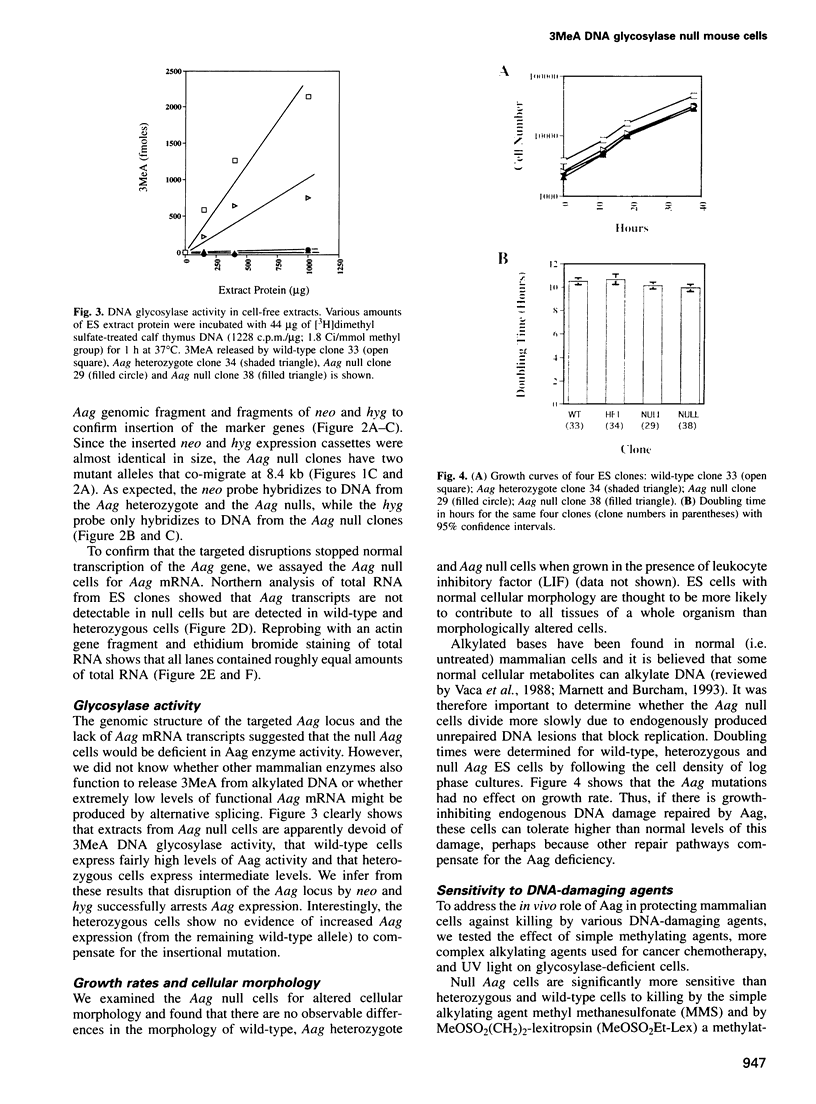
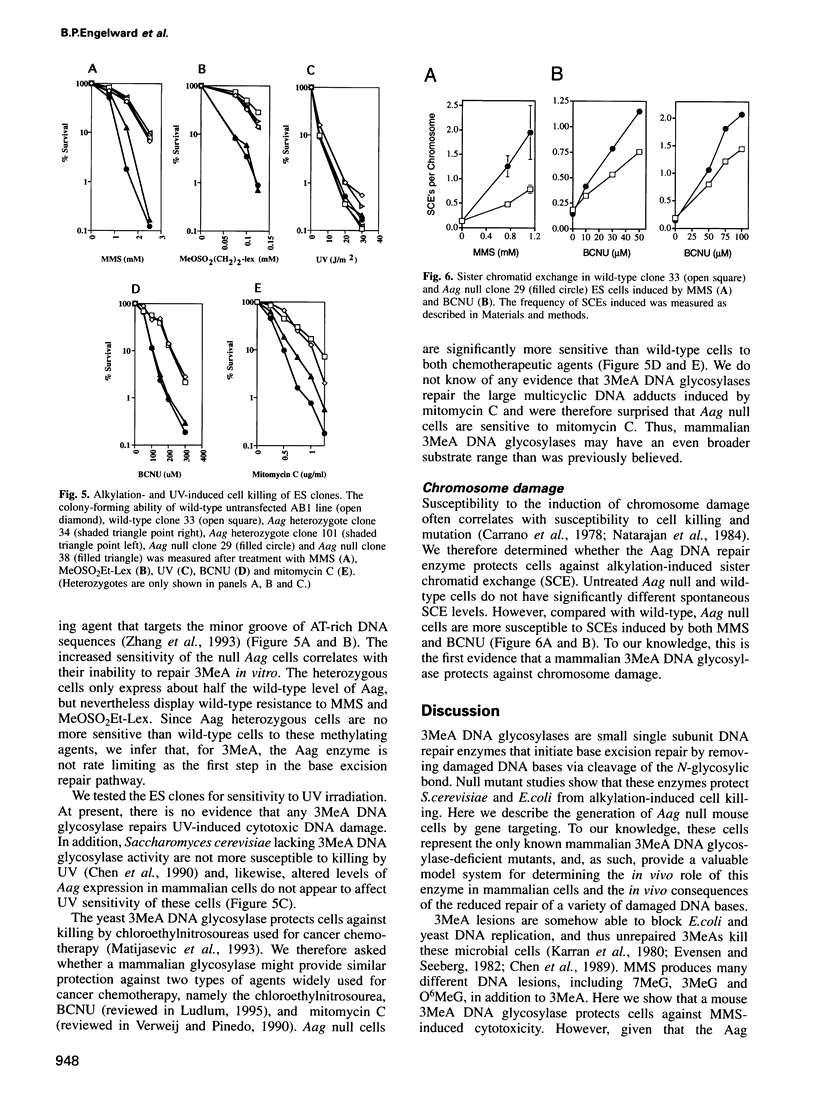
Abstract
In Escherichia coli, the repair of 3-methyladenine (3MeA) DNA lesions prevents alkylation-induced cell death because unrepaired 3MeA blocks DNA replication. Whether this lesion is cytotoxic to mammalian cells has been difficult to establish in the absence of 3MeA repair-deficient cell lines. We previously isolated and characterized a mouse 3MeA DNA glycosylase cDNA (Aag) that provides resistance to killing by alkylating agents in E. coli. To determine the in vivo role of Aag, we cloned a large fragment of the Aag gene and used it to create Aag-deficient mouse cells by targeted homologous recombination. Aag null cells have no detectable Aag transcripts or 3MeA DNA glycosylase activity. The loss of Aag renders cells significantly more sensitive to methyl methanesulfonate-induced chromosome damage, and to cell killing induced by two methylating agents, one of which produces almost exclusively 3MeAs. Aag null embryonic stem cells become sensitive to two cancer chemotherapeutic alkylating agents, namely 1,3-bis(2-chloroethyl)-1-nitrosourea and mitomycin C, indicating that Aag status is an important determinant of cellular resistance to these agents. We conclude that this mammalian 3MeA DNA glycosylase plays a pivotal role in preventing alkylation-induced chromosome damage and cytotoxicity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basu A. K., Hanrahan C. J., Malia S. A., Kumar S., Bizanek R., Tomasz M. Effect of site-specifically located mitomycin C-DNA monoadducts on in vitro DNA synthesis by DNA polymerases. Biochemistry. 1993 May 11;32(18):4708–4718. doi: 10.1021/bi00069a004. [DOI] [PubMed] [Google Scholar]
- Berdal K. G., Bjørås M., Bjelland S., Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T., Roy R., Yamamoto K., Kasai H., Nishimura S., Tano K., Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S., Birkeland N. K., Benneche T., Volden G., Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem. 1994 Dec 2;269(48):30489–30495. [PubMed] [Google Scholar]
- Bjelland S., Bjørås M., Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993 May 11;21(9):2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodell W. J., Tokuda K., Ludlum D. B. Differences in DNA alkylation products formed in sensitive and resistant human glioma cells treated with N-(2-chloroethyl)-N-nitrosourea. Cancer Res. 1988 Aug 15;48(16):4489–4492. [PubMed] [Google Scholar]
- Boiteux S., Huisman O., Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano A. V., Thompson L. H., Lindl P. A., Minkler J. L. Sister chromatid exchange as an indicator of mutagenesis. Nature. 1978 Feb 9;271(5645):551–553. doi: 10.1038/271551a0. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ibeanu G. C., Tano K., Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991 Aug 25;266(24):15710–15715. [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Dosanjh M. K., Chenna A., Kim E., Fraenkel-Conrat H., Samson L., Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh M. K., Roy R., Mitra S., Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase). Biochemistry. 1994 Feb 22;33(7):1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- Engelward B. P., Boosalis M. S., Chen B. J., Deng Z., Siciliano M. J., Samson L. D. Cloning and characterization of a mouse 3-methyladenine/7-methyl-guanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis. 1993 Feb;14(2):175–181. doi: 10.1093/carcin/14.2.175. [DOI] [PubMed] [Google Scholar]
- Erickson L. C., Laurent G., Sharkey N. A., Kohn K. W. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumour cells. Nature. 1980 Dec 25;288(5792):727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Fishel R., Lescoe M. K., Rao M. R., Copeland N. G., Jenkins N. A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993 Dec 3;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Gonzaga P. E., Brent T. P. Affinity purification and characterization of human O6-alkylguanine-DNA alkyltransferase complexed with BCNU-treated, synthetic oligonucleotide. Nucleic Acids Res. 1989 Aug 25;17(16):6581–6590. doi: 10.1093/nar/17.16.6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y., Carter C. A., Kirk M. C., Ludlum D. B. Release of 7-alkylguanines from N-(2-chloroethyl)-N'-cyclohexyl-N-nitrosourea-modified DNA by 3-methyladenine DNA glycosylase II. Cancer Res. 1991 Jan 15;51(2):499–503. [PubMed] [Google Scholar]
- Habraken Y., Carter C. A., Sekiguchi M., Ludlum D. B. Release of N2,3-ethanoguanine from haloethylnitrosourea-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Carcinogenesis. 1991 Oct;12(10):1971–1973. doi: 10.1093/carcin/12.10.1971. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Ludlum D. B. Release of chloroethyl ethyl sulfide-modified DNA bases by bacterial 3-methyladenine-DNA glycosylases I and II. Carcinogenesis. 1989 Mar;10(3):489–492. doi: 10.1093/carcin/10.3.489. [DOI] [PubMed] [Google Scholar]
- IYER V. N., SZYBALSKI W. MITOMYCINS AND PORFIROMYCIN: CHEMICAL MECHANISM OF ACTIVATION AND CROSS-LINKING OF DNA. Science. 1964 Jul 3;145(3627):55–58. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Klungland A., Fairbairn L., Watson A. J., Margison G. P., Seeberg E. Expression of the E.coli 3-methyladenine DNA glycosylase I gene in mammalian cells reduces the toxic and mutagenic effects of methylating agents. EMBO J. 1992 Dec;11(12):4439–4444. doi: 10.1002/j.1460-2075.1992.tb05544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P. W., Zijderveld A., Linders K., Rudnicki M. A., Jaenisch R., Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991 Aug 11;19(15):4293–4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Leach F. S., Nicolaides N. C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomäki P., Sistonen P., Aaltonen L. A., Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993 Dec 17;75(6):1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Thomas K. R., Capecchi M. R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988 Nov 24;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Marnett L. J., Burcham P. C. Endogenous DNA adducts: potential and paradox. Chem Res Toxicol. 1993 Nov-Dec;6(6):771–785. doi: 10.1021/tx00036a005. [DOI] [PubMed] [Google Scholar]
- Matijasevic Z., Boosalis M., Mackay W., Samson L., Ludlum D. B. Protection against chloroethylnitrosourea cytotoxicity by eukaryotic 3-methyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11855–11859. doi: 10.1073/pnas.90.24.11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijasevic Z., Sekiguchi M., Ludlum D. B. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Karran P., Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P., Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990 Sep 21;62(6):1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Mereau A., Grey L., Piquet-Pellorce C., Heath J. K. Characterization of a binding protein for leukemia inhibitory factor localized in extracellular matrix. J Cell Biol. 1993 Aug;122(3):713–719. doi: 10.1083/jcb.122.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A. T., Simons J. W., Vogel E. W., van Zeeland A. A. Relationship between cell killing, chromosomal aberrations, sister-chromatid exchanges and point mutations induced by monofunctional alkylating agents in Chinese hamster cells. A correlation with different ethylation products in DNA. Mutat Res. 1984 Aug;128(1):31–40. doi: 10.1016/0027-5107(84)90044-7. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990 Oct;9(10):3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. R., Laval J. Human cDNA expressing a functional DNA glycosylase excising 3-methyladenine and 7-methylguanine. Biochem Biophys Res Commun. 1991 May 15;176(3):1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N., Nicolaides N. C., Wei Y. F., Ruben S. M., Carter K. C., Rosen C. A., Haseltine W. A., Fleischmann R. D., Fraser C. M., Adams M. D. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994 Mar 18;263(5153):1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Perry P., Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974 Sep 13;251(5471):156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- Popp R. A., Stratton L. P., Hawley D. K., Effron K. Hemoglobin of mice with radiation-induced mutations at the hemoglobin loci. J Mol Biol. 1979 Jan 15;127(2):141–148. doi: 10.1016/0022-2836(79)90235-3. [DOI] [PubMed] [Google Scholar]
- Ramírez-Solis R., Davis A. C., Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- Roy R., Brooks C., Mitra S. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochemistry. 1994 Dec 20;33(50):15131–15140. doi: 10.1021/bi00254a024. [DOI] [PubMed] [Google Scholar]
- Samson L., Derfler B., Boosalis M., Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson L., Linn S. DNA alkylation repair and the induction of cell death and sister chromatid exchange in human cells. Carcinogenesis. 1987 Feb;8(2):227–230. doi: 10.1093/carcin/8.2.227. [DOI] [PubMed] [Google Scholar]
- Santerre A., Britt A. B. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2240–2244. doi: 10.1073/pnas.91.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M., Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B., Antoccia A., Basu A. K., Dosanjh M. K., Fraenkel-Conrat H., Gallagher P. E., Kuśmierek J. T., Qiu Z. H., Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow E. T., Foote R. S., Mitra S. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984 Jul 10;259(13):8095–8100. [PubMed] [Google Scholar]
- Thomas L., Yang C. H., Goldthwait D. A. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982 Mar 16;21(6):1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- Tybulewicz V. L., Crawford C. E., Jackson P. K., Bronson R. T., Mulligan R. C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991 Jun 28;65(7):1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Vaca C. E., Wilhelm J., Harms-Ringdahl M. Interaction of lipid peroxidation products with DNA. A review. Mutat Res. 1988 Mar;195(2):137–149. doi: 10.1016/0165-1110(88)90022-x. [DOI] [PubMed] [Google Scholar]
- Vickers M. A., Vyas P., Harris P. C., Simmons D. L., Higgs D. R. Structure of the human 3-methyladenine DNA glycosylase gene and localization close to the 16p telomere. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3437–3441. doi: 10.1073/pnas.90.8.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. N., Chan C. L., Eastman A., Bresnick E. Expression of human O6-methylguanine-DNA methyltransferase in a DNA excision repair-deficient Chinese hamster ovary cell line and its response to certain alkylating agents. Cancer Res. 1992 Jan 1;52(1):32–35. [PubMed] [Google Scholar]
- Zhang Y., Chen F. X., Mehta P., Gold B. Groove- and sequence-selective alkylation of DNA by sulfonate esters tethered to lexitropsins. Biochemistry. 1993 Aug 10;32(31):7954–7965. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- te Riele H., Maandag E. R., Clarke A., Hooper M., Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990 Dec 13;348(6302):649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]






