Abstract
Background and Objective
Many physiological and pathophysiological conditions are attributable in part to cytoskeletal regulation of cellular responses to signals. Moesin (membrane-organizing extension spike protein), an ERM (ezrin, radixin and moesin) family member, is involved in lipopolysaccharide (LPS)-mediated events in mononuclear phagocytes; however, its role in signaling is not fully understood. The aim of this study was to investigate the LPS-induced moesin signaling pathways in macrophages.
Material and Methods
Macrophages were stimulated with 500 ng/mL LPS in macrophage serum-free medium. For blocking experiments, cells were pre-incubated with anti-moesin antibody. Moesin total protein and phosphorylation were studied with western blotting. Moesin mRNA was assessed using quantitative real-time PCR. To explore binding of moesin to LPS, native polyacrylamide gel electrophoresis (PAGE) gel shift assay was performed. Moesin immunoprecipitation with CD14, MD-2 and Toll-like receptor 4 (TLR4) and co-immunoprecipitation of MyD88–interleukin-1 receptor-associated kinase (IRAK) and IRAK–tumor necrosis factor receptor-activated factor 6 (TRAF6) were analyzed. Phosphorylation of IRAK and activities of MAPK, nuclear factor κB (NF-κB) and IκBα were studied. Tumor necrosis factor α, interleukin-1β and interferon β were measured by ELISA.
Results
Moesin was identified as part of a protein cluster that facilitates LPS recognition and results in the expression of proinflammatory cytokines. Lipopolysaccharide stimulates moesin expression and phosphorylation by binding directly to the moesin carboxyl-terminus. Moesin is temporally associated with TLR4 and MD-2 after LPS stimulation, while CD14 is continuously bound to moesin. Lipopolysaccharide-induced signaling is transferred downstream to p38, p44/42 MAPK and NF-κB activation. Blockage of moesin function interrupts the LPS response through an inhibition of MyD88, IRAK and TRAF6, negatively affecting subsequent activation of the MAP kinases (p38 and ERK), NF-κB activation and translocation to the nucleus.
Conclusion
These results suggest an important role for moesin in the innate immune response and TLR4-mediated pattern recognition in periodontal disease.
Keywords: macrophage, lipopolysaccharide, signal transduction, moesin
Cytoskeletal proteins are involved in many cellular functions, including regulation of the actin cytoskeleton, control of cell shape, adhesion and motility, and modulation of signaling pathways (1). Many physiological and pathophysiological conditions are attributable in part to cytoskeletal regulation of cellular responses to external and internal cell signals. Moesin (membrane-organizing extension spike protein) is a previously described cytoskeletal protein, which belongs to the ezrin–radixin–moesin (ERM) family (2,3). The ERM proteins are expressed in several cell types, and recent data suggest unique roles for these proteins (4). Phosphorylation of moesin transforms the protein from an inactive into an active stage and mediates its binding to surrounding cytoskeletal proteins or F-actin (5,6). Previous publications suggested a role for the cytoskeletal protein moesin in lipopolysaccharide (LPS)-induced immune responses (7,8). Inhibition of moesin expression ablates LPS responsiveness, and blockage of its function using an anti-moesin antibody inhibits the release of tumor necrosis factor α (TNF-α) by LPS-stimulated monocytes/macrophages (8). Homozygous moesin knockout mice exhibit a threefold reduction in neutrophil infiltration in response to LPS injection (7). However, a complete understanding of signaling cascades associated with moesin and LPS is lacking.
Inflammatory cytokines and other soluble mediators are expressed in LPS-stimulated macrophages through activation of transcription factors, including nuclear factor κB (NF-κB) and activator protein-1 (AP-1; 9–11). The LPS recognition pathway includes LPS binding protein (LBP), a serum glycoprotein that first binds to the lipid A moiety of LPS (12–14) followed by binding of the LPS–LBP complex to CD14 (13,15,16). Mice with a targeted deletion of the gene encoding CD14 are hyporesponsive to LPS and resistant to the lethal effects of LPS (16); however, mice lacking CD14 still respond to high concentrations of LPS (17). CD14 is a glycosylphosphatidylinositol (GPI)-anchored molecule that lacks a cytoplasmic signaling domain and is therefore incapable of downstream signaling (18). The mammalian homologues of the Drosophila Toll protein, identified as Toll-like receptor (TLR) proteins, mediate the response to LPS (19). The TLR proteins possess leucine-rich extracellular repeats that recognize the LBP–CD14 complex (20), and the intracellular domain resembles the interleukin-1β (IL-1β) receptor, hence the term Toll/IL-1 receptor homology domain (TIR; 21–23). The TIR domain in the cytoplasmic portion of the molecule is essential for triggering activation of MAP kinases (MAPK) and the transcription factor, nuclear factor κB (NF-κB; 24–26). Toll-like receptors utilize IL-1β signaling components, including the adaptor protein MyD88, interleukin-1 receptor-associated kinase (IRAK) and TNF receptor-activated factor 6 (TRAF6; 19,27,28). MyD88 contains a death domain (DD), a highly conserved protein-binding domain that facilitates interaction with another DD-containing signaling molecule, IRAK (29). Interleukin-1 receptor-associated kinase is phosphorylated, dissociates from MyD88 and binds to TRAF6, activating several downstream kinases (28–33). Following LPS stimulation, two signaling pathways have been described, the MyD88-dependent and -independent pathways (26,34–36). Activation of the MyD88-dependent pathway results in rapid NF-κB activation and release of proinflammatory cytokines, such as TNF-α and IL-1β, while activation of the MyD88-independent pathway results in rapid activation of interferon regulatory factor 3 (IRF3) leading to interferon β (IFN-β) release with delayed NF-κB activation (34,35,37). Anti-moesin antibody inhibits the release of TNF-α by LPS-stimulated monocytes. Moesin is the only Band 4 protein expressed on the surface of mononuclear phagocytes (38); its mRNA knockdown ablates LPS responsiveness (8).
Since LPS is a critical virulence factor produced by periodontopathogens, the identification of the signaling pathways through pattern recognition of LPS and therefore the specific bacteria will provide new insights into molecular mechanisms associated with the cellular cytoskeleton and an immune response to LPS stimulation. The role of moesin during this process is not clear. The aim of this study was to analyze moesin phosphorylation and binding activity as well as the molecular mechanism downstream of cell signaling events in macrophages in response to LPS.
Material and methods
Reagents and materials
The cell line THP-1 cells, Vita cell RPMI 1640 cell culture medium and fetal bovine serum (FBS) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Moesin and radixin recombinant proteins and C- and N-terminus truncated proteins were purchased from Promab (Albany, CA, USA). Macrophage serum-free medium (M-SFM) and TRIzol® were obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). TaqMan probes, sense primers and anti-sense primers of moesin and β-actin were obtained from Applied Biosystems (Foster City, CA, USA). Escherichia coli LPS (strain O55:B5) and moesin affinity-purified mouse monoclonal antibody (IgG1) clone 38/ 78 were purchased from Sigma (St Louis, MO, USA). The IRAK monoclonal antibody (recognizes active IRAK at 100 kDa) was from BD Pharmingen (San Diego, CA, USA). Rabbit anti-IRAK polyclonal antibody was from Upstate USA, Inc. (Charlottesville, VA, USA). MyD88 antibody (agarose-conjugated goat IgG), protein A/G plus agarose, affinity-purified TLR4 rabbit polyclonal antibody, affinity purified MD-2 rabbit polyclonal antibody, affinity-purified rabbit polyclonal phospho-moesin antibody, affinity-purified CD14 monoclonal antibody, TRAF6 monoclonal antibody, phospho-threonine specific antibody and actin monoclonal antibody, as well as horseradish peroxidase (HRP)-conjugated appropriate secondary antibodies, were all purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). SuperSignal West Pico chemiluminescent substrate was from Pierce (Rockford, IL, USA). NF-κB/ p65 ActivELISA Kit was purchased from Imgenex (San Diego, CA, USA). Tumor necrosis factor α, IL-1β and IFN-β ELISA kits were purchased from R&D Systems (Minneapolis, MN, USA). p38 and ERK1/2 (p44/42) MAP kinase activity kits and phospho-specific IκBα antibody were from Cell Signaling (Beverly, MA, USA). The p38 inhibitor SB202190 and p44/42 inhibitor PD98059 were purchased from Calbiochem (La Jolla, CA, USA). Polyvinylidene difluoride (PVDF) transfer membranes were purchased from Millipore (Bedford, MA, USA). Protein assay reagents and silver staining kits were purchased from Bio-Rad (Hercules, CA, USA). Unless otherwise specified, all other reagents were purchased from Sigma.
Cell culture, differentiation and stimulation
Macrophages were differentiated from THP-1 cells (human acute monocytic leukemia cell line, which could be differentiated into macrophages) by culture in Vita cell RPMI 1640 (supplemented with 10% fetal bovine serum, 2 mM L-glutamine adjusted to contain 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mM Hepes, 1.0 mM sodium pyruvate and 0.05 mM β-mercaptoethanol). Cells were then plated in six-well tissue culture plates and stimulated to differentiate and adhere with 20 ng/mL phorbol 12-myristate 13-acetate (PMA) for 24 h (39). For all procedures, cells were cultured at 37°C in an atmosphere of air containing 5% CO2. Differentiated THP-1 cells (approximately 8 × 105/mL) were stimulated with 500 ng/mL LPS in macrophage serum-free medium (Invitrogen Life Technologies). Peak response from LPS stimulation was based on dose–response experiments. For blocking experiments, cells were pre-incubated with anti-moesin antibody (10 μg/mL, final concentration) for 2 h.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis
To investigate moesin total protein and phosphorylation upon stimulation, stimulated cells were lysed and sonicated, and supernatant was collected by centrifugation at 16,000g. The protein content was determined by Bradford assay (40). Samples were boiled in Laemmle sample buffer (41), and 5 μg of protein per lane were separated on 10% SDS-PAGE gels in running buffer (25 mM Trizma base, [2-Amino-2-(hydroxymethyl)-1,3-propanediol, THAM, Tris base, Tris(hydroxymethyl)aminomethane, Trometamol] 192 mM glycine and 0.1% SDS). After electrophoresis, proteins were transferred to PVDF membranes in a Tris–glycine–20% methanol buffer (25 mM Trizma base, 192 mM glycine and 20% methanol); membranes were blocked with 5% skim milk in Tris-buffered saline (10 mM Tris, pH 7.6, and 150 mM NaCl) with 0.1% Tween-20 (TBS-T) followed by incubation with either moesin monoclonal antibody (0.5 μg/ mL; Sigma) or phospho-specific moesin antibody (1:1000 dilution; Santa Cruz Biotechnology) in 5% skim milk in TBS-T. After washing with TBS-T, membranes were incubated with the appropriate HRP-conjugated secondary antibody (1:10,000 dilution) in 5% skim milk in TBS-T, and developed using enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL, USA). Autoradiograms were quantified using the Bio-Rad imaging system.
Preparation of RNA, reverse transcription and quantitative real-time PCR (Q-PCR)
Total RNA was isolated with TRIzol® using standard procedures (Invitrogen) and quantified by spectrometry at 260 and 280 nm (Bio-Rad). Complementary DNA first strand synthesis was carried out using approximately 50 ng of total RNA primed with random hexamers and reverse transcribed using AML-V reverse transcriptase (Applied Biosystems). Thermal cycler conditions were 25°C for 10 min, 48°C for 30 min and 95°C for 5 min (ABI 9700; Applied Biosystems). Quantitative real-time PCR was performed using primers and TaqMan probes labeled with MGBFAM dye specific for human moesin (Assays-on-demand; Applied Biosystems). Standards for moesin and β-actin were obtained from a 10-fold serial dilution of cDNA from THP-1 cells. Human β-actin was selected as an endogenous control and was amplified using preformulated VIC-TAMRA labeled TaqMan probes (Endogenous Control; Applied Biosystems). Quantification was performed in an automated thermal cycler (ABI Prism 7000; Applied Biosystems) according to standard protocols. Experiments were performed in duplicate for each data point.
Native PAGE gel shift assay for moesin and LPS
Gel shift assays were performed as previously described (42). Briefly, recombinant proteins (30 μg/mL) for moesin and radixin (N- and C-terminal) were incubated with sonicated LPS (0–500 μg/mL) in phosphate-buffered saline (PBS) lacking Ca2+ and Mg2+ with 1 mM EDTA at 37°C for 30 min. Glycerol and bromophenol blue were added, and the mixture was electrophoresed using 4–20% non-denaturing PAGE at 100–150 V for 2 h in a running buffer containing 192 mM glycine, 24 mM Tris, pH 8.3, without detergents. Silver staining was performed according to the manufacturer‘s instructions (Bio-Rad).
Immunoprecipitation
Cell lysates (0.5–1 mg) were precleared with normal mouse IgG and protein A/ G agarose beads at 4°C for 1 h, followed by overnight immunoprecipitation with goat anti-moesin polyclonal antibody (1 μg/mL) and protein A/G agarose beads (20 μL). Beads were collected by centrifugation for 30 s, washed three times with lysis buffer, and resuspended in sample buffer, then boiled for 5 min followed by SDSPAGE. Western blots were developed with anti-TLR4 (1:500 dilution), anti-MD-2 (1:750 dilution), anti-CD14 (1:500 dilution) or anti-β-actin antibody (1:250 dilutions). Horseradish peroxidase conjugate was used as the developing agent. Proteins were visualized on X ray film using the Pierce chemiluminescence system (Pierce Biotechnology). Results were confirmed in three independent experiments.
Co-immunoprecipitation assay for MyD88–IRAK and IRAK–TRAF6
One microgram of normal goat IgG was added together with 20 μL of protein plus A/G agarose to approximately 1 mL of whole cell lysate (0.5–1 mg/mL protein concentration) and incubated at 4°C for 60 min. Beads were pelleted by centrifugation at 1000g, and supernatant was transferred to a fresh microcentrifuge tube. Ten micograms of MyD88 antibody–agarose conjugate was then added and incubated at 4°C overnight with mixing. Beads were collected by centrifugation, and supernatant was carefully discarded. Beads were washed with lysis buffer, centrifuged, and the pellet was resuspended in 50 μL of SDS sample buffer. After boiling, samples were loaded on 10% gels. After electrophoresis, western blotting using anti-IRAK antibody (BD Pharmingen) at 1:500 dilution was performed. For IRAK–TRAF6 co-immunoprecipitation, 4 μg anti-IRAK rabbit IgG was incubated with cell lysate overnight at 4°C. Protein A/G beads were then incubated with the lysate for 2 h at 4°C. Beads were collected and washed as described above. After transfer, PVDF membranes were probed with TRAF6 monoclonal antibody (1:1000 dilution).
Phosphorylation assay for Interleukin-1 receptor-associated kinase (IRAK)
Interleukin-1 receptor-associated kinase rabbit IgG (8 μg/mL) was incubated with cell lysate overnight at 4°C, followed by addition of 20 μL of protein A/G agarose beads and incubation for another 2 h. Beads were washed with lysis buffer and resuspended in SDS sample buffer. After boiling, samples were loaded on 8% gels. After SDS-PAGE and transfer to PVDF, membranes were probed with phospho-threonine specific antibody (1:750 dilution).
Assay of MAPK activity
Macrophages were stimulated with LPS in 12-well tissue culture plates. For blocking experiments, cells were pre-incubated with anti-moesin monoclonal antibody or an isotype-matched control antibody. The p38 inhibitor SB202190 and p44/42 inhibitor PD98059 were used as controls (43–46). After LPS stimulation, media were aspirated, and cells were rinsed once with ice-cold PBS. Cells were lysed and transferred to microcentrifuge tubes. Cell lysates were sonicated, and supernatant was collected by microcentrifugation at 16,000g for 15 min at 4°C. The protein concentration of each sample was adjusted to 1 mg/mL, and samples were incubated with immobilized phospho-specific monoclonal antibodies to either p44/42 or p38. After overnight incubation, beads were microcentrifuged, the pellet was washed and suspended in 50 μL kinase buffer supplemented with 200 μM ATP and 2 μg Elk-1 fusion protein for p44/ 42 or 2 μg ATF-2 fusion protein for p38, followed by incubation for 30 min at 30°C. The reaction was terminated by adding 3× SDS sample buffer. Samples were boiled, and 30 μL was loaded on 10% SDS-PAGE gels. After transfer, PVDF membranes were blocked in TBS-T and 5% skim milk and incubated with the primary antibody, which corresponded to the specific kinase (1:1000 dilution) in TBS-T containing 5% BSA. After washing, membranes were incubated with HRP-conjugated anti-rabbit secondary antibody (1:2000 dilution) in blocking buffer, washed, and developed using enhanced chemiluminescence.
Analysis of NF-κB activation
Activation of the p65 subunit of NF-κB in macrophages was determined by ELISA (Imgenex; 47,48). The specificity of the assay was verified by including an excess of soluble oligonucleotides containing native or mutated NF-κB consensus binding site. After LPS stimulation, cells were harvested, centrifuged (500 g for 15 min), and resuspended in hypotonic lysis buffer. Ten per cent of NP-40 (nonyl phenoxylpolyethoxylethanol) was added, and samples were blended by vortex mixing and centrifuged. The supernatant cytoplasmic extracts were collected; cold nuclear extraction buffer was added to the pellets, incubated on ice with intermittent vortex mixing and, after centrifugation (500 g for 15 min), the supernatant (nuclear extract) was collected. The NF-κB/p65 ActivELISA Kit measures free p65 in the nuclear extract.
Western blotting for IκBα
Equal amounts of protein from cell lysates were separated by 10% SDS-PAGE. After transfer, membranes were blocked in TBS-T supplemented with 5% skim milk, and membranes were probed with phospho-specific IκBα antibody (1:1000 dilution) in TBS-T with 5% BSA. After washing, membranes were incubated with HRP-conjugated secondary antibody, and proteins were detected using enhanced chemiluminescence.
ELISA for TNF-α, IL-1β and IFN-β
In order to test the impact of endotoxin activation on the MyD88-dependent and -independent pathways, macrophages were differentiated from THP-1 cells in 12-well tissue culture plates at a concentration of 8 × 106 cells per well. Non-adherent cells were aspirated, and the wells were washed twice with PBS. After the final wash, cells were stimulated with different concentrations of LPS, in duplicate, and the plates were incubated for 18 h at 37°C in an atmosphere of air containing 5% CO2. For blocking experiments, cells were pretreated with anti-moesin antibody for 2 h prior to LPS stimulation. Tumor necrosis factor α and IL-1β were measured as the resultant cytokines of the MyD88- dependent pathway, while IFN-β was analyzed as a measure of MyD88- independent pathway regulation. The release of TNF-α, IL-1β and IFN-β into the culture supernatant was assayed using a commercially available ELISA performed according to the manufacturer’s instructions (R&D Systems).
Statistical analysis
Data were evaluated by analysis of variance (ANOVA) with Bonferroni multiple-comparison test using the SPSS program (SPSS Inc., Chicago, IL, USA). Statistical differences were considered significant at an α level of 0.05. All reported experiments were performed at least three times.
Results
Characterization of moesin expression in macrophages stimulated with LPS
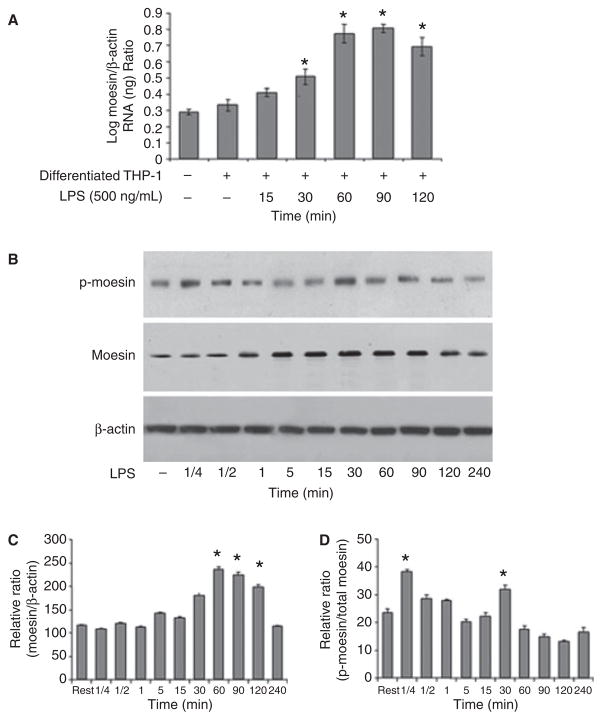
Previously, we have shown that total moesin protein is upregulated after LPS challenge; however, we did not analyze its conformational appearance (7). Expression analyses using Q-PCR revealed an increase of moesin mRNA levels within 30 min after challenging macrophages with LPS (Fig. 1A). In line with this, protein levels reached a maximum 60 min after exposing macrophages to LPS, as assessed by western blotting (Fig. 1B,C). Additionally, moesin exhibited rapid phosphorylation after only 15 s of LPS stimulation (Fig. 1B,D). This was followed by a significant dephosphorylation after 5 min. A second peak of phosphorylation was observed at 30 min that, again, was followed by dephosphorylation at 60 min (Fig. 1B,D), indicating that moesin is positively regulated and converts into an active form during the macrophage response to LPS.
Fig 1.
Expression and phosphorylation of moesin after lipopolysaccharide (LPS) stimulation. (A) Moesin is increasingly expressed in LPS-stimulated macrophages as determined by Q-PCR. Changes in expression are significant at 30, 60, 90 and 120 min compared with unstimulated cells (*p < 0.05). (B) Phosphorylation of moesin and total moesin protein are compared to β-actin by western blot. Phosphorylation of moesin is rapid, appearing at 15 s followed by dephosphorylation and a second phosphorylation at 30 min. The second phosphorylation event coincides with the increase in moesin protein. (C) Total moesin protein per cell is expressed as a ratio of moesin to β-actin. There is a significant increase in total moesin protein observed at 60, 90 and 120 min (*p < 0.05). (D) Relative ratio of phospho-moesin to total moesin indicates the phosphorylation events at 15 s and 30 min (*p < 0.05).
Interaction of moesin and LPS in macrophages
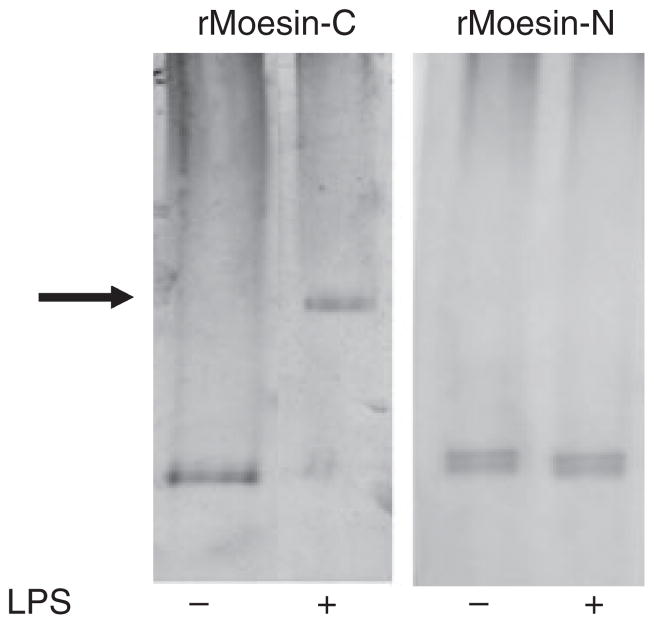
Since moesin is present on cell membranes, we used a native PAGE gel shift assay to examine whether LPS can bind directly to this cytoskeletal protein. Two recombinant fragments of moesin were examined, the C-terminal and N-terminal domains. Recombinant radixin, an ERM family member protein (both C- and N-terminus), was used as control. Incubation of LPS with the C-moesin protein fragment resulted in a protein retardation in the gel, as shown in Fig. 2, suggesting a stable binding of LPS to the recombinant C-moesin domain. No shift was observed when LPS was incubated with N-moesin or with C- or N-radixin (data not shown), indicating a specific interaction of LPS with the C-terminal domain of moesin.
Fig 2.
Native PAGE gel shift analysis of moesin–LPS complex. Carboxyl and amine halves of recombinant moesin (rMoesin-C, rMoesin-N) and radixin (30 μg) were incubated in the absence and presence of LPS at 37°C for 30 min. A shift in the electrophoresis mobility (arrow) was only detected when rMoesin-C was incubated with LPS (500 μg/mL). No shift was observed for rMoesin-N or for either the carboxyl or the amine half of recombinant radixin (data not shown).
Moesin associates with proteins specifically involved in LPS-activated pathways
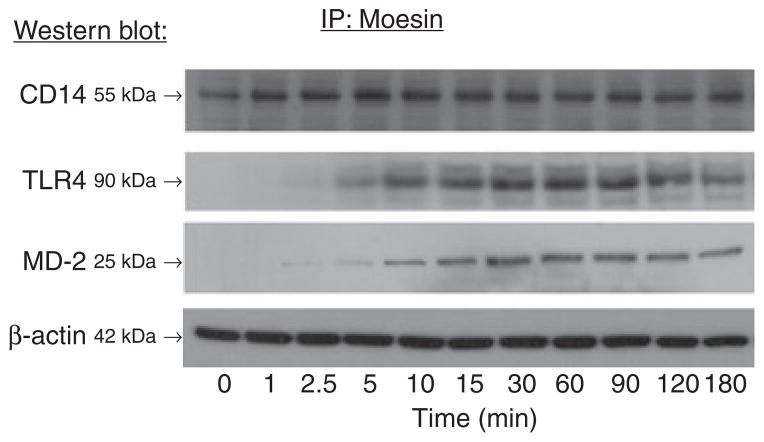
To evaluate the participation of moesin in propagating the LPS signal, immunoprecipitation with anti-moesin antibody was performed after stimulating macrophages with LPS. Afterwards, moesin-bound proteins were identified using western blotting with anti-CD14, anti-TLR4 and anti-MD-2 antibodies. As shown in Fig. 3, CD14 co-precipitated with moesin at all time points, with or without LPS stimulation. In contrast, TLR4 and MD-2 co-precipitated with moesin only after the cells were stimulated with LPS, revealing a continuous binding of moesin to CD14 and a specific association of moesin with TLR4 and MD-2.
Fig 3.
Immunoprecipitation (IP) and western blotting. Cell lysates from control and LPS-treated samples were immunoprecipitated with anti-moesin antibody, followed by western blotting using antibodies against CD14, TLR4 or MD-2. Each sample was separated by SDS-PAGE prior to immunoprecipitation, and western blotting was performed for β-actin as a control for equal loading. The results show that CD14 co-precipitated with moesin in both the control and LPS-stimulated conditions, with a stronger signal in stimulated conditions. There was no association observed between TLR4 or MD-2 and moesin in the resting cell lysates. After 5 min of stimulation, both TLR4 and MD-2 were observed to co-precipitate with moesin.
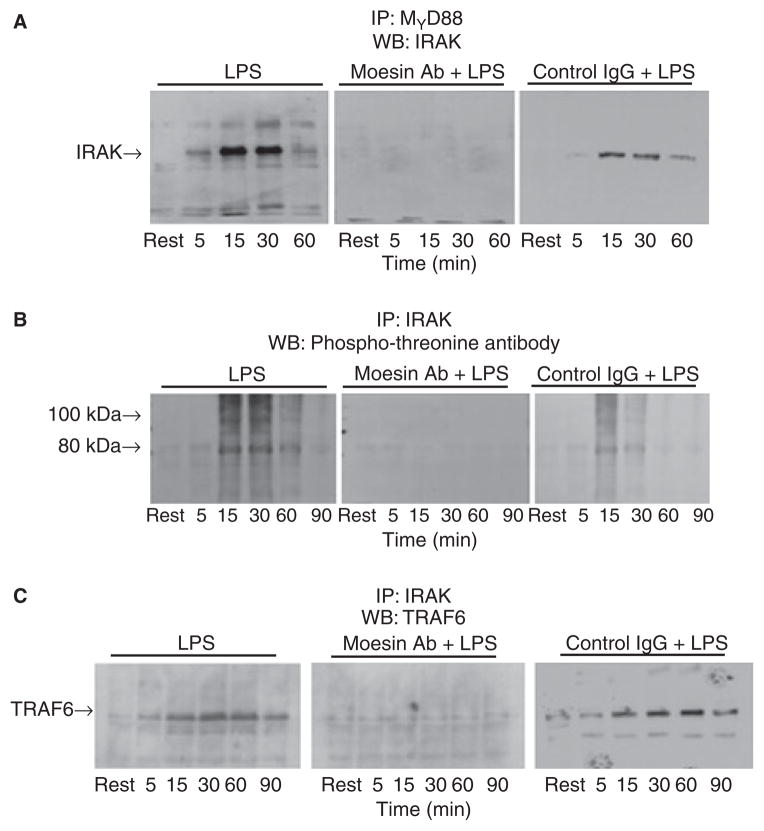
Inhibition of moesin function interferes with LPS-induced recruitment of IRAK to MyD88 and subsequent activation of TRAF6
MyD88 is essential for the recruitment and phosphorylation of IRAK (19). We, therefore, investigated the inhibition of this interaction in LPS-treated cells with or without blocking moesin function using an anti-moesin antibody. As shown in Fig. 4A, anti-moesin antibody inhibited the association of IRAK with MyD88. Cells stimulated with LPS exhibited phosphorylation of IRAK (Fig. 4B), but cells pre-incubated with anti-moesin antibody exhibited total inhibition of IRAK phosphorylation. Moreover, for proximal signals to transduce, IRAK binds and activates TRAF6 (31). Immunoprecipitation with IRAK antibody followed by western blotting for TRAF6 was performed. Cells pretreated with anti-moesin antibody showed inhibition of IRAK binding to TRAF6 comparable to the resting conditions (Fig. 4C). No inhibition of LPS-induced signaling events was observed when the cells were incubated with an isotype-matched control IgG.
Fig 4.
Inhibition of IRAK, MyD88 and IRAK activation and phosphorylation by anti-moesin antibody. Stimulated differentiated THP-1 cells, with and without prior treatment with anti-moesin antibody or an isotype-matched control antibody, were lysed (as described in the ‘Material and methods’ section) and subjected to immunoprecipitation (IP) with anti- MyD88 goat IgG followed by western blotting (WB) for IRAK (A), immunoprecipitation with IRAK rabbit IgG followed by western blotting with an antibody that recognizes threonine phosphorylation (B), or immunoprecipitation with IRAK rabbit IgG followed by western blotting with anti-TRAF6 (C). In cells pretreated with anti-moesin antibody, recruitment of IRAK to MyD88 was inhibited and subsequent IRAK phosphorylation was blocked (A and B, respectively). Furthermore, IRAK binding to TRAF6 was also inhibited (C). No effects were observed when the differentiated THP-1 cells were pre-incubated with the isotype-matched control antibody prior to LPS stimulation.
Lipopolysaccharide-induced p38 and p44/42 MAP kinase activity in macrophages with impaired moesin function
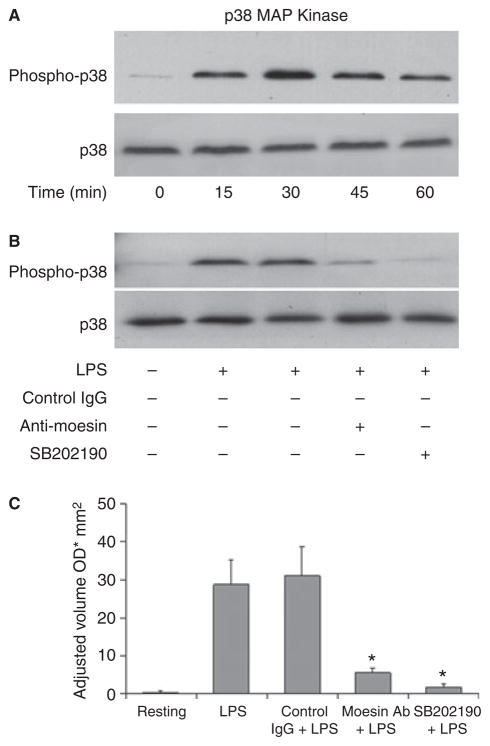
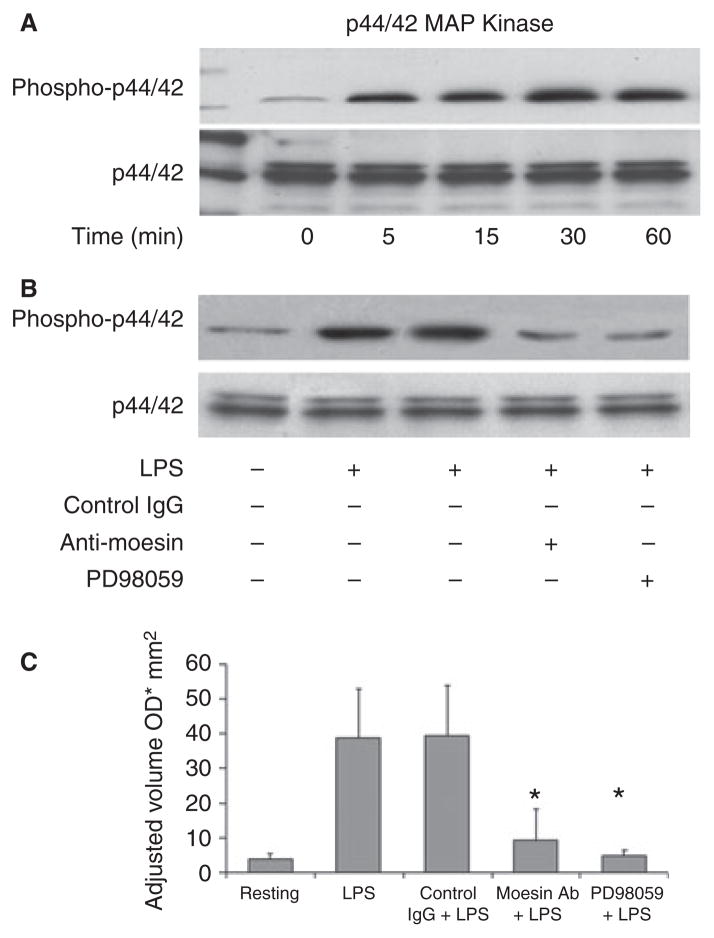
In order to determine which further signaling cascade is being inhibited by anti-moesin antibody, the activation and phosphorylation of p38 and p44/ 42 MAPK in response to LPS were analyzed. Results revealed that both p38 and p44/42 MAPK were activated and reached their maximal phosphorylation at 30 min (Figs 5A,6A), but phosphorylation was blocked with anti-moesin antibody (Figs 5B,6B). Specific inhibitors of p38 and p44/42 were used as positive controls (Figs 5B,6B), and densitometric scans were used to quantify the western blots (Figs 5C,6C).
Fig 5.
Effect of anti-moesin antibody on p38 MAPK activity. (A) Normal p38 activation by LPS (500 ng/mL) was initially analyzed, and maximal activation was observed at 30 min. (B) Differentiated THP-1 cells were pretreated with anti-moesin antibody (10 μg/mL), isotype- matched control antibody (10 μg/mL) or the p38 inhibitor SB202190 (25 μM). Cells pretreated with anti-moesin antibody or SB202190 prior to LPS stimulation showed significantly less p38 activation. (C) The gels were quantified by densitometry, and density was expressed as adjusted volume (OD/mm2).
Fig 6.
Effect of anti-moesin antibody on p44/42 MAPK activity. (A) Normal p44/42 activation by LPS (500 ng/mL) was initially analyzed, and maximal activation was observed at 30 min. (B) For blocking experiments, macrophages were pretreated with anti-moesin antibody (10 μg/mL), isotype-matched control antibody (10 μg/mL) or the p44/42 inhibitor PD98059 (25 μM). Cells pretreated with anti-moesin antibody or PD98059 prior to LPS stimulation showed significantly less p44/42 activation. (C) The gels were quantified by densitometry, and density expressed as adjusted volume (OD/mm2).
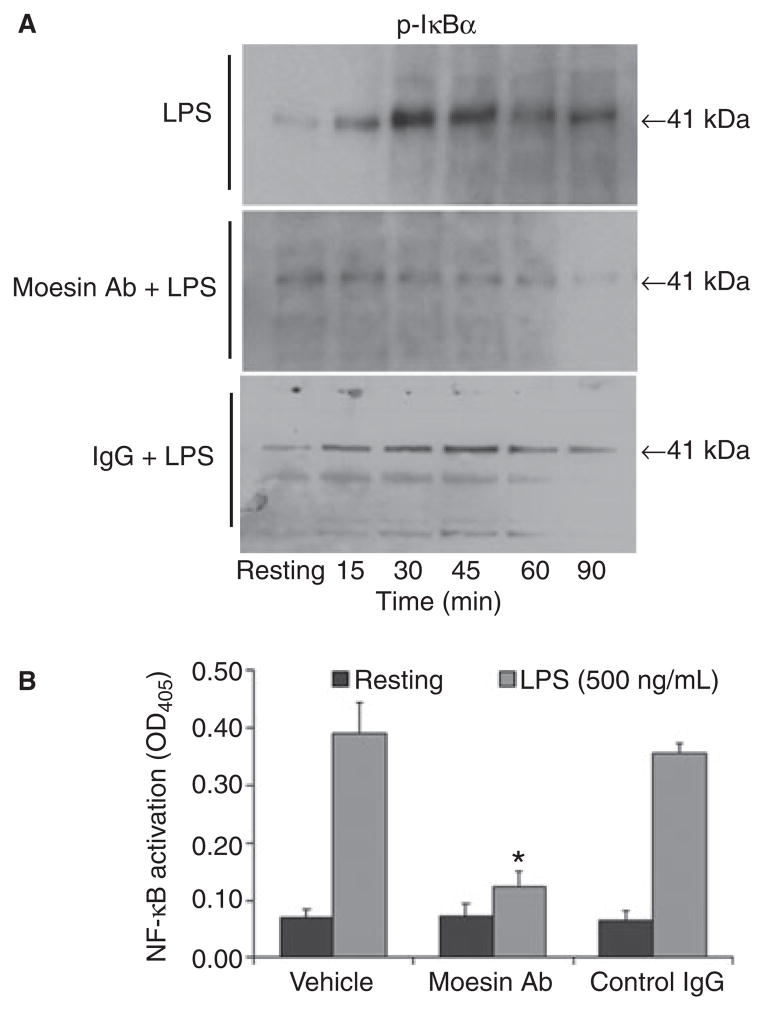
Inhibition of IκBα phosphorylation and NF-κB activation in LPS-stimulated macrophages with impaired moesin function
To assess NF-κB activation downstream to moesin phosphorylation and association with LPS, TLR4 and MD- 2, the NF-κB ELISA was used to measure free p65 in the nuclear extract. As seen in Fig. 7, LPS challenge resulted in IκBα phosphorylation, followed by the translocation and activation of the NF-κB dimers, but phosphorylation of IκBα was blocked by anti-moesin antibody, and NF-κB activity was significantly inhibited.
Fig 7.
Phosphorylation of IκBα, and translocation and activation NF-κB. Macrophages were stimulated with LPS (500 ng/mL). For blocking experiments, cells were treated with anti-moesin antibody (10 μg/mL) prior to LPS stimulation. Cytoplasmic and nuclear extractions were performed according to the reagent supplier’s recommendations (Imgenex). The phosphorylated form of IκBα was detected in cytoplasmic extracts by western blotting. Translocation and activation of NF-κB were measured using the active ELISA assay of nuclear extracts. (A) Phosphorylation of IκBα stimulated by LPS. No phosphorylation was observed in cells pretreated with anti-moesin antibody. (B) Activation of NF-κB and its translocation into the nucleus upon LPS stimulation. When cells were pretreated with anti-moesin antibody, no translocation was observed. No effects were observed when the differentiated THP-1 cells were pre-incubated with an isotype-matched control IgG. *p < 0.05 compared to vehicle and control.
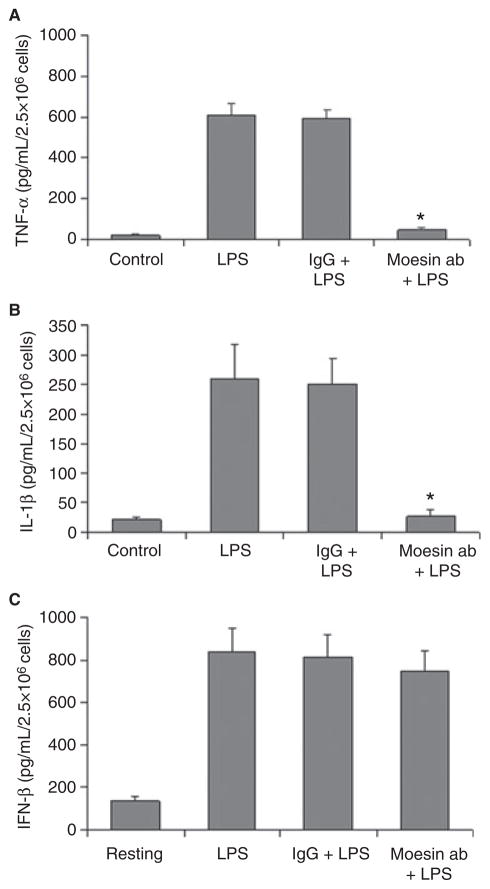
Role of moesin in MyD88-dependent and independent pathways
To examine the effect of anti-moesin antibody on the MyD88-independent pathway, an ELISA for IFN-β was performed (Fig. 8). No inhibition of the IFN-β secretion was observed when the cells were pre-incubated with anti-moesin antibody (Fig. 8C), suggesting that moesin-mediated signaling does not involve the MyD88-independent pathway. However, consistent with previous reports (7,8,38), we observed a significant reduction in TNF-α and IL-1β secretion when cells were pre-incubated with anti-moesin antibody prior to LPS stimulation (p < 0.05), whereas cells pre-incubated with an isotype-matched control IgG showed no change (Fig. 8A,B). Taken together, the above observations indicate a crucial mediator role for active moesin in signaling events associated with LPS in macrophages. Interestingly, moesin seems to be specifically involved with pathways associated with MyD88 and its downstream signaling transduction.
Fig 8.
Effect of moesin on TNF-α and IL-1β secretion in LPS-stimulated cells. Cells were cultured as described in the ‘Material and methods’ section and were pretreated with anti-moesin antibody or an isotype-matched control (10 μg/mL) followed by LPS stimulation (500 ng/mL) for 18 h. Control cells received no antibodies or LPS. Concentrations of TNF-α, IL-1β and IFN-β were determined by ELISA. Data are presented as means of three experiments, with standard deviations. Significance was determined by ANOVA with Bonferroni’s correction for multiple comparisons. There was a significant inhibition of the production of TNF-α and IL-1β in cells pretreated with anti-moesin antibody (*p < 0.05). However, there was no effect on anti-moesin antibody on the secretion of IFN-β.
Discussion
Cellular structure and function depend on the existence of precisely localized protein complexes associated with the plasma membrane and cytoskeletal actin (1). Ezrin–radixin–moesin proteins have been proposed to link transmembrane proteins to the actin cytoskeleton and, more importantly, have been implicated in signaling pathways which incorporate extracellular signals into intracellular mechanisms (3). Previous data suggested an involvement of cytoskeletal proteins in cellular signaling mechanisms, which create the innate immune response against microbial pathogens (7,8). To convert into an active stage, moesin is phosphorylated at threonine-558 by different kinases, such as Rho-activated kinase (6,49); its N- and C-domains are involved in plasma protein and actin filament binding, respectively (50). Lipopolysaccharide is the principle component of the outer membrane of gram-negative bacteria, and its recognition by monocytes/macrophages orchestrates an immediate and active adaptive immune response through the expression of a variety of inflammatory cytokines, including TNF-α and IL-1β (51–53). Production of these inflammatory cytokines contributes to the efficient control of growth and dissemination of invading pathogens. The regulation of cytokine expression is driven by a complex network of receptor and cytoskeletal proteins, kinases and transcription factors, which include CD14, TLR4, MD-2, MAPKs such as ERK1, ERK2 (54), p38 MAPK (55), and c-Jun N-terminal kinases (JNK), NF-κB and others. In the present study, we systematically analyzed the role of the cytoskeletal protein moesin in LPS-induced signaling pathways that ultimately result in the expression of proinflammatory cytokines in macrophages. Upon LPS stimulation, moesin is not only increasingly expressed and phosphorylated, but also can bind LPS directly at its carboxyl-terminus. Furthermore, moesin is temporally associated with TLR4 and MD-2 after LPS stimulation, whereas CD14 is continuously bound to moesin in differentiated THP-1 cells. To analyze the impact of moesin on LPS-induced proximal intracellular signaling, a previously tested blocking anti-moesin antibody was used to inhibit moesin function (38). The data clearly established that inhibition of moesin interrupts LPS response pathways through a blockage of LPS intermediate signaling components, including MyD88, IRAK and TRAF6. This not only negatively affects subsequent activation of the MAP kinases p38 and ERK but also activation of NF-κB and its translocation to the nucleus, where NF-κB regulates TNF-α and IL-1β production. Interestingly, anti-moesin antibody did not block the secretion of IFN-β, which is considered to be a MyD88-independent signaling pathway, suggesting that moesin-mediated intracellular signaling is a specific pathway and depends on the activation of MyD88 (35–37).
Moesin was first described as a structural protein that serves as a link between the plasma membrane and the cytoskeleton (56). Moesin can regulate cell shape, motility, adhesion, vesicular trafficking and signal transduction (1). Analyses of its protein structure revealed that moesin could be incorporated into either the plasma membrane and/or the cytoplasm. Threonine-558 phosphorylation of moesin at its C-terminal portion is a crucial step which ensures activation and free movement of the protein in response to stimuli (57–59). Keresztes et al. (60) suggested that upon adhesion of porcine neutrophils to a plastic surface, activated moesin is translocated to the plasma membrane and the extracellular surface. Ariel et al. (61) reported that moesin could be expressed on the cell surface not only by stimulation with phytohemagglutinin( PHA) or PMA, but also with calyculin A, a specific inhibitor of phosphatases 1 and 2A, which is capable of inducing moesin phosphorylation (59). In addition, moesin and other ERM family members contain at their N-terminus a so-called FERM (band four-point-one, ezrin, radixin, moesin homology domain) with a sequence of three subdomains, F1–F3 (62,63). The cluster of acidic amino acids, especially in the pleckstrin homology domain, has been shown to be important in interactions of ERM proteins with lipid layers of the membrane and is involved in signal transduction (13,62,64–67). Our results suggest that moesin activation mechanisms are consistent with moesin function in macrophages, since moesin was found to be a dynamic molecule with the apparent capability of quick activation (phosphorylation) in response to LPS stimulation. We postulate, based on those results, that phosphorylation of moesin induces a conformational change that exposes cryptic binding sites for additional proteins downstream of LPS signaling.
We detected an association of moesin with CD14, TLR4 and MD-2 proteins. Interestingly, CD14 is continuously linked to moesin, whereas TLR4 and MD-2 are transiently bound to moesin only after stimulation with LPS. In addition, we have confirmed and localized the direct binding of moesin (C-terminus) to LPS upon stimulation. Given the foregoing, it appears that moesin is involved in the early recognition and response to LPS challenge. Previous understanding of the innate recognition of bacterial LPS is based on the discovery that LPS binds to LBP in serum (12,13), which, in turn, rapidly catalyzes the transfer of LPS to membrane-bound CD14 (mCD14) or soluble CD14 (sCD14). Although CD14 has been identified as an LPS receptor (18), it is a GPI-anchored protein and thus lacks transmembrane and intracellular domains (68–70). Several binding studies have shown that CD14-blocking monoclonal antibodies only partially inhibit LPS binding, suggesting the existence of alternative receptors (71). Thus, additional proteins, such as moesin, might act in concert with the LPS–CD14 complex to initiate the signaling process leading to LPS-induced cellular activation.
The TLR proteins possess leucine-rich extracellular repeats that recognize the LBP–CD14 complex (20). In addition, MD-2, an adaptor protein, which forms a complex with the extracellular domain of TLR4, supports effective LPS recognition (72). Our data implicate a role for moesin in the formation of the LPS–receptor complex. Lipopolysaccharide binding to moesin causes activation by phosphorylation, which might expose moesin-binding domains for binding to TLR4 and MD-2 and initiates LPS signaling. Since TLR4–MD-2 is tightly connected to the innate immune response, the association of moesin with TLR4–MD-2 reveals a new mechanism and illustrates additional diversity and redundancy in the biological activity of these two proteins. Previous studies attempted to identify proteins involved in LPS recognition and binding. Akashi et al. (73) stated that their results using mechanically solubilized cell lysates clearly showed physical association between LPS and TLR4–MD-2; however, these were co-immunoprecipitation experiments, and direct binding was not actually demonstrated. The only evidence suggesting that there is a direct interaction between LPS and membrane proteins forming a membrane receptor complex was in transfected cells, in which an interaction was demonstrated between TLR, MD-2 and CD14 (alone or various combinations) with cross-linking to LPS (74). In these studies, LPS cross-linked to TLR4 and MD-2 only when co-expressed with CD14. Here, we demonstrate direct binding of LPS to moesin using a gel shift assay and localize the binding site to the C-terminus of the moesin molecule.
Our new observation of a physical association between moesin and CD14, TLR4 and MD-2 emphasizes the functional importance of moesin in the activation of an LPS receptor cluster. Bacterial recognition systems in mammalian cells are complex and require several components to relay signals. Triantafilou & Triantafilou (74) hypothesized that different receptors are recruited to the site of ligation to form an activation cluster, which is followed by multiple signaling cascades. Thus, the existence of a moesin–CD14–TLR4–MD-2 receptor cluster is consistent with the variety of signaling cascades that can be triggered by LPS.
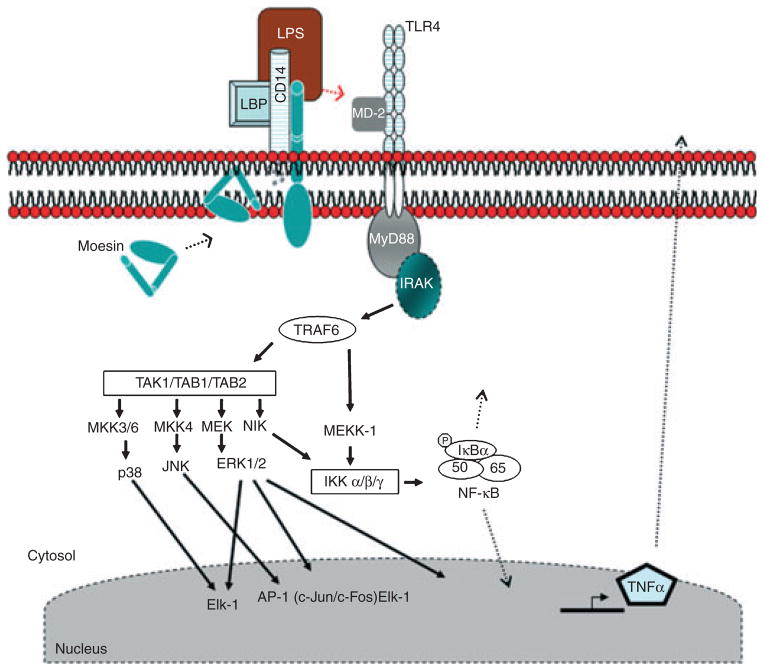
Based upon these findings, we suggest a new model for early LPS signaling events, in which moesin participates with CD14, TLR4 and MD-2 in LPS recognition (Fig. 9). In this model, LPS initially binds to CD14, which is associated in the membrane with moesin, inducing phosphorylation of moesin. The CD14–moesin complex then associates with the signal transducing proteins, TLR4 and MD-2. After the initial phosphorylation (unfolding) of moesin induced by LPS challenge, the observed dephosphorylation of moesin may be the result of its binding to TLR4/MD-2. Essentially, moesin functions as an LPS-presenting protein to the TLR4–MD-2 complex. The four-molecule cluster recruits IRAK through the adaptor protein MyD88, and IRAK autophosphorylates. Phospho-IRAK dissociates from MyD88 and binds to TRAF6, which then phosphorylates IKK (IκB kinase) and MAPK. These kinases in turn phosphorylate downstream kinases, leading to the activation of p38 and p44/42 as well as NF-κB, among others, ultimately leading to the transcription and secretion of proinflammatory cytokines (e.g. TNF-α).
Fig 9.
Proposed model for LPS recognition and signaling. In the absence of stimulation, our model shows CD14 and moesin associated in the cell membrane in macrophages. Following LPS stimulation, moesin phosphorylation is significantly increased and the moesin binds to TLR4 and MD-2. This receptor cluster then activates the adaptor protein MyD88. The MyD88 recruits and activates IRAK through their respective death domains. The IRAK subsequently autophosphorylates, dissociates from MyD88 and interacts with TRAF6. Active TRAF6 then activates and phosphorylates MEKK-1 or MKK3/6, MKK4, MEK and NIK. Both NIK and MEKK-1 are activators of IKK, which in turn phosphorylates IκBα. The phosphorylation of IκBα results in its dissociation and degradation, freeing NF-κB. The NF-κB translocates to the nucleus, where transcription begins. TAB1, TAB2 and TAK1 are activators of p38, JNK and ERK1/2 (p44/42). The activation of these MAPKs leads to the production of proinflammatory cytokines, such as TNF-α. (LBP, LPS-binding protein; MEK, MAP kinase/ERK kinase; MKK, MAP kinase kinase; MEKK, MAP kinase kinase; NIK, B-inducing kinase).
In conclusion, the activation of moesin in monocytes/macrophages is dynamic and appears to be regulated positively by LPS. We propose that the innate recognition of LPS involves the dynamic association of multiple receptors to form a cluster of molecules. The binding site and specific residues of moesin that bind CD14, TLR and MD-2 remain to be elucidated. Complete identification of the diverse receptor clusters that are responsible for bacterial recognition will enhance current understanding and therapeutic intervention associated with antibacterial therapy in periodontics.
Acknowledgments
This work was supported by the USPHS grants DE-16191 (T.E.V.D., S.A., K.H.Z. and A.K.), DE15566 (T.E.V.D. and A.K.), DE14079 (S.A.) and DE12482 (S.A.).
References
- 1.Gautreau A, Louvard D, Arpin M. ERM proteins and NF2 tumor suppressor: the Yin and Yang of cortical actin organization and cell growth signaling. Curr Opin Cell Biol. 2002;14:104–109. doi: 10.1016/s0955-0674(01)00300-3. [DOI] [PubMed] [Google Scholar]
- 2.Amieva MR, Furthmayr H. Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell-cell contacts. Exp Cell Res. 1995;219:180–196. doi: 10.1006/excr.1995.1218. [DOI] [PubMed] [Google Scholar]
- 3.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 4.Masumoto J, Sagara J, Hayama M, Hidaka E, Katsuyama T, Taniguchi S. Differential expression of moesin in cells of hematopoietic lineage and lymphatic systems. Histochem Cell Biol. 1998;110:33–41. doi: 10.1007/s004180050262. [DOI] [PubMed] [Google Scholar]
- 5.Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautreau A, Louvard D, Arpin M. Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J Cell Biol. 2000;150:193–203. doi: 10.1083/jcb.150.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amar S, Oyaisu K, Li L, Van Dyke T. Moesin: a potential LPS receptor on human monocytes. J Endotoxin Res. 2001;7:281–286. [PubMed] [Google Scholar]
- 8.Iontcheva I, Amar S, Zawawi KH, Kantarci A, Van Dyke TE. Role for moesin in lipopolysaccharide-stimulated signal transduction. Infect Immun. 2004;72:2312–2320. doi: 10.1128/IAI.72.4.2312-2320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muroi M, Muroi Y, Yamamoto K, Suzuki T. Influence of 3’ half-site sequence of NFkappa B motifs on the binding of lipopolysaccharide-activatable macrophage NF-kappa B proteins. J Biol Chem. 1993;268:19534–19539. [PubMed] [Google Scholar]
- 10.Fujihara M, Muroi M, Muroi Y, Ito N, Suzuki T. Mechanism of lipopolysaccharide-triggered junB activation in a mouse macrophage-like cell line (J774) J Biol Chem. 1993;268:14898–14905. [PubMed] [Google Scholar]
- 11.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 12.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 13.Schumann RR, Leong SR, Flaggs GW, et al. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 14.Gegner JA, Ulevitch RJ, Tobias PS. Lipopolysaccharide (LPS) signal transduction and clearance. Dual roles for LPS binding protein and membrane CD14. J Biol Chem. 1995;270:5320–5325. doi: 10.1074/jbc.270.10.5320. [DOI] [PubMed] [Google Scholar]
- 15.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 16.Haziot A, Ferrero E, Kontgen F, et al. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 17.Wurfel MM, Monks BG, Ingalls RR, et al. Targeted deletion of the lipopolysaccharide (LPS)-binding protein gene leads to profound suppression of LPS responses ex vivo, whereas in vivo responses remain intact. J Exp Med. 1997;186:2051–2056. doi: 10.1084/jem.186.12.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- 19.Medzhitov R, Preston-Hurlburt P, Kopp E, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 20.Poltorak A, Ricciardi-Castagnoli P, Citterio S, Beutler B. Physical contact between lipopolysaccharide and toll-like receptor 4 revealed by genetic complementation. Proc Natl Acad Sci U S A. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhary PM, Ferguson C, Nguyen V, et al. Cloning and characterization of two Toll/Interleukin-1 receptor-like genes TIL3 and TIL4: evidence for a multi-gene receptor family in humans. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 23.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 25.Means TK, Golenbock DT, Fenton MJ. The biology of Toll-like receptors. Cytokine Growth Factor Rev. 2000;11:219–232. doi: 10.1016/s1359-6101(00)00006-x. [DOI] [PubMed] [Google Scholar]
- 26.Sato S, Takeuchi O, Fujita T, Tomizawa H, Takeda K, Akira S. A variety of microbial components induce tolerance to lipopolysaccharide by differentially affecting MyD88-dependent and -independent pathways. Int Immunol. 2002;14:783–791. doi: 10.1093/intimm/dxf046. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Natoli G, Saccani S, Levrero M, Mantovani A. The human toll signaling pathway: divergence of nuclear factor kappaB and JNK/SAPK activation upstream of tumor necrosis factor receptor-associated factor 6 (TRAF6) J Exp Med. 1998;187:2097–2101. doi: 10.1084/jem.187.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raschi E, Testoni C, Bosisio D, et al. Role of the MyD88 transduction signaling pathway in endothelial activation by antiphospholipid antibodies. Blood. 2003;101:3495–3500. doi: 10.1182/blood-2002-08-2349. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Q, Akashi S, Miyake K, Petty HR. Lipopolysaccharide induces physical proximity between CD14 and toll-like receptor 4 (TLR4) prior to nuclear translocation of NF-kappa B. J Immunol. 2000;165:3541–3544. doi: 10.4049/jimmunol.165.7.3541. [DOI] [PubMed] [Google Scholar]
- 31.Yamin TT, Miller DK. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- 32.Swantek JL, Tsen MF, Cobb MH, Thomas JA. IL-1 receptor-associated kinase modulates host responsiveness to endotoxin. J Immunol. 2000;164:4301–4306. doi: 10.4049/jimmunol.164.8.4301. [DOI] [PubMed] [Google Scholar]
- 33.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 34.Akira S, Hoshino K, Kaisho T. The role of Toll-like receptors and MyD88 in innate immune responses. J Endotoxin Res. 2000;6:383–387. [PubMed] [Google Scholar]
- 35.Kawai T, Takeuchi O, Fujita T, et al. Lipopolysaccharide stimulates theMyD88- independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 37.Hoebe K, Du X, Georgel P, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 38.Tohme ZN, Amar S, Van Dyke TE. Moesin functions as a lipopolysaccharide receptor on human monocytes. Infect Immun. 1999;67:3215–3220. doi: 10.1128/iai.67.7.3215-3220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchiya S, Kobayashi Y, Goto Y, et al. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Hailman E, Lichenstein HS, Wurfel MM, et al. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ajizian SJ, English BK, Meals EA. Specific inhibitors of p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways block inducible nitric oxide synthase and tumor necrosis factor accumulation in murine macrophages stimulated with lipopolysaccharide and interferon-gamma. J Infect Dis. 1999;179:939–944. doi: 10.1086/314659. [DOI] [PubMed] [Google Scholar]
- 44.Guha M, O’Connell MA, Pawlinski R, et al. Lipopolysaccharide activation of the MEK-ERK1/2 pathway in human monocytic cells mediates tissue factor and tumor necrosis factor alpha expression by inducing Elk-1 phosphorylation and Egr-1 expression. Blood. 2001;98:1429–1439. doi: 10.1182/blood.v98.5.1429. [DOI] [PubMed] [Google Scholar]
- 45.Manthey CL, Wang SW, Kinney SD, Yao Z. SB202190, a selective inhibitor of p38 mitogen-activated protein kinase, is a powerful regulator of LPS-induced mRNAs in monocytes. J Leukoc Biol. 1998;64:409–417. doi: 10.1002/jlb.64.3.409. [DOI] [PubMed] [Google Scholar]
- 46.Rutault K, Hazzalin CA, Mahadevan LC. Combinations of ERK and p38 MAPK inhibitors ablate tumor necrosis factor-alpha (TNF-alpha) mRNA induction. Evidence for selective destabilization of TNF-alpha transcripts. J Biol Chem. 2001;276:6666–6674. doi: 10.1074/jbc.M005486200. [DOI] [PubMed] [Google Scholar]
- 47.Hajishengallis G, Martin M, Schifferle RE, Genco RJ. Counteracting interactions between lipopolysaccharide molecules with differential activation of toll-like receptors. Infect Immun. 2002;70:6658–6664. doi: 10.1128/IAI.70.12.6658-6664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mancuso G, Midiri A, Beninati C, et al. Mitogen-activated protein kinases and NF-kappa B are involved in TNF-alpha responses to group B streptococci. J Immunol. 2002;169:1401–1409. doi: 10.4049/jimmunol.169.3.1401. [DOI] [PubMed] [Google Scholar]
- 49.Yonemura S, Matsui T, Tsukita S. Rhodependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J Cell Sci. 2002;115:2569–2580. doi: 10.1242/jcs.115.12.2569. [DOI] [PubMed] [Google Scholar]
- 50.Tsukita S, Yonemura S. ERM (ezrin/radixin/ moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- 51.Beutler B, Krochin N, Milsark IW, Luedke C, Cerami A. Control of cachectin (tumor necrosis factor) synthesis: mechanisms of endotoxin resistance. Science. 1986;232:977–980. doi: 10.1126/science.3754653. [DOI] [PubMed] [Google Scholar]
- 52.Nathan CF. Secretory products of macrophages. J Clin Invest. 1987;79:319–326. doi: 10.1172/JCI112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raetz CR. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein SL, Sanghera JS, Lemke K, DeFranco AL, Pelech SL. Bacterial lipopolysaccharide induces tyrosine phosphorylation and activation of mitogenactivated protein kinases in macrophages. J Biol Chem. 1992;267:14955–14962. [PubMed] [Google Scholar]
- 55.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 56.Lankes WT, Furthmayr H. Moesin: a member of the protein 4.1-talin-ezrin family of proteins. Proc Natl Acad Sci U S A. 1991;88:8297–8301. doi: 10.1073/pnas.88.19.8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivetic A, Ridley AJ. Ezrin/radixin/moesin proteins and Rho GTPase signalling in leucocytes. Immunology. 2004;112:165–176. doi: 10.1111/j.1365-2567.2004.01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura F, Amieva MR, Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem. 1995;270:31377–31385. doi: 10.1074/jbc.270.52.31377. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura F, Amieva MR, Hirota C, Mizuno Y, Furthmayr H. Phosphorylation of 558T of moesin detected by site-specific antibodies in RAW264.7 macrophages. Biochem Biophys Res Commun. 1996;226:650–656. doi: 10.1006/bbrc.1996.1410. [DOI] [PubMed] [Google Scholar]
- 60.Keresztes M, Lajtos Z, Fischer J, Dux L. Moesin becomes linked to the plasma membrane in attached neutrophil granulocytes. Biochem Biophys Res Commun. 1998;252:723–727. doi: 10.1006/bbrc.1998.9730. [DOI] [PubMed] [Google Scholar]
- 61.Ariel A, Hershkoviz R, Altbaum-Weiss I, Ganor S, Lider O. Cell surface-expressed moesin-like receptor regulates T cell interactions with tissue components and binds an adhesion-modulating IL-2 peptide generated by elastase. J Immunol. 2001;166:3052–3060. doi: 10.4049/jimmunol.166.5.3052. [DOI] [PubMed] [Google Scholar]
- 62.Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117:1547–1552. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- 63.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 64.Niggli V. Structural properties of lipidbinding sites in cytoskeletal proteins. Trends Biochem Sci. 2001;26:604–611. doi: 10.1016/s0968-0004(01)01927-2. [DOI] [PubMed] [Google Scholar]
- 65.Simons PC, Pietromonaco SF, Reczek D, Bretscher A, Elias L. C-terminal threonine phosphorylation activates ERM proteins to link the cell’s cortical lipid bilayer to the cytoskeleton. Biochem Biophys Res Commun. 1998;253:561–565. doi: 10.1006/bbrc.1998.9823. [DOI] [PubMed] [Google Scholar]
- 66.Sechi AS, Wehland J. The actin cytoskeleton and plasma membrane connection: PtdIns(4,5)P(2) influences cytoskeletal protein activity at the plasma membrane. J Cell Sci. 2000;21:3685–3695. doi: 10.1242/jcs.113.21.3685. [DOI] [PubMed] [Google Scholar]
- 67.Edwards SD, Keep NH. The 2.7 A crystal structure of the activated FERM domain of moesin: an analysis of structural changes on activation. Biochemistry. 2001;40:7061–7068. doi: 10.1021/bi010419h. [DOI] [PubMed] [Google Scholar]
- 68.Lynn WA, Liu Y, Golenbock DT. Neither CD14 nor serum is absolutely necessary for activation of mononuclear phagocytes by bacterial lipopolysaccharide. Infect Immun. 1993;61:4452–4461. doi: 10.1128/iai.61.10.4452-4461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Troelstra A, Antal-Szalmas P, de Graaf-Miltenburg LA, et al. Saturable CD14-dependent binding of fluorescein-labeled lipopolysaccharide to human monocytes. Infect Immun. 1997;65:2272–2277. doi: 10.1128/iai.65.6.2272-2277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Triantafilou M, Triantafilou K, Fernandez N. Rough and smooth forms of fluorescein-labelled bacterial endotoxin exhibit CD14/LBP dependent and independent binding that is influencedby endotoxin concentration. Eur J Biochem. 2000;267:2218–2226. doi: 10.1046/j.1432-1327.2000.01222.x. [DOI] [PubMed] [Google Scholar]
- 71.Shimazu R, Akashi S, Ogata H, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276:21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 73.Akashi S, Saitoh S, Wakabayashi Y, et al. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. J Exp Med. 2003;198:1035–1042. doi: 10.1084/jem.20031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23:301–304. doi: 10.1016/s1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]