Abstract
The thrombospondins (TSPs) are a family of matricellular proteins that regulate cellular phenotype through interactions with a myriad of other proteins and proteoglycans. We have identified a novel interaction of the members of the TSP gene family with stromal interaction molecule 1 (STIM1). This association is robust since it is preserved in Triton X-100, can be detected with multiple anti-TSP-1 and anti-STIM1 antibodies, and is detected in a wide range of cell types. We have also found that STIM1 co-immunoprecipitates with TSP-4 and cartilage oligomeric matrix protein (COMP), and that a recombinant version of the N-terminal domain of STIM1 binds to the signature domain of TSP-1 and COMP. The association of the TSPs with STIM1 is observed in both the presence and absence of calcium indicating that the calcium-dependent conformation of the signature domain of TSPs is not required for binding. Thus, this interaction could occur in the ER under conditions of normal or low calcium concentration. Furthermore, we observed that the expression of COMP in HEK 293 cells decreases STIM1-mediated calcium release activated calcium (CRAC) channel currents and increases arachidonic acid calcium (ARC) channel currents. These data indicate that the TSPs regulate STIM1 function and participate in the reciprocal regulation of two channels that mediate calcium entry into the cell.
Keywords: Thrombospondin, Stromal interaction molecule 1, Calcium signaling, Arachidonate-regulated calcium channel
1. Introduction
The thrombospondins (TSPs) are a family of five matricellular proteins that function during a wide range of physiological and pathological processes, including development, inflammation, angiogenesis and neoplasia (Adams and Lawler, 2011). The TSPs are transiently associated with the cell surface where they interact with a variety of membrane proteins, including proteoglycans, integrins, CD36, and CD47 (Adams and Lawler, 2011). Through these varied interactions, TSPs regulate extracellular matrix structure and cellular phenotype during tissue development and remodeling. For example, TSP-1 increases the association of CD36 with vascular endothelial growth factor receptor-2 (VEGFR-2) while decreasing the association of CD47 with VEGFR-2 in endothelial cells (Kaur et al., 2010; Kazerounian et al., 2011). As a result, TSP-1 orchestrates fundamental changes in the way that endothelial cells respond to VEGF (Kaur et al., 2010; Kazerounian et al., 2011; Chu et al., 2013).
Each subunit of the TSP-1 trimer consists of multiple domains: amino- and carboxyl-terminal globular domains, a region of sequence homology to procollagen (PHR), and three types of repeated sequence motifs, designated type 1, type 2, and type 3 repeats (Lawler and Hynes, 1986). Since the type 1 repeats were first identified in TSP-1 as a distinct structural motif, they have been designated thrombospondin repeats or TSRs (Lawler and Hynes, 1986; Tucker, 2004). The five members of the thrombospondin gene family can be divided into two subgroups, based on their structures (Bornstein et al., 1991; Oldberg et al., 1992; Vos et al., 1992; Lawler et al., 1993a,b; Efimov et al., 1994; Newton et al., 1994). TSP-1 and -2 (subgroup A) have the complete set of structural domains described above and are trimeric. By contrast, the subgroup B TSPs, TSP-3, and -4, and cartilage oligomeric matrix protein (COMP), lack both the TSRs and the PHR but contain an additional type 2 repeat (Oldberg et al., 1992; Vos et al., 1992; Lawler et al., 1993b). The subgroup B proteins are also different from the subgroup A members in that they form pentamers instead of trimers (Vos et al., 1992; Efimov et al., 1994). The type 2 repeats, the type 3 repeats and the carboxyl-terminal domains have the highest level of conservation amongst the TSPs and are collectively known as the signature domain. The structure of all or part of the signature domains of TSP-1 and -2, and COMP have been determined by X-ray crystallography revealing that the C-terminal domain forms a β-sandwich and that the type 3 repeats and portions of the type 2 repeats are closely associated with the surfaces of the β-sandwich (Kvansakul et al., 2004; Carlson et al., 2005; Tan et al., 2009). Binding sites for about 30 calcium ions are included in this structure. These sites are primarily located in the type 3 repeats which fold to form a contiguous series of calcium-binding sites, but calcium-binding sites are also present in the type 2 repeats and the C-terminal β-sandwich. The ability of the signature domain of the TSPs to bind numerous calcium ions suggests that the TSPs may play a role in calcium homeostasis. We hypothesize that the direct interaction of the signature domain with calcium channel(s) or other calcium-binding proteins might facilitate such functionality.
In this study, we reveal the association of TSP-1, TSP-4 and COMP with stromal interaction molecule 1 (STIM1). STIM1 is a transmembrane protein that functions in the endoplasmic reticulum (ER) to detect calcium depletion (Putney, 2011; Shuttleworth, 2012; Soboloff et al., 2012). A calcium-binding site in the N-terminal domain of STIM1 detects that the level of calcium in the ER is low and orchestrates the opening of store-operated calcium release activated calcium (CRAC) channels so that ER calcium can be replenished (Putney, 2011; Soboloff et al., 2012). Whereas most of the STIM1 is located in the ER, lower levels (~15–25%) are present on the plasma membrane where STIM1 functions to promote the opening of arachidonate-regulated calcium (ARC) channels, which are independent of store depletion (Putney, 2011; Shuttleworth, 2012; Soboloff et al., 2012). Here, we establish that over-expression of COMP promotes ARC channel currents while concurrently decreasing CRAC channel currents.
2. Materials and methods
2.1. Antibodies
The mouse monoclonal anti-STIM1 antibody clone 44 was obtained from BD Biosciences (Lexington, KY) and 5A2 was purchased from Novus Biologicals (Littleton, CO). The rabbit polyclonal anti-COMP was purchased from Kamiya Biomedical Company (Seattle WA). Preparation of the rabbit polyclonal anti-TSP-1 antibody R1 was described previously (Saumet et al., 2005), as were the mouse anti-TSP-1 monoclonal antibodies MA-I and MA-IV (Lawler et al., 1985). Preparation of the rabbit polyclonal anti-TSP-1 antibody R3 was described previously (Incardona et al., 1996). Rabbit polyclonal antibodies to peptides within the sequences of TSP-4 (Lawler et al., 1995) and COMP (Hecht et al., 1998) were also prepared by the Lawler lab and designated 1259 and F8, respectively.
2.2. Cell culture and platelet preparation
HEK 293 cells were grown in Dulbecco’s Modification of Eagle’s Medium plus 10% fetal bovine serum and stably transfected with COMP pcDNA3.1(+) (Invitrogen) following protocols supplied with the transfection reagent Fugene (Roche). Individual clones were isolated and analyzed for expression levels by SDS-PAGE and western blotting. Culture conditions for transient transfects with Flp-In™-HEK 293 cells (Invitrogen) were as previously described (Thompson and Shuttleworth, 2012). Where indicated, cells were transfected using an Amaxa Nucleofector II following the manufacturer’s guidelines using 0.5 μg COMP pcDNA3.1 either with or without 0.5 μg Orai1 M070 as appropriate, along with 0.2 μg EGFP to identify transfected cells. Whole-cell currents were recorded 40–48 h after transfection, and 18–24 h after plating the cells on poly-l-lysine-coated coverslips.
Jurkat cells were cultured in RPMI-1640 plus 10% fetal bovine serum. MDA-MB-231 cells were maintained in DMEM/F12 medium plus 10% fetal bovine serum. Human dermal microvascular endothelial cells were cultured in Vitrogen-coated dishes and maintained in EBM-2 media (Clonetics Corp., San Diego, CA) containing 20% fetal bovine serum (FBS), 1 μg/ml hydrocortisone acetate, 5 × 105 dibutyryl-cAMP, 200 units/ml penicillin, 100 units/ml streptomycin and 250 μg/ml amphotericin. All cell types were incubated at 37 °C with 5% CO2. Human chondrocytes were kindly provided by Drs. K. Posey and J. Hecht (University of Texas). Platelet rich plasma (Brigham & Women’s Hospital) was centrifuged at 2500 rpm for 20 min and the collected platelets were washed twice in 10 volumes of pH 6.5 buffer that contained 0.102 M NaCl, 3.9 mM K2HPO4, 3.9 mM Na2HPO4, 22 mM NaH2PO4, and 5 mM glucose prior to lyses.
2.3. Immunoprecipitation and western blotting
Adherent HEK 293, endothelial cells and MDA-MB-231 cells were briefly washed with PBS and incubated in 0.25% trypsin to detach the cells. Jurkat cells and platelets were collected by centrifugation and washed with PBS. The cells were treated with lysis buffer at pH 7.4 containing 20 mM HEPES, 150 mM NaCl, 5 mM EDTA, 1% Brij 99 or Triton X-100, and protease inhibitors (HALTS, Pierce). After 30 min at 4 °C, insoluble material was removed by centrifugation at 16,000 ×g (15 min, 4 °C) and was either used immediately for immunoprecipitation experiments or stored at −80 °C. To preclear the samples, 1 ml of cell lysate (400–1000 μg of protein), 5 μg of non-immune IgG and 20 μl (pellet volume) of Protein A or G Sepharose beads (Pharmacia Biotech) were mixed in a microcentrifuge tube for 1 h at 4 °C. After removal of the Sepharose beads by centrifugation, 5 μg of antibody (R1, MA-IV, or STIM1) and 20 μl (pellet volume) of Protein A or G beads were added and the samples were incubated for 2–3 h at 4 °C with gentle rocking. The beads were washed 4 times with lysis buffer, and the precipitated immunocomplexes were eluted in 40 μl of 2× SDS-PAGE loading buffer, boiling for 4 min. The eluted samples were separated by SDS-PAGE either in the presence or in the absence of 1% dithiothreitol and western blotting was performed. In some experiments, 30–40 μl of cell lysate was also blotted.
To determine if TSP-1 associates with STIM1 in the plasma membrane, MDA-MB-231 cells were incubated with the anti-TSP-1 polyclonal antibody R1 (~2 μg/ml) for 1 h at 4 °C. This step allowed anti-TSP-1 antibody to bind only to TSP-1 that is expressed at the plasma membrane. The cells were washed in cold PBS three times and then disrupted in Triton X-100 lysis buffer. The cell lysates were spun down at 14,000 rpm for 15 min and were then incubated with Protein A Sepharose beads for 2–3 h on a rocking platform at 4 °C. Beads were washed 3× using lysis buffer and boiled with SDS sample buffer and the eluted proteins were resolved on a reducing SDS-PAGE. The samples were western blotted for TSP-1 and STIM1.
2.4. Mass spectroscopy analysis
Human platelets (5 × 109 cells/10 ml) were washed with cold PBS and lysed in buffer containing 20 mM HEPES pH 7.40, 150 mM NaCl, 5 mM EDTA, 1% Brij 99, and protease inhibitors (HALTS, Pierce). After 30 min at 4 °C, insoluble material was removed by centrifugation at 16,000 ×g (15 min, 4 °C). The platelet lysates were pre-cleared by adding 20 μg of non-immune mouse IgG (Sigma) and 200 μl of Protein G-Sepharose (Amersham Pharmacia Biotech) and rocking gently at 4 °C for 60 min. Immunoprecipitation was performed by combining 20 μg of the anti-TSP-1 mouse monoclonal MA-IV and 200 μl of Protein G-Sepharose. The samples were incubated for 16 h at 4 °C with gentle rocking. Immune complexes were collected by centrifugation, washed four times in lysis buffer, and separated by SDS-PAGE in the presence of a reducing agent. Coomassie Blue stained bands were subjected to in-gel reduction, carboxyamidomethylation and tryptic digestion (Promega). Multiple peptide sequences were determined in a single run by microcapillary reverse-phase chromatography which was directly coupled to a Finnigan LCQ quadrupole ion trap mass spectrometer equipped with a custom nano-electrospray source. The Harvard Microchemistry Facility completed this analysis on a fee-for-service basis (Miao et al., 2001b).
2.5. Preparation of recombinant N-terminal domain of STIM1
A recombinant version of the N-terminal domain of STIM1 (amino acids 1–184 of human STIM1) was prepared by PCR using a template of RNA isolated from MDA-MB-231 breast cancer cells. STIM1 was prepared using the forward primer 873hSTIM1f (GAT GAT CCC GGG CTC AGC CAT AGT CAC AGT GAG AAG) and the reverse primer 874hSTIM1r (GAT ACC GGT AGT CAA GAG AGG AGG CCC AAA GAG). The PCR product was then sequenced and cloned between the XmaI and the AgeI sites of the vector pMT/BiP V5-HisA (Invitrogen, Carlsbad, CA). The recombinant protein included the vector-derived sequence RSPWPG at the N-terminal and the sequence TGHHHHHH at the C-terminal. Vector transfection, cell selection, and protein expression and purification were performed as described previously (Miao et al., 2001a).
Calcium-dependent conformational changes in the recombinant N-terminal domain of STIM1 were detected using limited tryptic or chymotryptic digestion at an enzyme-to-substrate ratio of 1:500 (w/w). The calcium concentration was adjusted by the addition of varying concentrations of EGTA and digestion was performed at 0 °C for 20 h. Digestion was stopped by adding SDS sample buffer and boiling, prior to SDS-PAGE. Proteolytic fragments were visualized by staining with Coomassie Blue.
2.6. Solid-phase binding assay
The solid-phase binding assay was performed in FluoroNunc Maxisorb 96 well plates (Nunc). The wells were incubated overnight at 4 °C with 200 μl of various proteins at 50 μg/ml dissolved in coating buffer (pH 9.6) containing 15 mM Na2CO3, 35 mM NaCO3 and 0.02% NaN3. The wells were washed three times with 300 μl of assay buffer (pH 7.6), containing 10 mM Tris, 0.14 M NaCl, 0.5 mM CaCl2, 0.02% NaN3 and 0.05% Tween20. The recombinant N-terminal domain of STIM1 was fluorescently labeled, using the EZ-Label Fluorescein Protein Labeling Kit from Pierce. One hundred microliters of varying concentrations of labeled protein were incubated overnight in the wells at 4 °C. The wells were washed twice with assay buffer and a plate reader was used to read the fluorescence. Known amounts of labeled STIM1 were included in each assay to construct a standard curve. Wells were coated with BSA as a negative control.
2.7. Measurement of cellular calcium
For intracellular calcium measurements, HEK 293 cells were suspended at a density of 5 × 106 cells/ml and loaded with 2 μM acetoxymethyl (AM) ester of Fura-2 (Molecular Probes, Eugene, Oregon) for 45 min at 37 °C in Ringer’s buffer containing 135 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5.6 mM glucose, 10 mM HEPES (pH 7.4), and 0.1% BSA. After loading, the cells were washed multiple times in the loading buffer to remove free Fura-2. The cells were then suspended in calcium free Ringer’s buffer plus 0.1 mM EGTA. The fluorescence was measured using a Photon Technology International Deltascan 4000 spectrofluorimeter. Baseline Fura-2AM fluorescence was monitored by alternately exciting the dye at 340 and 380 nm, and collecting the emission at wavelength 510 nm for 1 min. Store depletion was induced by the addition of thapsigargin at a concentration of 2 μM and measurements made for 5 min. Following store depletion extracellular calcium was replenished with the addition of 1.8 mM calcium chloride to the cell suspension and data collected for an additional 5 min. Changes in intracellular calcium are expressed as the ratio of Fura-2AM fluorescence due to excitation at 340 nm and 380 nm.
To concurrently measure cellular calcium at multiple concentrations of exogenous calcium, HEK 293 cells were removed from T175 flasks with trypsin and centrifuged at 500 ×g for 7 min. Cells were then resus-pended in 4 ml Fluo-4NW and 2.5 mM Probenecid (Invitrogen, Grand Island, NY) following the manufacturer’s protocol. The Flou-4NW re-agent was then removed by centrifugation at 500 ×g for 7 min, rinsing the cells once in Ca2+-free HBSS (Ca2+ free HBSS Invitrogen #14170112 supplemented with 1 mM MgCl2, 5.5 mM glucose, 2.5 mM Probenecid) and then resuspending in the same buffer. Each group of cells (HEK 293 and HEK 293 expressing COMP) was then aliquoted using a multichannel pipettor at densities of 15–35 K per well on 384 well, clear flat bottom, poly-l-lysine coated microplates (Greiner Bio-One). Thapsigargin (2 μM, Sigma-Aldrich, St. Louis, MO.) or an equal volume of Ca2+-free HBSS was then added to each well, incubated for 10 min at room temperature and then placed in a Flipr plate reader (Molecular Devices). Ca2+ dose–response data was generated by adding external Ca2+ to each well to final concentrations of 20 μM to 5 mM. The wells with no Ca2+ added were considered baseline. Each Ca2+ dose was replicated in 4 wells. External Ca2+ was added by the Flipr. The arbitrary fluorescent units were normalized to baseline and reported as F/Fo where F = fluorescence and Fo = mean fluorescence for the first 5 time points. F/Fo versus time curves were generated and the store-operated Ca2+ (SOC) response estimated by calculating the area under the curve.
2.8. Electrophysiology
CRAC channel currents induced by depletion of ER Ca2+ stores and ARC channel currents induced by bath addition of arachidonic acid (8 μM) were measured by patch-clamp as previously described (Thompson and Shuttleworth, 2011). Briefly, whole-cell current recordings were performed on cells at room temperature (20–22 °C), using 250 ms voltage pulses to −80 mV delivered every 2 s from a holding potential of 0 mV. To obtain current–voltage relationships, 10 ms pulses to potentials between −100 mV and +80 mV were applied at 20 mV intervals. The standard internal (pipette) solution contained 140 mM CsC2H3O2, 3.5 mM CaCl2, 3.72 mM MgCl2, 10 mM EGTA, and 10 mM HEPES (pH 7.2). The calculated free Ca2+ concentration of this solution was 100 nM. The standard extracellular solution contained 140 mM NaCl, 5 mM CsCl, 1.2 mM MgCl2, 10 mM CaCl2, 10 mM glucose, and 10 mM HEPES (pH 7.4). For arachidonic acid-activated ARC channel current measurements, the initial currents obtained before activation of the channel were used for leak subtraction. For store-operated CRAC channel measurements, the pipette solution was changed to a Ca2+-free solution containing the potent InsP3 receptor agonist adenophostin A (2 μM), and leak-subtraction of the measured currents was obtained at the end of each experiment by application of an external solution containing La3+ (100 μM).
2.9. Statistical analysis
Statistics were carried out using an unpaired or paired Student’s t-test available in the GraphPad software. A p value <0.05 was considered significant. Data are expressed as mean ± standard deviation.
3. Results
3.1. STIM1 associates with TSP-1
We used a proteomic approach to identify proteins that associate with the signature domain of TSP-1 in human blood platelets. TSP-1 was immunoprecipitated from Brij 99 solubilized human blood platelets using a monoclonal antibody to the N-terminal domain, designated MA-IV. We postulated that an antibody to this domain would be least likely to displace proteins that are bound to the signature domain because the two domains are at opposite ends of the molecule. The immunoprecipitates were electrophoresed and various molecular weight ranges were excised and analyzed by mass spectroscopy. The greatest number of peptides identified include those from fibrinogen (18 peptides), the integrin αIIbβ3 (6 peptides), coagulation factor XIII (6 peptides) and GRP78/BiP (4 peptides), and all are known to associate with TSP-1, validating our experimental approach (Bale and Mosher, 1986; Prabakaran et al., 1996; Panetti et al., 1999). Notably, several novel binding partners, such as STIM1 (6 peptides), were identified. We next performed immunoblotting of TSP-1 immunoprecipitates for STIM1 to confirm the mass spectrometry data. Platelets were solubilized in TBS containing either 1% Brij 99 or Triton X-100 and immunoprecipitations were preformed with the anti-TSP-1 monoclonal antibody MA-IV or the anti-TSP-1 polyclonal antibody R1. Immunoblotting of these immunoprecipitates with a monoclonal anti-STIM1 antibody (clone 44) identified an 84,000-dalton band that corresponds to the molecular weight of intact STIM1 (results with R1 are shown in Fig. 1A, lane 2). No STIM1 was detected when the immunoprecipitations were performed with isotype matched IgG (for MA-IV) or pre-immune serum (Fig. 1A, lanes 1, 3 and 5). In addition, an extremely weak band with a molecular weight of 68,000 Da was variably detected in these immunoprecipitates (Fig. 1A, lane 2). We also performed the immunoprecipitation with an anti-STIM1 antibody, designated 5A2, followed by western blotting with a rabbit polyclonal antiserum that was raised against a fusion protein containing the type 1 repeats of TSP-1 designated R3. These experiments specifically identified the 180,000-dalton subunit of TSP-1 in the STIM1 immunoprecipitates (Fig. 1B). Taken together, these data demonstrate a specific interaction between STIM1 and TSPs, and are consistent with a recent report by Ambily et al. (2014) in which TSP-1 was detected in STIM1 immunoprecipitations from human platelets.
Fig. 1.
STIM1 associates with TSP-1. A. Triton X-100 extracts of human blood platelets were immunoprecipitated with the anti-TSP-1 polyclonal antibody (R1, lanes 2, 4 and 6) or pre-immune serum (lanes 1,3 and 5) in the presence of EDTA (lanes 1 and 2), calcium (lanes 3 and 4), or calcium with the calpain inhibitor ALLN (lanes 5 and 6). STIM1 was detected in the immunoprecipitates with an anti-STIM1 monoclonal antibody (clone 44). B. Triton X-100 extracts of human blood platelets were immunoprecipitated with the anti-STIM1 monoclonal (clone 5A2, lane 3) or isotype matched non-immune IgG (lane 1). The platelet lysate is shown in lane 5, and lanes 2 and 4 are empty. TSP-1 was detected in the immunoprecipitates with the anti-TSP-1 polyclonal antibody R3. C. Triton X-100 extracts of Jurkat cells were immunoprecipitated with the anti-TSP-1 polyclonal antibody (R1, lane 2) or pre-immune serum (lane 1) in the presence of EDTA and blotted for STIM1. The cell lysate is shown in lane 4, and lane 3 is empty. D. Triton X-100 extracts of Jurkat cells were immunoprecipitated with the anti-TSP-1 polyclonal antibody (R1, lane 2) or pre-immune serum (lane 1) in the presence of EDTA and blotted for STIM2. E. Triton X-100 extracts of endothelial cells (lane 3) were immunoprecipitated with the anti-TSP-1 polyclonal antibody (R1, lane 2) or pre-immune serum (lane 1) in the presence of EDTA and blotted for STIM1. F. Triton X-100 extracts of MDA-MB-231 cells were immunoprecipitated with the anti-TSP-1 polyclonal antibody (R1, lane 2) or pre-immune serum (lane 1) in the presence of EDTA and blotted for STIM1. The incubation with pre-immune serum and R1 was also done with intact MDA-MB-231 cells prior to solubilization. The cells were then washed, solubilized and immunocomplexes were brought down with Protein A Sepharose beads. The pre-immune sample (lane 3), R1 sample (lane 4) and cell lysate (lane 5) were blotted for STIM1. The pre-immune sample (lane 6), the R1 sample (lane 7) and the lysate (lane 8) were also blotted for TSP-1 to establish the presence of membrane associated TSP-1.
Depletion of calcium from STIM1 results in the aggregation of STIM1 molecules to form protein aggregates or puncta in the ER membrane (Putney, 2011; Soboloff et al., 2012). Since the immunoprecipitations that were performed in this study were done in the presence of EDTA, we were concerned that the association of STIM1 with TSP-1 might reflect non-specific trapping of TSP-1 in STIM1 protein aggregates. To address this concern, we performed immunoprecipitation on extracts in which the EDTA was replaced with 500 μM CaCl2. After solubilization of the platelets in the presence of calcium and immunoprecipitation with R1, the anti-STIM1 antibody clone 44 detected only a weak band at 68,000 Da (Fig. 1A, lane 4). The presence of a fragment of STIM1 in these immunoprecipitates raised the possibility that STIM1 was rapidly degraded after the platelets were solubilized in the presence of calcium. Since platelets contain calcium-dependent proteases, designated calpains, the calpain inhibitor ALLN (20 μM) was included in the calcium-containing solubilization buffer to inhibit the potential degradation of the immunoprecipitated proteins. In the presence of ALLN, an 84,000-dalton band that co-migrates with intact STIM1 was brought down by immunoprecipitating TSP-1 with R1 in the presence of calcium (Fig. 1A, lane 6). These studies indicated that TSP-1 interacts with STIM1 in the presence and absence of calcium, and that STIM1 was a substrate for calpain following solubilization.
To determine if STIM1 associates with TSP-1 in other cells, we immunoprecipitated TSP-1 from Jurkat cells, endothelial cells and various tumor cells. An 84,000-dalton STIM1 band was detected in the R1 immunoprecipitates from all of the cell types examined (Fig. 1C, E and F). We also found that STIM2 immunoprecipitates with TSP-1 from Jurkat cell lysates (Fig. 1D). Two bands were observed in the immunoprecipitates from endothelial cells (Fig. 1E, lane 2). Given that only the 84,000-dalton band was visualized in the whole cell lysates, it appears that STIM1 was proteolytically cleaved after solubilization of these cells (Fig. 1E, lane 3).
STIM1 has been reported to be a transmembrane protein in the ER and in the plasma membrane (Putney, 2011; Shuttleworth, 2012; Soboloff et al., 2012). To determine which pool of STIM1 binds TSP-1, we incubated intact MDA-MB-231 cells with the anti-TSP-1 antibody prior to solubilization to investigate if the interaction of TSP-1 with STIM1 occurred in the plasma membrane. Following incubation with R1, the cells were washed three times in PBS and then solubilized in 1% Triton X-100 lysis buffer. No STIM1 was detected in the R1 immunoprecipitation in these experiments, which suggested that TSP-1 and STIM1 do not remain associated on the plasma membrane in MDA-MB-231 (Fig. 1F, lanes 3, 4 and 5). To confirm that TSP-1 was immunoprecipitated, the blot was reprobed with an antibody to TSP-1 (Fig. 1F, lanes 6, 7 and 8).
3.2. STIM1 associates with TSP-4 and COMP
Since the signature domain of the TSPs is highly conserved in all members of the TSP gene family, we probed other members of the TSP family for their ability to associate with STIM1. We were particularly interested in members of the subgroup B TSPs, which display the greatest differences to TSP-1. HEK 293 cells were transfected with either full-length expression vectors for TSP-4 or COMP (Lawler et al., 1995; Chen et al., 2000). The cells were lysed with Triton X-100 and immunoprecipitation of TSP-4 or COMP was performed followed by western blotting using an anti-STIM1 antibody. Subpanels A and B of Fig. 2 demonstrate that the 84,000-dalton STIM1 band co-immunoprecipitates with both TSP-4 and COMP, respectively. Moreover, STIM1 is detected in the anti-COMP immunoprecipitates from cultured human chondrocytes (Fig. 2B).
Fig. 2.

STIM1 associates with TSP-4 and COMP. A. Triton X-100 solubilized 293 cells engineered to express human TSP-4 were immunoprecipitated with the anti-TSP-4 polyclonal antibody (designated 1259, lane 2) or pre-immune serum (lane 1) and blotted for STIM1. B. Triton X-100 extracts of 293 cells engineered to express human COMP (lanes 1 and 2), or primary human costochondral chondrocytes (lane 3) were immunoprecipitated with the anti-COMP polyclonal antibody (designated F8, lanes 2 and 3) or pre-immune serum (lane 1) and blotted for STIM1. The cell lysates for the primary human costrochondral chondrocytes (lane 4) are also shown. C. Lysates of 293 cells transfected with empty vector (control) or the human COMP expression vector (+COMP) were blotted for COMP and β-actin as indicated. D. Lysates of 293 cells transfected with empty vector (control) or the human COMP expression vector (+COMP) were blotted for STIM1 and β-actin as indicated. E. Lysates of 293 cells transfected with empty vector (control) or the human Orai1 expression vector (+Orai1) were blotted for STIM1 and β-actin as indicated.
3.3. The N-terminal domain of STIM1 binds to the signature domain of the TSPs
The first 184 amino acids of STIM1 comprise the portion of the protein that is exposed in the lumen of the ER or the external side of the plasma membrane (Stathopulos et al., 2006, 2008; Johnstone et al., 2010; Soboloff et al., 2012). This domain contains the calcium binding site and the SAM motif (Stathopulos et al., 2006, 2008; Johnstone et al., 2010; Soboloff et al., 2012). The transmembrane and cytoplasmic domains extend to the C-terminal of the protein. Since the TSPs and STIM1 were most likely to interact in the lumen of the ER or outside of the cell, we asked if a recombinant version of the N-terminal of STIM1 might also bind to TSPs. Expression of the N-terminal domain in S2 cells yielded a 22,000-dalton protein that binds the anti-STIM1 antibody clone 44. We used limited chymotryptic digestion to show that the N-terminal domain binds to calcium. The 22,000-dalton STIM1 band was almost completely digested by chymotrypsin (1:500 w/w) in the absence of calcium (Fig. 3). Increasing the amount of calcium in the digest progressively protected STIM1 from degradation so the level of the 22,000-dalton band increased as the calcium concentration increased. The midpoint of this transition (0.5 mM) represented an estimate of the association constant of calcium for STIM1. The value of 0.5 mM total calcium agreed with the value of 0.2 to 0.6 mM reported previously (Stathopulos et al., 2006). Taken together, the data indicated that the recombinant N-terminal domain of STIM1 faithfully recapitulated the function of the wild-type protein.
Fig. 3.

Limited chymotryptic digestion of a recombinant version of the N-terminal domain of STIM1 with varying concentrations of calcium. A purified recombinant N-terminal domain of STIM1 was dialyzed against Tris-buffered saline containing 2 mM CaCl2 (lane 1). EDTA was added to each sample so that the final Ca2+ concentrations would be between 2 mM and 0.01 mM (lanes 2 through 9, as indicated below each lane) along with chymotryptic digestion, which was carried out at an enzyme/substrate ratio of 1:500 (w/w) for 20 h at 0 °C. The digestion was halted with the addition of a reduced SDS sample buffer. The polypeptides were separated using SDS-PAGE. The relative quantity of the 22,000-dalton polypeptide was shown above the lanes with the 2 mM sample estimated at 1.0. Proteolytic fragments were visualized by staining with Coomassie Blue. The molecular mass of the markers (M) is marked on the left.
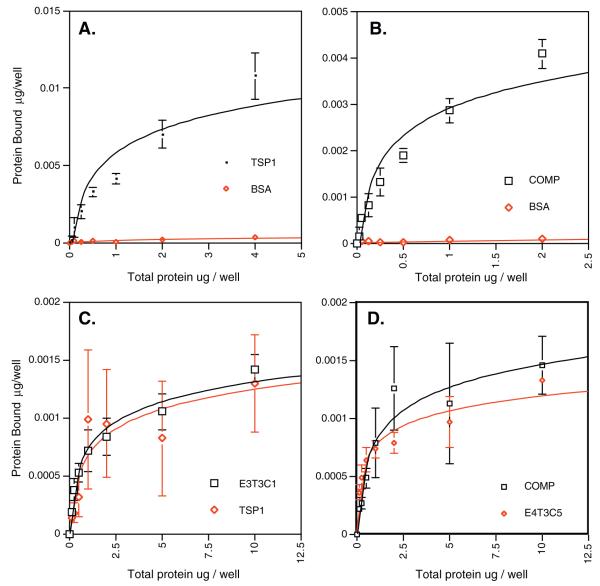
Having established the co-association between the TSPs and STIM1, we next undertook experiments to determine if this association is due to direct binding. A solid-phase binding assay demonstrated the direct interaction of the N-terminal domain of STIM1 with TSPs (Fig. 4). The wells of the plate were coated with TSP-1 or COMP, and BSA was used as a negative control. The addition of increasing amounts of labeled N-terminal domain of STIM1 resulted in increased binding to the TSPs with little or no binding to BSA observed (Fig. 4A and B). The binding of fluorescently labeled N-terminal domain of STIM1 to TSP-1 or COMP was inhibited by the non-labeled N-terminal of the STIM1 domain, indicating that the observed binding was not an artifact of the labeling procedure. Binding of the N-terminal domain of STIM1 to the TSPs was also observed when the assay was performed in the presence of EDTA (Fig. 4C and D). This result was consistent with the earlier experiments in which STIM1 and TSP-1 co-immunoprecipitate in the presence or absence of calcium. Moreover, the N-terminal domain of STIM1 also bound to recombinant versions of the signature domains of TSP-1 (E3T3C1) and COMP (E4T3C5) in a saturable manner (Fig. 4C and D). These data demonstrate that STIM1 co-association with TSPs is the result of direct binding.
Fig. 4.
Binding of the N-terminal domain of STIM1 to TSPs. A. Human platelet TSP-1 and BSA were adsorbed by the wells of an Immulon II plate overnight in 2 mM CaCl2. Fluorescently labeled, recombinant N-terminal domain of STIM1 was incubated in the wells overnight in 2 mM CaCl2. B. Recombinant COMP and BSA were adsorbed to the wells of an Immulon II plate overnight in 2 mM CaCl2. The fluorescently labeled, recombinant N-terminal domain of STIM1 was incubated in the wells overnight in 2 mM CaCl2. C. Human platelet TSP-1 and E3T3C1 were adsorbed by the wells of a FluoroNunc Maxisorb plate overnight in the presence of 5 mM EDTA. Fluorescently labeled, recombinant N-terminal domain of STIM1 was incubated in the wells overnight in 5 mM EDTA. D. Human recombinant COMP and E4T3C5 were adsorbed by the wells of a FluoroNunc Maxisorb plate overnight in the presence of 5 mM EDTA. The fluorescently labeled, recombinant N-terminal domain of STIM1 was incubated in the wells overnight in 5 mM EDTA. In all cases, the wells were washed, and the bound protein then was quantified using a fluorescent plate reader. Each point shown is the mean and standard deviation of four determinations.
3.4. COMP over-expression affects STIM1 function
To determine whether or not TSPs affect STIM1 function, we over-expressed COMP in HEK 293 cells, which express endogenous STIM1, but not endogenous TSPs. We chose COMP for these studies because it lacks the N-terminal domain and the type 1 repeats that are found in other TSPs and have been shown to mediate numerous cellular functions (Adams and Lawler, 2011). Interestingly, stable over-expression of COMP in HEK 293 cells resulted in a variable (25 ± 15%, n = 8, p = 0.0038) increase in total STIM1 expression (Fig. 2D).
STIM1 is known to play a key role in both the store-dependent and the store-independent entry of Ca2+ into cells as mediated by the CRAC (Ca2+-release activated Ca2+) channels and ARC (arachidonate-regulated Ca2+) channels, respectively, both of which are encoded by the Orai family of proteins. Critically, while the activity of the CRAC channels is specifically regulated by the major pool of cellular STIM1 that lies within the ER membrane, it is the minor pool of STIM1 resident in the plasma membrane that is responsible for regulating the activation of the ARC channels. Thapsigargin blocks SERCA pump function, depletes ER calcium, and causes STIM1 to form puncta below the plasma membrane where it enables the activation of the CRAC channels. Addition of thapsigargin to HEK 293 cells in the absence of extracellular calcium results in increased cytoplasmic calcium as measured by Fura-2 fluorescence (Fig. 5A), reflecting the release of calcium from ER stores. Subsequent addition of calcium-containing buffers to the cell suspension results in rapid influx of calcium (Fig. 5A). Over-expression of COMP increases the amount of calcium influx induced by thapsigargin as measured by the difference between the treated and untreated samples (Fig. 5A). This significant (p = 0.0012) increase in calcium influx in response to thapsigargin was observed over a broad range of calcium concentrations (Fig. 5B).
Fig. 5.
Expression of COMP increases calcium influx induced by thapsigargin. A. Control transfected HEK 293 cells (blue line) and COMP expressing HEK 293 cells (red line) were loaded with Fura-2 and treated with 2 μM thapsigargin. After 300 s, 1.8 mM CaCl2 was added to the sample and the fluorescence was monitored for an additional 300 s. B. To concurrently measure cellular calcium at multiple concentrations of exogenous calcium, control transfected HEK cells (black lines) and COMP expressing HEK 293 (green lines) were loaded with Fluo-4NW and 2.5 mM Probenecid. Thapsigargin (2 μM, black squares and green triangles) or an equal volume of Ca2+-free HBSS (black and green circles) was then added to each well, incubated for 10 min at room temperature and then placed in a Flipr plate reader. Ca2+ dose–response data was generated by adding external Ca2+ to each well to final concentrations of 20 μM to 5 mM. The wells with no Ca2+ added were considered baseline. Each Ca2+ dose was replicated in 4 wells. External Ca2+ was added by the Flipr. The arbitrary fluorescent units were normalized to baseline and reported as F/Fo where F = fluorescence and Fo = mean fluorescence for the first 5 time points. F/Fo versus time curves were generated and the store-operated Ca2+ (SOC) response estimated by calculating the area under the curve (AUC).
The modest increase in cytoplasmic calcium that we detected in COMP expressing cells with Fura-2 and Fluo-4 may be due to a broad spectrum of cellular responses, including changes in calcium channel activity, changes in plasma membrane calcium pump activity, and changes in calcium buffering within the ER or mitochondria. To determine if COMP affects calcium channel activity, we directly measured CRAC and ARC channel currents. Expression of COMP in the Flp-In-HEK 293 cells resulted in the reduction of endogenous store-operated CRAC channel currents to virtually negligible values (from 0.33 ± 0.01 pA/pF at −80 mV, to 0.07 ± 0.01 pA/pF, n = 5) (Fig. 6). In contrast, currents through the arachidonic acid-stimulated ARC channels in the same COMP-expressing cells were increased by almost 70% from 0.40 ± 0.04 pA/pF at −80 mV (n = 6), to a value of 0.68 ± 0.03 pA/pF (n = 5). To explore this effect further, we repeated these experiments in cells engineered to have a 2- to 3-fold increase in Orai1 — a protein that is an essential component of both CRAC and ARC channels (Mignen et al., 2008). As before, expression of COMP in these Orai1-expressing cells reduced the magnitude of CRAC channel currents following store depletion, from a value of 0.34 ± 0.03 pA/pF at −80 mV (n = 6), to only 0.17 ± 0.08 pA/pF (n = 6) (Fig. 7). However, in the same Orai1-expressing cells, expression of COMP markedly increased ARC channel currents from a value of 0.37 ± 0.03 pA/pF at −80 mV (n = 6), to 0.91 ± 0.13 pA/pF at −80 mV (n = 5) — an approximate 2.5-fold increase (Fig. 7). The over-expression of Orai1 did not affect the level of STIM1 protein (Fig. 2E). We could not quantify the levels of Orai1 and 3 proteins because we were unable to identify highly sensitive antibodies that can be used to western blot these proteins. Together, these data indicate that the expression of COMP in these cells results in a specific, and significant, increase in the magnitude of the inward Ca2+-selective currents via the ARC channels, while profoundly inhibiting those through the corresponding CRAC channel currents.
Fig. 6.
Measurement of CRAC and ARC channel currents in the presence and absence of COMP. CRAC channel currents (top left) induced by depletion of ER Ca2+ stores and ARC channel currents (top right) induced by bath addition of arachidonic acid (8 μM) in control transfected (black circles) and COMP expressing HEK 293 cells (red circles) were measured by patch-clamp as previously described in Materials and methods. For arachidonic acid-activated ARC channel current measurements, the initial currents obtained before activation of the channel were used for leak subtraction. For store-operated CRAC channel measurements, the pipette solution was changed to a Ca2+-free solution containing the potent InsP3 receptor agonist adenophostin A (2 μM), and leak-subtraction of the measured currents was obtained at the end of each experiment by application of an external solution containing La3+ (100 μM).
Fig. 7.
Measurement of CRAC and ARC channel currents in the presence and absence of COMP in Orai1-over-expressing cells. CRAC channel currents (top left) induced by depletion of ER Ca2+ stores and ARC channel currents (top right) induced by bath addition of arachidonic acid (8 μM) in control transfected (black circles) and COMP expressing HEK 293 cells (red circles) were measured by patch-clamp as previously described in Materials and methods. For arachidonic acid-activated ARC channel current measurements, the initial currents obtained before activation of the channel were used for leak subtraction. For store-operated CRAC channel measurements, the pipette solution was changed to a Ca2+-free solution containing the potent InsP3 receptor agonist adenophostin A (2 μM), and leak-subtraction of the measured currents was obtained at the end of each experiment by application of an external solution containing La3+ (100 μM).
4. Discussion
In these studies, we have identified a novel interaction of the members of the TSP gene family with STIM1. This association is robust since it is preserved in Triton X-100, can be detected with multiple anti-TSP-1 and anti-STIM1 antibodies, and is detected in a wide range of cell types. This interaction could occur in the ER under conditions of normal or low calcium concentration. Furthermore, we observed that the expression of COMP in HEK 293 cells affects STIM1-mediated CRAC and ARC channel function. CRAC channel current is regulated by clustering of STIM1 in the ER membrane in locations that are juxtaposed with Orai1 in the plasma membrane (Putney, 2011; Shuttleworth, 2012; Soboloff et al., 2012). We have observed that over-expression of COMP results in a decrease in CRAC channel current. By contrast, over-expression of COMP results in an increase in ARC channel current, which is mediated by an association of STIM1 with Orai1 and 3 in the plane of the plasma membrane. These observations are consistent with data showing that these two routes of calcium entry into the cell are reciprocally regulated (Mignen et al., 2001). Given the demonstrated interaction between COMP and STIM1, the most logical explanation for these effects on the discrete Orai channel mediated Ca2+ entries would seem to be that expression of COMP induces a redistribution of at least a component of total cellular STIM1, from the majority ER-resident pool to the pool constitutively residing in the plasma membrane. Critically, because the latter represents only a minor pool (~15–25%) of total cellular STIM1, the redistribution of only a relatively minor component of the ER pool to the plasma membrane would be required to induce a significant increase in the activity of the ARC channels. In addition, the increase in ARC channel current (2.5-fold in Orai1 over-expressing cells) is considerably greater than the increase in total STIM1 (25 ± 15%) that we observed in COMP-expressing HEK 293 cells. These data suggest that COMP and perhaps other TSPs promote ARC channel currents, and hence cellular responsiveness to arachidonic acid-mediated signal transduction, by stimulating the transport of STIM1 from the ER to the plasma membrane. The reciprocal regulation of CRAC and ARC channels may serve to protect the cell from excessive calcium entry (Taylor, 2002).
We have also observed that expression of COMP decreases CRAC channel current suggesting that COMP inhibits the ability of STIM1 to open these channels. It is possible that the binding of STIM1 to COMP in the ER suppresses its ability to form puncta and open CRAC channels. The binding of STIM1 to COMP in the ER membrane could inhibit puncta formation by buffering the calcium-binding site in STIM1 or by decreasing the mobility of STIM1 in the ER membrane. The signature domains of the TSPs bind approximately 30 calcium ions through a cooperative mechanism with an average dissociation constant that is comparable to that of the calcium-binding site of STIM1 (Lawler and Simons, 1983; Chen et al., 1996, 2000; Stathopulos et al., 2006). It is possible that the association of TSPs with STIM1 provides a source of calcium ions that can maintain STIM in the calcium bound conformation and inhibit puncta formation at reduced calcium concentrations.
STIM1 in the plasma membrane was initially reported to function as a cell adhesion molecule that binds B lineage lymphocyte precursors in bone marrow (Oritani and Kincade, 1996). TSP-1 has been reported to be expressed in the bone marrow and to support adhesion of hematopoietic progenitor cells (Long and Dixit, 1990). It has recently been proposed that STIM1 functions as a receptor for TSP-1 during collagen-induced platelet aggregation (Ambily et al., 2014). Taken together, the data raise the possibility that STIM1 may join a long list of membrane proteins, including integrins, proteoglycans, CD36 and CD47, that are receptors for TSPs.
The data presented here imply that the TSPs affect cellular calcium signaling through an interaction with STIM1 in the ER and plasma membrane. This interaction may mediate cellular responses to a wide range of physiological and pathological stimuli that utilize calcium and/or arachidonic acid as second messengers. It is interesting to note that TSP-1, STIM1 and Orai1 have been reported to promote tumor cell migration, invasion and metastasis (Yang et al., 2009; Yee et al., 2009; Chen et al., 2011). Thrombospondins have also been shown to activate the unfolded protein response in the ER through binding another ER transmembrane protein, ATF6α (Lynch et al., 2012). The unfolded protein response may lead to expansion of the ER and the increased STIM1 expression that we observed. Whereas the vast majority of studies have sought functions for the TSPs at the plasma membrane and in the extracellular matrix, it is now clear that they also have important function within the ER prior to secretion.
Acknowledgments
This manuscript is dedicated to the memory of Paul Bornstein who highlighted the importance and dynamic nature of the interface between the cell and its extracellular environment. The authors wish to thank Evey Fernadez and Sami Lawler for help in preparing the manuscript and Neal Smith for expert technical assistance. Human chondrocytes were provided by Drs. Karen Posey and Jacqui Hecht and MDA-MB-231 cells were provided by Dr. Shideh Kazerounian. This work was supported by grants HL68003 from the National Heart, Lung, and Blood Institute (JL) and GM040457 from the National Institute of General Medical Sciences (TS) of the National Institutes of Health.
Abbreviations
- TSP
thrombospondin
- STIM1
stromal interaction molecule 1
- ARC channel
arachidonate-regulated calcium channel
- CRAC channel
calcium release activated calcium channel
- COMP
cartilage oligomeric matrix protein
- VEGF
vascular endothelial growth factor
- VEGFR-2
VEGF receptor 2
- SOC
store operated calcium
- ER
endoplasmic reticulum
- PHR
procollagen homology region
References
- Adams JC, Lawler J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011;3:a009712. doi: 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambily A, Kaiser WJ, Pierro C, Chamberlain EV, Li Z, Jones CI, Kassouf N, Gibbins JM, Authi KS. The role of plasma membrane STIM1 and Caentry in platelet aggregation. STIM1 binds to novel proteins in human platelets. Cell. Signal. 2014;26:502–511. doi: 10.1016/j.cellsig.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale MD, Mosher DF. Thrombospondin is a substrate for blood coagulation factor XIIIa. Biochemistry. 1986;25:5667–5673. doi: 10.1021/bi00367a048. [DOI] [PubMed] [Google Scholar]
- Bornstein P, O’Rourke K, Wilstrom L, Wolf FW, Katz R, Li P, Dixit VM. A second, expressed thrombospondin gene (Thbs2) exists in the mouse genome. J. Biol. Chem. 1991;266:12821–12824. [PubMed] [Google Scholar]
- Carlson CB, Bernstein DA, Annis DS, Misenheimer TM, Hannah BL, Mosher DF, Keck JL. Structure of the calcium-rich signature domain of human thrombospondin-2. Nat. Struct. Mol. Biol. 2005;12:910–914. doi: 10.1038/nsmb997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Aeschlimann D, Nowlen J, Mosher DF. Expression and initial characterization of recombinant mouse thrombospondin 1 and thrombospondin 3. FEBS Lett. 1996;387:36–41. doi: 10.1016/0014-5793(96)00460-7. [DOI] [PubMed] [Google Scholar]
- Chen H, Deere M, Hecht JT, Lawler J. Cartilage oligomeric matrix protein is a calcium-binding protein, and a mutation in its type 3 repeats causes conformational changes. J. Biol. Chem. 2000;275:26538–26544. doi: 10.1074/jbc.M909780199. [DOI] [PubMed] [Google Scholar]
- Chen YF, Chiu WT, Chen YT, Lin PY, Huang HJ, Chou CY, Chang HC, Tang MJ, Shen MR. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15225–15230. doi: 10.1073/pnas.1103315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LY, Ramakrishnan DP, Silverstein RL. Thrombospondin-1 modulates VEGF signaling via CD36 by recruiting SHP-1 to VEGFR2 complex in microvascular endothelial cells. Blood. 2013;122:1822–1832. doi: 10.1182/blood-2013-01-482315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Lustig A, Engel J. The thrombospondin-like chains of cartilage oligomeric matrix protein are assembled by a five-stranded a-helical bundle between residues 20 and 83. FEBS Lett. 1994;341:54–58. doi: 10.1016/0014-5793(94)80239-4. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Deere M, Putnam E, Cole W, Vertel B, Chen H, Lawler J. Characterization of cartilage oligomeric matrix protein (COMP) in human normal and pseudoachondroplasia musculoskeletal tissues. Matrix Biol. 1998;17:269–278. doi: 10.1016/s0945-053x(98)90080-4. [DOI] [PubMed] [Google Scholar]
- Incardona F, Lawler J, Cataldo D, Panet A, Legrand Y, Foidart JM, Legrand C. Heparin-binding domain, type 1 and type 2 repeats of thrombospondin mediate its interaction with human breast cancer cells. J. Cell. Biochem. 1996;62:431–442. doi: 10.1002/(sici)1097-4644(19960915)62:4<431::aid-jcb1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Johnstone LS, Graham SJ, Dziadek MA. STIM proteins: integrators of signalling pathways in development, differentiation and disease. J. Cell. Mol. Med. 2010;14:1890–1903. doi: 10.1111/j.1582-4934.2010.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Martin-Manso G, Pendrak ML, Garfield SH, Isenberg JS, Roberts DD. Thrombospondin-1 inhibits VEGF receptor-2 signaling by disrupting its association with CD47. J. Biol. Chem. 2010;285:38923–38932. doi: 10.1074/jbc.M110.172304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerounian S, Duquette M, Reyes MA, Lawler JT, Song K, Perruzzi C, Primo L, Khosravi-Far R, Bussolino F, Rabinovitz I, Lawler J. Priming of the vascular endothelial growth factor signaling pathway by thrombospondin-1, CD36, and spleen tyrosine kinase. Blood. 2011;117:4658–4666. doi: 10.1182/blood-2010-09-305284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvansakul M, Adams JC, Hohenester E. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J. 2004;23:1223–1233. doi: 10.1038/sj.emboj.7600166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J, Hynes RO. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium binding sites and homologies with several different proteins. J. Cell Biol. 1986;103:1635–1648. doi: 10.1083/jcb.103.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J, Simons ER. Cooperative binding of calcium to thrombospondin. The effect of calcium on the circular dichroism and limited tryptic digestion of thrombospondin. J. Biol. Chem. 1983;258:12098–12101. [PubMed] [Google Scholar]
- Lawler J, Derick LH, Connolly JE, Chen JH, Chao FC. The structure of human platelet thrombospondin. J. Biol. Chem. 1985;260:3762–3772. [PubMed] [Google Scholar]
- Lawler J, Duquette M, Urry L, McHenry K, Smith TF. The evolution of the thrombospondin gene family. J. Mol. Evol. 1993a;36:509–516. doi: 10.1007/BF00556355. [DOI] [PubMed] [Google Scholar]
- Lawler J, Duquette M, Whittaker C, Adams JC, McHenry K, DeSimone D. Identification and characterization of thrombospondin-4, a new member of the thrombospondin gene family. J. Cell Biol. 1993b;120:1059–1067. doi: 10.1083/jcb.120.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J, McHenry K, Duquette M, Derick L. Characterization of human thrombospondin-4. J. Biol. Chem. 1995;270:2809–2814. doi: 10.1074/jbc.270.6.2809. [DOI] [PubMed] [Google Scholar]
- Long MW, Dixit VM. Thrombospondin functions as a cytoadhesion molecule for human hematopoietic progenitor cells. Blood. 1990;75:2311–2318. [PubMed] [Google Scholar]
- Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, Prywes R, Lorenz JN, Mori K, Lawler J, Robbins J, Molkentin JD. A thrombospondin-dependent pathway for a protective ER stress response. Cell. 2012;149:1257–1268. doi: 10.1016/j.cell.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao WM, Seng WL, Duquette M, Lawler P, Laus C, Lawler J. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-beta-dependent and -independent mechanisms. Cancer Res. 2001a;61:7830–7839. [PubMed] [Google Scholar]
- Miao WM, Vasile E, Lane WS, Lawler J. CD36 associates with CD9 and integrins on human blood platelets. Blood. 2001b;97:1689–1696. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- Mignen O, Thompson JL, Shuttleworth TJ. Reciprocal regulation of capacitative and arachidonate-regulated noncapacitative Ca2+ entry pathways. J. Biol. Chem. 2001;276:35676–35683. doi: 10.1074/jbc.M105626200. [DOI] [PubMed] [Google Scholar]
- Mignen O, Thompson JL, Shuttleworth TJ. Both Orai1 and Orai3 are essential components of the arachidonate-regulated Ca2+-selective (ARC) channels. J. Physiol. 2008;586:185–195. doi: 10.1113/jphysiol.2007.146258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton G, Weremowicz S, Morton CC, Copeland NG, Gilbert DJ, Jenkins NA, Lawler J. Characterization of human and mouse cartilage oligomeric matrix protein. Genomics. 1994;24:435–439. doi: 10.1006/geno.1994.1649. [DOI] [PubMed] [Google Scholar]
- Oldberg A, Antonsson P, Lindblom K, Heinegard D. COMP is structurally related to the thrombospondins. J. Biol. Chem. 1992;267:22346–22350. [PubMed] [Google Scholar]
- Oritani K, Kincade PW. Identification of stromal cell products that interact with pre-B cells. J. Cell Biol. 1996;134:771–782. doi: 10.1083/jcb.134.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panetti TS, Kudryk BJ, Mosher DF. Interaction of recombinant procollagen and properdin modules of thrombospondin-1 with heparin and fibrinogen/fibrin. J. Biol. Chem. 1999;274:430–437. doi: 10.1074/jbc.274.1.430. [DOI] [PubMed] [Google Scholar]
- Prabakaran D, Kim PS, Dixit VM, Arvan P. Oligomeric assembly of thrombospondin in the endoplasmic reticulum of thyroid epithelial cells. Eur. J. Cell Biol. 1996;70:134–141. [PubMed] [Google Scholar]
- Putney JW. The physiological function of store-operated calcium entry. Neurochem. Res. 2011;36:1157–1165. doi: 10.1007/s11064-010-0383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumet A, Slimane MB, Lanotte M, Lawler J, Dubernard V. Type 3 repeat/C-terminal domain of thrombospondin-1 triggers caspase-independent cell death through CD47/alphavbeta3 in promyelocytic leukemia NB4 cells. Blood. 2005;106:658–667. doi: 10.1182/blood-2004-09-3585. [DOI] [PubMed] [Google Scholar]
- Shuttleworth TJ. STIM and Orai proteins and the non-capacitative ARC channels. Front. Biosci. (Landmark Ed) 2012;17:847–860. doi: 10.2741/3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: an initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Tan K, Duquette M, Joachimiak A, Lawler J. The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding. FASEB J. 2009;23:2490–2501. doi: 10.1096/fj.08-128090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CW. Controlling calcium entry. Cell. 2002;111:767–769. doi: 10.1016/s0092-8674(02)01197-2. [DOI] [PubMed] [Google Scholar]
- Thompson JL, Shuttleworth TJ. Orai channel-dependent activation of phospholipase C-delta: a novel mechanism for the effects of calcium entry on calcium oscillations. J. Physiol. 2011;589:5057–5069. doi: 10.1113/jphysiol.2011.214437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JL, Shuttleworth TJ. A plasma membrane-targeted cytosolic domain of STIM1 selectively activates ARC channels, an arachidonate-regulated store-independent Orai channel. Channels (Austin) 2012;6:370–378. doi: 10.4161/chan.21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP. The thrombospondin type 1 repeat superfamily. Int. J. Biochem. Cell Biol. 2004;36:969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Vos HL, Devarayalu S, De Vries Y, Bornstein P. Thrombospondin-3 (Thbs3), a new member of the thrombospondin gene family. J. Biol. Chem. 1992;267:12192–12196. [PubMed] [Google Scholar]
- Yang S, Zhang JJ, Huang XY. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell. 2009;15:124–134. doi: 10.1016/j.ccr.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res. Treat. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]