Abstract
Sexual reproduction in flowering plants offers a number of remarkable aspects to developmental biologists. First, the spore mother cells – precursors of the plant reproductive lineage – are specified late in development, as opposed to precocious germline isolation during embryogenesis in most animals. Second, unlike in most animals where meiosis directly produces gametes, plant meiosis entails the differentiation of a multicellular, haploid gametophyte, within which gametic as well as non-gametic accessory cells are formed. These observations raise the question of the factors inducing and modus operandi of cell fate transitions that originate in floral tissues and gametophytes, respectively. Cell fate transitions in the reproductive lineage imply cellular reprogramming operating at the physiological, cytological and transcriptome level, but also at the chromatin level. A number of observations point to large-scale chromatin reorganization events associated with cellular differentiation of the female spore mother cells and of the female gametes. These include a reorganization of the heterochromatin compartment, the genome-wide alteration of the histone modification landscape, and the remodeling of nucleosome composition. The dynamic expression of DNA methyltransferases and actors of small RNA pathways also suggest additional, global epigenetic alterations that remain to be characterized. Are these events a cause or a consequence of cellular differentiation, and how do they contribute to cell fate transition? Does chromatin dynamics induce competence for immediate cellular functions (meiosis, fertilization), or does it also contribute long-term effects in cellular identity and developmental competence of the reproductive lineage? This review attempts to review these fascinating questions.
Keywords: chromatin, Sporogenesis, Gametogenesis, Pluripotency, reprogramming
Introduction
The life cycle of living organisms is marked by two major events that allow for the mathematical stability of genetic information across generations: meiosis enables allelic reshuffling and halves the chromosome number; fertilization allows for the regeneration of a diploid organism through the union of two haploid gametes. In multicellular organisms, key processes to these phase transitions are the differentiation of specialist cells committed to meiosis and fertilization, respectively. The differentiation of meiocytes marks the separation of the ‘germplasm’ from the soma, a developmental concept expressed in the 19th century by August Weissmann (Weismann, 1892); however, the definition of a germline (carrying the germplasm) is inherently based on a notion of cell lineage with a deterministic fate, and cannot easily be transposed across all multicellular organisms. There is a variety of developmental strategies across the animal and plant kingdoms regarding the specification of meiotic precursor cells (early vs. late), the fate of the meiotic products (unicellular vs. pluricellular) and the fertilization process itself (single vs. double) (Kondrashov, 1997). Far from the ambition to systematically review the evolutionary diversity of these processes, summarizing the main facts, terminology and key differences between land plants and animals provides a conceptual background. Thus, in the first section we describe the reproductive lineage of flowering plants from the precursor cells to the mature gametes, in particular highlighting the successive cellular transitions that imply novel cell fate establishment. Chromatin organization provides an instructive template to genome expression. Hence, a legitimate question is whether cellular reprogramming during fate transitions in the reproductive lineage is associated with local or global changes in chromatin organization and composition, collectively referred to as chromatin dynamics. We will briefly review our understanding of chromatin dynamics in plants before reviewing the multiple waves of large-scale chromatin events associated with cell fate transitions in the female reproductive lineage. We then discuss the possible roles of chromatin dynamics in this developmental context. Notably, we present the viewpoint that cellular reprogramming driven by chromatin dynamics has both immediate and long-term effects, namely: (i) the execution of forthcoming cellular functions (meiosis, gamete formation, fertilization); and (ii) the establishment of long-term developmental competence.
Successive Cellular Transitions in the Female Reproductive Lineage
The reproductive lineage initiates with the specification of meiocyte precursor cells that undergo a cellular transition from a mitotic to a meiotic fate, a process entailing the differentiation of gametes. Those precursor cells are referred to as primordial germ cells (PGCs) in animals and spore mother cells (SMCs) in plants. In animals, the germline is specified early during embryogenesis through the cytoplasmic isolation of maternal determinants before embryonic differentiation (e.g. insects, worms and some amphibians) or via the inductive signals of a pre-differentiated embryonic tissue (e.g. the epiblast in mammals and birds) (Extavour and Akam, 2003). PGCs proliferate and give rise to meiotic-competent cells, generically called meiocytes (e.g. primary spermatocytes and oocytes in animals). By contrast to animal PGCs, plant SMCs are not embryonically set aside. Instead, they differentiate in the adult plant that has undergone a developmental transition from vegetative to reproductive effort (Figure 1). SMCs differentiate in dedicated tissues of the male and female sexual organs of the flower, in pluripotent somatic niches of the anther and ovule primordium, respectively. SMCs originate either directly from the selection of a hypodermal cell (e.g. female SMCs in Arabidopsis) or indirectly following asymmetric division of one or several hypodermal, archesporial cells [e.g. female SMCs in Zea mays (maize) and male SMCs in most flowering plants; Feng et al., 2013; Kelliher et al., 2014; Schmidt et al., 2015]. SMC differentiation produces meiotic-competent cells, the meiocytes, also called sporocytes (male SMCs are also referred to as microsporocytes, microspore mother cells or pollen mother cells; female SMCs are also called megasporocytes or megaspore mother cells). Here, we use SMC from the initial hypodermal stage up to the functional meiocyte. SMC development is hence marked by: (i) specification and (ii) differentiation.
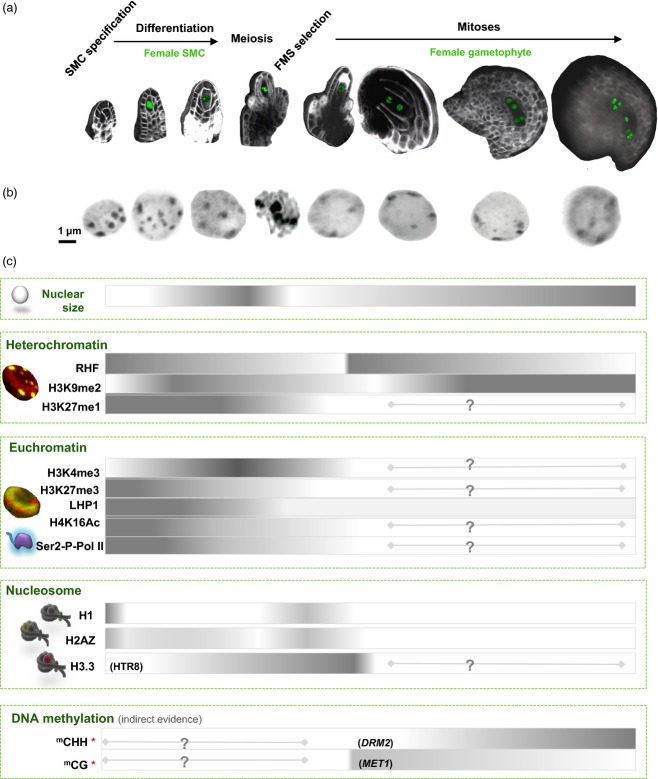
Figure 1.
Late establishment of the reproductive lineage in flowering plants. Flowering plants do not set aside their germline during embryogenesis, unlike most animals. Instead, the meiotic precursor cells, or spore mother cells (SMCs), differentiate de novo in very young flower buds in female and male floral organs: the ovule and anther primordia, respectively (left). In most flowering plants, only one female SMC (also called megaspore mother cell) is formed per ovule primordium, whereas several male SMCs (also called pollen mother cells) are formed in anther locules. Meiosis produces haploid spores (not shown on this scheme) that develop mitotically into multicellular, haploid gametophytes (right), within which the gametes differentiate de novo. The female gametophyte (also called embryo sac) is enclosed in maternal sporophytic integuments of the ovule. It comprises seven cells at maturity: two gametes – the egg cell (pink) and the central cell (yellow); and five accessory cells – three antipodals (blue) and two synergids (green). The central cell contains a single dihaploid nucleus resulting from the fusion of two polar nuclei. The male gametophyte is enclosed in the pollen grain and comprises a vegetative cell filling the grain (vegetative nucleus, yellow) and two sperm cells engulfed in the vegetative cell (sperm nuclei, blue). The images correspond to pseudo-colored, three-dimensional reconstructions (powered by imaris; Bitplane AG, http://www.bitplane.com) of confocal serial optical sections through whole-mount Arabidopsis reproductive organs: ovule primordium, anther, and mature ovule stained for cell membranes using FM4-64 dye and pollen grain stained for DNA using propidium iodide.
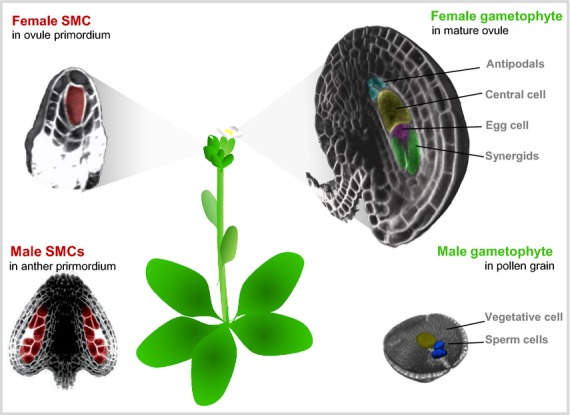
Meiosis marks the second developmental transition towards functional gametes. Meiosis universally fulfills the function of reducing the genome to its haploid chromosomal complement. Yet, the fate of meiotic products greatly differs between kingdoms. In animals, the gametes directly differentiate from the meiotic product. By contrast, in plants the haploid spore is pluripotent and gives rise to the multicellular gametophyte through mitotic divisions. The female gametophyte develops within, and is strongly influenced by, the ovule composed of maternal integuments (sporophytic tissue). The development of the typical Polygonum type of female gametophyte shared by 70% of flowering plants (Maheshwari, 1950) is characterized by three cycles of mitosis without cytokinesis before the partitioning of the multinucleate cytoplasm in very distinct cell types: two gametes (the egg cell and the central cell) and five accessory cells (two synergids and three antipodals) (Figure 1). The two gametes themselves share very distinct post-fertilization fates, respectively forming the totipotent zygote and the endosperm, a terminally differentiated structure that does not contribute genetic material to the next generation.
From this simplified developmental sketch it becomes clear that SMC and gamete formation imply the establishment of novel cell fate, in the anther and ovule primordia, and in the gametophytes, respectively. Key questions emerging are: (i) what are the upstream regulators of cell fate transition (instructors) and (ii) what are the downstream factors operating cellular reprogramming (effectors). In Arabidopsis, maize and Oryza sativa (rice), transcriptome profiling studies have shown that both SMC (Schmidt et al., 2011; Kubo et al., 2013; Kelliher and Walbot, 2014) and female gametophyte (Yu et al., 2005; Johnston et al., 2007; Wuest et al., 2010; Schmidt et al., 2012; Chettoor et al., 2014) differentiation correlates with important rewiring of the transcriptional program. The surrounding somatic cells play a central role in either promoting or restricting the developmental competence of the SMC and gametophyte (for comprehensive reviews, see Feng et al., 2013; Schmidt et al., 2015). The molecular pathways involved in this soma-to-reproductive lineage interaction include intercellular signaling components (Sheridan et al., 1999; Zhao et al., 2002; Lieber et al., 2011) and non-cell autonomous, small RNA-mediated gene regulation (Olmedo-Monfil et al., 2010; Tucker et al., 2012). Yet, in addition, it recently emerged that cellular differentiation in the reproductive lineage is associated with cell-specific alterations of chromatin organization, structure and composition.
Defining Chromatin Dynamics During Cellular Differentiation
Chromatin dynamics encompass the qualitative (distribution pattern) and quantitative changes in chromatin composition and modification, chromatin mobility, and chromosome organization in the nucleus. Beyond a packaging role, chromatin provides instructions for genome expression that operate at two levels. At the gene level, biochemical modifications of the DNA, nucleosomal and linker histones influence access to enzymatic complexes, affecting transcription, replication and DNA repair. At the nuclear level, spatial organization of chromatin domains compartmentalizes specific nuclear functions and reciprocally influences gene expression (reviewed in Misteli 2005, Schneider and Grosschedl 2007).
Transcriptional activity is a read-out of local chromatin state integrating biochemical modifications of histone tails, cytosine methylation and specific histone variants. Chromatin states are characterized by a functional indexing of combinatorial histone modifications and DNA methylation, with four main chromatin states that preferentially mark active genes, repressed genes, repeat elements and intergenic regions (Roudier et al., 2011). Transposable elements (TEs) are enriched in H3K9me2, H3K27me1 and H4K20me1, defining transcriptionally repressive heterochromatin states. Chromatin states at genic regions are based on a distinct set of modifications (e.g. H3K27me2,me3, H3K4me2, H3K36me3, H3KAc9), which are differentially combined in correlation with the expression status of each gene (Roudier et al., 2011). In a manner remarkably reminiscent of cell lineage marking in animals, H3K27me3 and H3K4me3 distribution patterns can be tissue-specific. H3K27me3 and H3K4me3 are typical hallmarks set by Polycomb-group (PcG) and Trithorax-like (Trx) protein complexes that define mitotically heritable, transcriptionally repressive and permissive chromatin states, respectively (reviewed in Kohler and Hennig, 2010). Furthermore, as in animals, DNA methylation in plants also largely influences transcription (Zhang et al., 2006; Zilberman et al., 2007). Cytosine methylation occurs in different sequence contexts. CG methylation is enriched in TEs and repeats, and largely targets genic regions as well (Cokus et al., 2008; Lister et al., 2008). In contrast, CHG and CHH methylation are almost exclusively found in heterochromatin (Cokus et al., 2008; Lister et al., 2008). Although it is clear that DNA methylation patterns are dynamic during plant development (Gehring and Henikoff, 2007), few studies address specific cell fate transitions. For instance, DNA methylation landscapes drastically change during male germline development and embryogenesis, as measured in immunostaining and/or methylome profile analyses in different species (Oakeley et al., 1997; Janousek et al., 2000; Huang et al., 2010; Calarco et al., 2012; Ibarra et al., 2012; Solis et al., 2012). In addition, nucleosome composition, with respect to specific variants of histones H2A and H3 influence transcriptional competence. Similar to its animal counterpart, H3.3 is enriched within transcriptionally active loci, and H3.3 variants distribution changes genome-wide during transcriptome reprogramming (Stroud et al., 2012; Wollmann et al., 2012; Nie et al., 2014; Shu et al., 2014).
At the microscopic level, plant chromatin is organized in euchromatin and heterochromatin domains, discrete heterochromatic chromocenters are formed comprising repeat elements (centromeric repeats, transposable elements, rDNA), and accordingly are enriched in repressive chromatin modifications (e.g. H3K9me1, me2, H3K27me1, DNA methylation; Fransz et al., 2006; Jasencakova et al., 2003; Mathieu et al., 2005; Naumann et al., 2005; Pecinka et al., 2004). The euchromatin compartment is characterized by both transcriptionally repressive (e.g. H3K27me2, me3 and H3K9me2) and permissive histone modifications (e.g. H3K4me2, me3, H3K9Ac and H4K16Ac; Fransz et al., 2002; Fuchs et al., 2006), with rare overlapping spatial distribution at the gene level (Roudier et al., 2011). The distribution pattern of euchromatin modifications differs slightly between flowering plant species, depending on the genome size, and number and distribution of DNA repeats and transposons (Houben et al., 2003), and differs with non-flowering plants (Fuchs et al., 2008). At interphase, chromosome arms are deployed in euchromatin and form distinct territories (Fransz et al., 2002). Although chromosomes are organized at random in plant species that do not share the polarized Rabl configuration (Pecinka et al., 2004; Schubert and Shaw, 2011), they frequently interact together. Specifically, patterns of inter- and intrachromosomal interactions are found to correlate with chromatin indexing, suggesting a functional relationship (Grob et al., 2013). Although the causality remains to be resolved, chromosomal interactions may provide a platform for the coordinated regulation of loci that function simultaneously (or not). In addition, transcription may influence, or be influenced by, the spatial localization of gene locis in the nucleus relative to the nuclear periphery, chromosome territories or heterochromatin domains. A few studies have provided exciting evidence for the dynamic spatial re-localization of genomic loci in response to environmental and developmental cues, involving long-distance intrachromosomal loops, and spatial repositioning at the nuclear periphery, at the periphery of chromosome territories, and in nuclear bodies (Makarevich et al., 2008; Crevillen et al., 2013; Feng et al., 2014; Liu et al., 2014) (Wegel et al., 2005; Costa and Shaw, 2006). Consistent with this view of dynamically relocated genomic regions, Wang et al. have recently identified insulated small ‘strips’ of kilobase-size domains enriched in H3K27me3 and H3.3, prone to frequent intrachromosomal interactions (Wang et al., 2015). Fitting the idea of dynamic chromosomal arrangements in relation to transcriptional competence, global mobility properties of the plant chromatin change during cellular differentiation: root meristematic cells retain a high chromatin mobility, whereas it decreases as cellular differentiation proceeds, a process largely modulated by histone acetylation (Rosa et al., 2014). Thus, a picture emerges where the plant chromatin is a highly dynamic matrix providing a functional template for instructing, interpreting, or both, the chromatin indexing level of gene regulation. This working model now needs to be challenged by combined efforts employing chromosome capture technologies, chromatin profiling and gene positioning analysis approaches in a cell-specific yet genome-wide manner.
Multiple Waves of Chromatin Dynamics: Pre- and Post-meiosis, and Pre- and Post-Cellularization in the Gametophyte
What about the organization of chromatin in cells of the reproductive lineage, particularly at cellular transitions? Does cell fate establishment (SMC fate and gametic fate) correlate with global or local re-instruction of the chromatin landscape? Because of the small number of target cells and their relative inaccessibility in the female reproductive lineage, technical approaches are currently lacking for reading the epigenome at the gene level, for instance using chromatin immunoprecipitation or DNA bisulfite sequencing. Our knowledge of chromatin organization in the SMC and developing gametophyte is currently essentially global, coming from cytogenetic and chromatin reporter studies, yet it revealed unanticipated dynamics. Robust methods have been developed for quantifying and inspecting the distribution of histone modifications at high optical resolution at the single-cell level in whole-mount ovules (Arabidopsis) or thick ovary sections (maize) (Garcia-Aguilar et al., 2010; She et al., 2013). These approaches enabled the quantitative probing of the chromatin landscape and its organization at a global nuclear scale in relation to cellular fate transition and differentiation in the female reproductive lineage.
Chromatin dynamics during SMC formation
Female SMC formation (specification and differentiation) is characterized by global changes in chromatin organization in Arabidopsis and maize. Although these changes are specific to the SMC itself in Arabidopsis (She et al., 2013), they extend to surrounding nucellar cells in maize (Garcia-Aguilar et al., 2010). Remarkably, SMC differentiation in Arabidopsis ovule primordia is characterized by progressive and biphasic modifications of chromatin composition and organization, targeting heterochromatin and euchromatin in two consecutive phases. Global eviction of somatic linker H1 histones specifically in the SMC, and not in the surrounding somatic cells, was observed in both reporter and immunostaining analyses, and is the first detectable event in the establishment of SMC fate (She et al., 2013). It marks the onset of a gradual chromatin decondensation characterized by a reduction in heterochromatin content (Figure 2). Yet, the remaining heterochromatin domains are enriched in H3K9me2, a mark typically associated with TE silencing (Fransz et al., 2006; Roudier et al., 2011). Thus, the detected alterations of heterochromatin in SMC is unlikely to affect TE activity, as is the case in the vegetative cell of pollen, for instance (Slotkin et al., 2009). Furthermore, the observation that centromeric repeats remain gathered at conspicuous foci marked by the centromeric histone CENH3 (She et al., 2013) suggests that chromatin decondensation affects a subset of the SMC chromatin only. This is in sharp contrast with the global decondensation observed in de-differentiated plant cells (Tessadori et al., 2007) and mutants lacking the chromatin remodeler DECREASE in DNA METHYLATION 1, which broadly alters the organization of centromeric repeats (Soppe et al., 2002; Probst et al., 2003). In a second phase, a quantitative redistribution of the PcG and Trx hallmarks is observed, with twice less repressive H3K27me3 mark and 2.5 times more permissive H3K4me2 mark in the SMC compared with the surrounding soma, respectively. Concurrently with these alterations the SMC chromatin is remodeled with, notably, the transient eviction of H2A.Z, a specific variant thought to influence nucleosome stability in animals (Marques et al., 2010) and transcriptional responsiveness in plants (Kumar and Wigge, 2010), and with the specific incorporation of H3.3, a class of variant deposited at transcriptionally active loci (Stroud et al., 2012; Wollmann et al., 2012; She et al., 2013). Yet, despite this apparent permissive chromatin state, RNA Pol II activity is progressively dampened and H4 acetylation dramatically decreases during SMC differentiation (Figure 2), possibly suggesting a poised chromatin state. The mechanisms driving these large-scale events of chromatin reprogramming in the SMC are not fully elucidated. Whereas the eviction of H1 is mediated by the proteasome-degradation pathway, the signaling mechanisms remain unknown. Furthermore, non-canonical enzymes are likely to be involved: for instance, mutations in the major H3K9 methylase KRYPTONITE (KYP) and in the major H3K27 demethylase REF6 have no influence on H3K9me2 and H3K27me3 dynamics in the female SMC, respectively. By contrast, H3K4me3 deposition in the SMC is largely, yet not entirely, mediated by the SET DOMAIN GROUP 2 (SDG2) enzyme (She et al., 2013). In addition, ATP-dependent chromatin remodelers, yet to be identified, are likely to be involved in the substitution/incorporation of specific core nucleosome variants, such as H2AZ and H3.3 (She et al., 2013).
Figure 2.
The female reproductive lineage is marked by several cellular transitions associated with large-scale chromatin dynamics.(a) The female reproductive lineage is initiated with spore mother cell (SMC) specification in ovule primordia. The SMC differentiates progressively with visible changes at the cytological and chromatin level, along with ovule organogenesis (tegument growth). The mature SMC executes meiosis and produces four haploid spores (not shown on this scheme), only one of which, the functional megaspore (FMS), is selected, whereas the remaining three degenerate. The last stages of mature embryo sac formation are shown Figure 3. The FMS enters a mitotic phase producing eight haploid nuclei in a syncytium. SMC differentiation, meiosis, mitotic gametophytic development and gamete differentiation are cell fate transitions associated with large-scale chromatin dynamics underlying cellular reprogramming. The images show Arabidopsis ovules (grey) enclosing the female reproductive lineage (green nuclei). The images were elaborated from 3D reconstructions (imaris; Bitplane AG) of serial confocal optical sections recording the membrane-specific FM4-64 dye overlain with images of GFP-tagged nuclei (photoshop; Adobe, http://www.adobe.com). GFP images correspond to chromatin reporters specifically expressed in the SMCs and gametophytic cells [KNU-nlsYFP in the SMCs (Tucker et al., 2012); H1.1::H1.1-GFP in meiotic SMCs (She et al., 2013); AKV::H2B-YFP in FMSs and developing gametophyte (Pillot et al., 2010b; Schmidt et al., 2011)]. Note that this panel is purely illustrative. Putative DNA methylation dynamics are indirectly inferred from genetic or reporter gene expression analyses (see discussion in the main text).(b) Representative pictures of SMCs and gametophytic nuclei are shown (3D reconstructions from confocal images of nuclei stained with propidium iodide, as described by She et al. (2013)].(c) The panels schematically represent quantitative changes in the chromatin of SMCs and gametophytic nuclei, compared with the surrounding somatic cells in Arabidopsis. The gradient in each panel can be read as temporal dynamics, starting at the specification of the SMCs from a somatic cell of the ovule primordium. Chromatin dynamics is characterized by: changes in nuclear size; heterochromatin content (RHF) and histone modifications; euchromatic histone modifications and active RNA Pol II; and histone variants and putative changes in DNA methylation. The color intensity is indicative of the trend for each feature, and cannot be compared between histone marks, for example. For all except DNA methylation, these representations are based on quantitative studies in whole-mount tissues using immunostaining and reporter analyses (She et al., 2013).
What we describe here is only a snapshot of large-scale events, mostly regarding histone composition and modifications. Additional dynamics are likely to be found for the distribution of DNA methylation; however, we are presently lacking sequence-specific probes. Support for this expectation includes: (i) the known interplay between linker histone composition and DNA methylation (Wierzbicki and Jerzmanowski, 2005; Zemach et al., 2013); and (ii) genetic analyses providing support for the functional requirement of DNA methylation in female SMC fate establishment in maize and Arabidopsis (Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010). In Arabidopsis, METHYLTRANSFERASE 1 (MET1), a homolog of the mammalian enzyme Dnmt1, is the major DNA methyltransferase maintaining CG methylation (Finnegan et al., 1996; Kankel et al., 2003), but is also capable of restoring methylation patterns de novo (Zubko et al., 2012). DOMAIN REARRANGED METHYLTRANSFERASE 2 (DRM2) and CHROMOMETHYLTRANFERASE 3 (CMT3) catalyze methylation at non-CG sites during the establishment and maintenance processes, respectively (Lindroth et al., 2001; Cao and Jacobsen, 2002). DNA methylation at non-CG sites is reinforced by a small RNA-directed process (RdDM, RNA-directed DNA methylation; for a review see Matzke and Mosher, 2014). Maize lines deficient in genes dmt102 and dmt103 encoding homologs of CMT3 and DRM2, as well as RdDM-deficient Arabidopsis lines, produce ectopic reproductive lineages, suggesting a role for non-CG methylation in germ cell fate (Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010). Because AGO9, a central player in the RdDM-mediated control of SMC fate, is expressed primarily in the nucellus, the current model is that the DNA methylation landscape in the SMC may possibly be influenced by small RNAs produced in neighboring cells.
In conclusion, chromatin organization and composition is highly dynamic during SMC differentiation, and probably underlies cellular reprogramming at this critical somatic → reproductive cell fate transition. The changes are largely specific to the SMC and do not affect the surrounding somatic cells, at least in Arabidopsis, yet are likely to involve regulatory interactions between the SMC and soma through mobile signals such as small RNAs. The histone modification signatures consistent with a transcriptionally competent yet poised chromatin, and the global relaxation of chromatin organization, suggest a global epigenetic reprogramming phase. We now face the challenge to determine the precise dynamics of the epigenome, at the DNA methylation and histone modification levels, during SMC specification, and in comparison with its surrounding somatic niche.
Chromatin dynamics during Meiosis
The chromatin landscape achieved in Arabidopsis female SMCs just prior to prophase I is not the final set-up. Meiotic execution itself entails additional dynamics of histone modifications, particularly during prophase I. Before chromosomes form visible bivalents, both H1 and H2AZ are reloaded (She et al., 2013). Prophase I progression is further characterized by: (i) further enrichment in H3K4me3 along the entire chromosomes; (ii) nearly undetectable levels of H3K27me3; (iii) low levels of H3K9me1; and (iv) massive enrichment of H3K9me2 in discrete heterochromatic foci (She et al., 2013). Because of the lack of similar analyses in the female SMCs of other plant species, interspecific comparison is currently not possible. Yet the distribution of those histone modifications on meiotic chromosomes is likely to reflect distinct genomic organization relative to repeat sequences and gene density. In particular, in crop species harboring a larger genome, a regional distribution of histone modifications is observed in male meiocytes (Higgins et al., 2012). Interestingly, in maize, mutations in AGO104, homologous to Arabidopsis AGO9, induce defects in centromere condensation, switching the meiotic cycle into a mitotic (somatic) cycle, and producing unreduced gametes. Similar to AGO9, AGO104 is expressed in nucellus tissue surrounding the female SMCs, suggesting a non-cell autonomous effect (Singh et al., 2011). In rice, another AGO protein, MEL1, homologous to AGO5, is also involved in meiosis progression (Nonomura et al., 2007). These examples invoke a key role for small RNA-dependent silencing mechanisms during meiosis. Additional chromatin remodeling and chromosome dynamics drive bivalent formation, crossing over and recombination, chromosome movements and involve a complex remodeling machinery as well as non-histone proteins. This very specific phase of chromatin dynamics has been nicely reviewed elsewhere (e.g. Pawlowski, 2010; Mainiero and Pawlowski, 2014).
Chromatin dynamics during FMS differentiation and gametophyte mitotic divisions
Upon meiosis completion, a tetrad of haploid spores is formed by cellularization of the meiotic syncytium, and a single apical spore differentiates into a pluripotent functional megaspore (FMS). FMS selection correlates with a new wave of dynamic changes in chromatin organization and modifications, which resembles the pre-meiotic wave, albeit with distinctive features (Figure 2). Like female SMCs, FMSs show a decondensed chromatin with reduced heterochromatin content, associated with a rapid re-eviction of H1 and of H2A.Z, and with drastically reduced levels of H3K27me1, H3K27me3, H4K16ac and the active form of RNA Pol II. This suggests low transcriptional activity relative to the surrounding nucellar nuclei. In contrast to the SMCs, however, the euchromatic permissive mark H3K4me3, as well as the heterochromatic repressive mark H3K9me2, are also depleted in the FMSs (She et al., 2013). The initiation of mitoses in the FMSs is likely to involve DNA methylation dynamics, as suggested by: (i) the loss of H1 and H2AZ, for which a mechanistic link with DNA methylation has been shown (Wierzbicki and Jerzmanowski, 2005; Coleman-Derr and Zilberman, 2012; Zemach et al., 2013); (ii) the action of somatically derived small RNAs, suggested by the developmental arrest of the FMSs in ovules lacking the function of AGO5 (Tucker and Koltunow, 2014); and (iii) the expression of the DNA methyltransferase MET1 (Jullien et al., 2012).
Yet chromatin dynamics goes beyond global alterations of histone modifications. In fact, ATP-dependent chromatin remodeling in the newly formed FMSs is likely to represent a major control checkpoint towards the mitotic phase of gametophyte development because targeted silencing of different members of the SWI2/SNF2 chromatin remodeling enzyme family – including CHR11 – in the FMSs leads to developmental arrest at the FMS stage or after one mitosis (Huanca-Mamani et al., 2005).
During the three mitotic cycles that resume the female gametophyte, high levels of H3K9me2 are re-established in well-defined heterochromatic foci in all nuclei. Similarly, in euchromatin, a GFP-tagged variant of the LHP1/TFL2 protein, a reader of the PcG-mediated H3K27me3 signature (Turck et al., 2007; Exner et al., 2009) is detected in two- and four-nucleate gametophytes (Figure S1) as well as in polar nuclei, synergid and egg cell nuclei of mature embryo sacs (Pillot et al., 2010b). Consistently, the PcG complex member SWINGER, driving H3K27me3 deposition, is detected in all gametophyte nuclei from the FMS stage onwards (Wang et al., 2006). Thus, following the massive reduction of different chromatin modifications in the FMSs, the mitotic phase of gametophyte development entails the renewed deposition of histone marks in both heterochromatic and euchromatic compartments. Interestingly, H3K9me2 deposition was indistinguishable among the eight nuclei of the gametophytic syncytium, whereas LHP1 seems strongly depleted specifically from the upmost chalazal nuclei sharing an antipodal fate (Pillot et al., 2010b; Figure S1); however, a more exhaustive developmental atlas of chromatin marks during this syncytial phase of development is necessary to confirm such a pre-patterning phenomenon.
Cell-specific chromatin patterns in the mature gametophyte
Cellularization marks the onset of another wave of large-scale chromatin changes that distinguishes the different cell types. Noticeably, a global dimorphism in chromatin and transcriptional states is established between the two female gametes (Figure 3; Pillot et al., 2010b). In the egg cell, numerous H3K9me2 small foci rearrange into large prominent foci coinciding with heterochromatic chromocenters. Concomitantly, high levels of LHP1/TFL2 are established, whereas the active form of RNA Pol II (Ser2-P-Pol II) becomes barely detectable. By contrast, in the central cell, cellularization followed by the fusion of the polar nuclei entails a strong depletion of both H3K9me2 and LHP1/TFL2, together with the loose distribution of heterochromatin (Pillot et al., 2010b; Figure 3). Concurrently, high levels of active Pol II are detected. The same dimorphic epigenetic features were observed in maize female gametophytes (Garcia-Aguilar et al., 2010). Thus, the egg cell is characterized by a repressive chromatin and a relatively quiescent transcriptional state, whereas the central cell chromatin is depleted of repressive marks and displays active global transcriptional competence.
Figure 3.
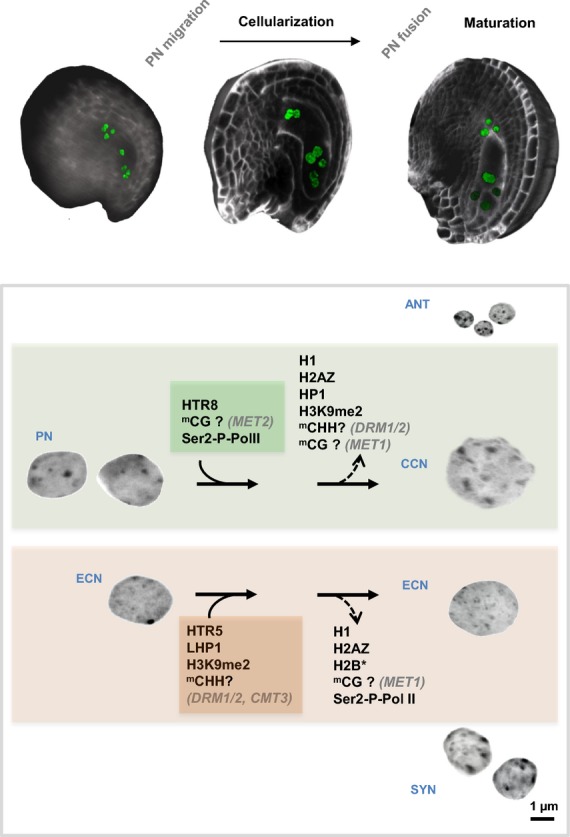
Epigenetic dimorphism between egg and central cell nuclei upon female gametophyte cellularization. After the formation of the eight-cell syncytial embryo sac, two polar nuclei (PN) migrate towards each other in a central position. This nuclear movement is followed shortly by cellularization of the gametophyte (see also Figure 1, cellularized gametophyte), an event that marks the onset of synergids (SYN), antipodals (ANT) and gamete differentiation. Egg cell nucleus (ECN) differentiation is characterized by the deposition of repressive epigenetic features and the depletion of permissive ones, whereas the opposite trend is observed for the central cell nucleus (CCN). The two gametes also differ in the repertoire of histone variant H3.3 expressed. As in Figure 2, the data are based on immunostaning and reporter analysis (She et al., 2013; : Pillot et al., 2010a,b), and the differences in DNA methylation profiles are inferred from genetic and reporter analyses (Pillot et al., 2010a,b; Jullien et al., 2012).
Chromatin dimorphism in the female gametes seems to be associated with a different status of DNA methylation, although we are lacking a specific, genome-wide readout of the DNA methylome in these cells. Genetic evidence is twofold. First, the central cell and egg cell chromatin states depend on the cytosine demethylase DEMETER-LIKE 1–3 (DML1–DML3), and the DNA methyltransferase CMT3, respectively (Pillot et al., 2010a,b). Interestingly, a cmt3 mutation has no impact before cellularization, suggesting an active targeting mechanism upon egg cell differentiation (Pillot et al., 2010b). The question to be resolved refers to the expression pattern of CMT3. CMT3 transcripts are abundant in the egg cell (Wuest et al., 2010), but a reporter protein fusion failed to be detected in gametophytic cells (Jullien et al., 2012). By contrast, the DNA methyltransferase DRM2 that contributes de novo CHH methylation is detected in the egg cell using reporter fusions, together with its homolog DRM1, for which a function in DNA methylation remains to be demonstrated (Cao and Jacobsen, 2002), whereas the CG methylation maintenance enzyme MET1 is undetectable (Jullien et al., 2012). In the central cell, DRM1, CMT3 and MET1 remain undetectable by protein reporter fusions, whereas a DMR2 reporter displayed a weak signal (Jullien et al., 2012), raising the question of an alternative mechanism to maintain/deposit DNA methylation. Possibly, the MET2a and MET2b homologs expressed in the central cell could confer this function (Jullien et al., 2012). These MET2 isoforms showed undetectable levels in the egg cell, an observation to be interpreted with caution because the protein fusion to H2B used in this study to detect MET2 may be subject to turnover (Pillot et al., 2010b). The function of DRM1, DMR2 and MET2a,b in the female gametes is currently unknown.
In addition, chromatin remodeling events are likely to intervene at embryo sac maturation in order to establish a gamete-specific nucleosome composition. In support of this hypothesis, the uniform incorporation of an H2B reporter protein in the eight-nucleate syncitium shifted at, or shortly after, polar nuclei fusion, towards selective depletion in the egg apparatus (Pillot et al., 2010b). Moreover, each cell of the mature embryo sac expresses a specific histone H3 repertoire that drastically differs from the surrounding somatic cells: the gametophyte is globally characterized by a significant depletion of the replication-dependent H3.1 isoform; the egg cell expresses only the H3.3 variant HTR5, whereas a single H3.1, HTR3; and two H3.3, HTR8 and HTR14, are present in the central cell (Ingouff et al., 2010). In rice, two of the three putative H2A.Z ortholog transcripts are enriched in the egg cell transcriptome, as compared with pollen; however, their expression in the central cell and accessory cells has not been described (Anderson et al., 2013).
Collectively, these cytological and genetic analyses highlight a stark dimorphism of the chromatin and transcriptional status in the two female gametes. This epigenetic dimorphism also extends beyond histone modifications and is likely to involve unequal DNA methylation levels between the two gametes, with a global demethylation in the central cell owing to demethylases activity vs. non-CG DNA methylation in the egg cell. This model awaits confirmation at the molecular level, however. Furthermore, it is becoming clear that the molecular mechanisms leading to chromatin dimorphism in the gametes rely on both cell-autonomous (e.g. CMT3 in the egg) and non-cell autonomous pathways. In analogy to the situation in the male germline, and as an interpolation of genetic and epigenome profiling data in the fertilization products, it was proposed that small RNAs produced by the central cell may influence the epigenetic set-up of the egg (for a review, see Castel and Martienssen, 2013). In addition, a specific role for AGO9 in TE silencing has been described in the egg cell, again highlighting the importance of non-cell autonomous, mobile small RNA signals in organizing chromatin in gametes. This role is consistent with AGO9 binding to mainly 24-nt small RNAs targeting centromeric TE that might involve CMT3 (Duran-Figueroa and Vielle-Calzada, 2010; Olmedo-Monfil et al., 2010; Pillot et al., 2010a). Finally, the epigenetic landscape in the egg cell – indicating that silencing mechanisms extend beyond centromeric TE to euchromatic regions, and suggesting a global transcriptional quiescence – is reminiscent of the epigenetic reprogramming required for the acquisition of totipotency in the animal germline.
Structural and Instructive Roles of Chromatin Dynamics
A legitimate question regards the cause-and-effect relationship between the observed large-scale chromatin dynamics during SMC and the establishment of gametic fate. In this section we explore three possible, non-exclusive roles of chromatin dynamics in: (i) activating genes required during meiotic progression in the SMC, gamete maturation and fertilization in the embryo sac; (ii) providing an instructive template for structurally dependent chromosome dynamics at meiosis and at karyogamy; and (iii) setting a global transcriptional quiescence and poising of developmental genes necessary for pluri-/totipotency establishment.
Chromatin dynamics is biphasic in female SMCs, and may support distinct functions. The first phase of chromatin dynamics in female SMCs characterized by H1 eviction and chromatin decondensation theoretically favors a transcriptional competence function and enables the establishment of a novel transcriptome. In maize anthers, meiotic genes are activated in the (male) SMC initials prior to, and not concomitantly with, the meiotic S phase (Kelliher et al., 2014). Whether transcriptional activation in female SMCs precedes or partially overlaps with the meiotic S phase must be resolved. Meiotic gene expression requires ATP-dependent chromatin remodeling, as shown by the lack of DMC1 activation in ovules lacking SWRI function in the ACTIN RELATED PROTEIN 6 mutant (Qin et al., 2014); however, even considering the difficulty in categorizing meiotic-specific genes, transcriptome profiles have identified a large number of genes that are likely to play a role beyond meiotic execution itself (Zhou and Pawlowski, 2014). In fact, several lines of evidence argue in favor of a role for pre-meiotically expressed genes beyond meiosis. One comes from the analysis of mutant ovules lacking SDG2 function: although mutant sdg2 female SMCs show significantly lower H3K4me3 levels than in the wild type, they progress normally through meiosis but fail to develop beyond the FMS stage (in wild-type FMS, the dramatically low levels of H3K4me3 compared with the surrounding cells argues against the requirement of SDG2 after meiosis). Thus, it was concluded that H3K4me3 remodeling before meiosis contributes to establish a post-meiotic, developmental competence to form the gametophyte (She et al., 2013). More indirect evidence comes from the analysis of ectopic female SMCs produced in the ago9 mutant ovules that restore chromatin dynamics to that in the wild type (She et al., 2013), although they are thought to circumvent meiosis and engage directly in gametophytic fate (Olmedo-Monfil et al., 2010). Lastly, a family of RNA helicases highly expressed in the female SMCs also functions post-meiotically: the major developmental defect of the mneme (mem) mutant occurs in the mature embryo sac and early embryo (Schmidt et al., 2011). Remarkably, MEM is not expressed in the developing and mature embryo sac, yet a lack of MEM function in the female SMCs affects heterochromatin formation and LHP1 distribution in the mature gametes (Schmidt et al., 2011). These three lines of evidence strongly argue for a role of the pre-meiotic wave of chromatin dynamics in establishing a post-meiotic developmental competence linked with the chromatin set-up of the gametophyte.
Furthermore, the second phase of chromatin dynamics in Arabidopsis female SMCs – just at the onset of prophase I – is likely to contribute structurally relevant changes to meiotic chromosomes. The C-terminal tail of H1 linker histones has a known effect on chromatin condensation (Caterino and Hayes, 2011), thus H1 reloading at prophase I might assist chromosome condensation. Concurrent H2AZ reloading together with massive H3K4me3 enrichment and re-acetylation of histones possibly contributes to establish an instructive template for crossing over, as proposed for male SMCs (Perrella et al., 2010; Choi et al., 2013; Zhou and Pawlowski, 2014). Consistent with this idea, in the large genome of crop plants, cross-overs coincide with the interspersed distribution of histone modifications along the chromosomes in male SMCs at prophase I (Higgins et al., 2014). Although the dynamics of DNA methylation has not been probed in female SMCs, this epigenetic modification is also likely to play a role in instructing the cross-over machinery because plants compromised for maintenance in DNA methylation, such as the met1 mutant, show a drastic alteration of the recombination landscape (Mirouze et al., 2012; Yelina et al., 2012). Further processes of chromatin remodeling that involve non-histone proteins and take place in prophase I, and are mechanistically linked with the progress of meiosis, are reviewed in Tiang et al. (2012) and Zhou and Pawlowski (2014). Collectively, the dynamic changes in histone modifications in SMCs suggest that they template instructions onto the chromosomes for meiotic chromatin condensation, pairing, cross-over and recombination.
Chromatin dynamics in the female gametophyte is also biphasic, and similarly to the situation during SMC differentiation, may underlie distinct functions. The rapid re-eviction of H1 and H2AZ, and depletion of H3K4me3, H3K27me3, H3K27me1, H3K9me2 and H4K16Ac in the FMSs suggest a vast resetting of the histone repertoire. Whether this resetting is necessary to enter a mitotic activity remains to be determined. The arrest of FMSs lacking the chromatin remodeler CHR11 indicates a role for ATP-dependent remodeling in nuclear proliferation (Huanca-Mamani et al., 2005). In addition, FMSs lacking the nucleoporin subunit MOS7/Nup88 are incompetent for gametophytic development (Park et al., 2014). Nucleoporins are primarily known for their role in RNA export. Thus, FMSs lacking this MOS7/Nup88 may arrest because of the failure in translating transcripts – trapped in the nucleus – encoding proteins necessary for developmental progression. Alternatively, nucleoporins might also contribute to the organization of the nucleus by tethering chromatin domains, as shown in animals (Burns and Wente, 2014). Spatial organization of the chromatin in relation to transcriptional activity is poorly known in plants. Analysis of chromatin organization in mos7 mutant FMSs might elucidate possible higher order chromatin organization dynamics in relation to cellular reprogramming in the female gametophyte.
The second wave of chromatin dynamics occurring after cellularization probably has a direct cell-autonomous role in creating a chromatin status compatible with the expression of the specific gametic transcriptome, which differs from the surrounding somatic cells, as shown in Arabidopsis and monocot species (Wuest et al., 2010; Anderson et al., 2013). The chromatin landscape of mature female gametes is also thought to set the ground-state for their post-fertilization developmental competence. Gametogenesis in animals represents a reprogramming window, during which the sex-specific epigenetic information retained by each of the two parental gametes is erased, and reprogrammed in the zygote, or is retained and transmitted, creating in both cases a chromatin environment compatible with totipotency and further development. We have little evidence for such a reprogramming in plants, but we know that repressive histone marks (for instance H3K9me2), LHP1/TFL2 and transcriptional quiescence are conserved between egg cell and zygote nuclei, suggesting the inheritance of these marks (Pillot et al., 2010b); however, maternally inherited H3 histones undergo a rapid turnover in the zygote (Ingouff et al., 2010). Therefore, the inheritance of H3 marks is likely to be a dynamic process requiring intermediate steps at DNA or RNA levels. Alternatively, histone retention can occur at specific loci, beyond the global cytological level of resolution, as shown in animal systems (Ihara et al., 2014). Interfering with chromatin or DNA methylation-modifying activities in the female gametes results in embryonic defects. Abnormal patterns of cell divisions were observed in early embryos inheriting maternal loss-of-function mutations in genes controlling CG, non-CG methylation and H3K9me2 deposition (Xiao et al., 2006; Pillot et al., 2010b; Autran et al., 2011).
These defects are often transient, as mature embryos are correctly formed, suggesting that Arabidopsis embryonic cells maintain a degree of plasticity, and/or that the defects are compensated for by the paternal genome (Del Toro-De Leon et al., 2014). This suggests that the correct inheritance (and correct reprogramming) of maternal epigenetic information is important for early embryo patterning. Another classical example of an epigenetic maternal effect is imprinting, where parental alleles are differentially expressed in the fertilization product. In this context, DEMETER-mediated DNA demethylation in the central cell is likely to contribute to the activation of maternal alleles of imprinted genes (Raissig et al., 2011; Kohler et al., 2012).
Thus, although the exact function of chromatin dynamics in SMCs and gametes is far from being elucidated, the current model proposes that chromatin dynamics contributes to immediate cellular functions during meiosis and fertilization, but also establishes post-meiotic and post-fertilization developmental competence. Even though SMCs and gamete-specific transcriptome profiles demonstrate a transcriptional program distinct from the surrounding somatic tissue (reviewed in Schmidt et al., 2015), we still lack the fine temporal resolution needed to determine whether transcriptional rewiring precedes or follows chromatin dynamics. Clearly many questions will remain unanswered until the technical limitations hindering epigenome profiling in these relatively inaccessible and rare cell types are overcome.
Impact on Transgenerational Epigenetic Inheritance?
The plant germline is formed late in development, after the plant switches from a vegetative to a reproductive effort. This gives ample time for the somatic tissues to be exposed to diverse biotic and abiotic environmental stresses. There is a growing body of evidence for changes in the epigenetic landscape in plant cells in response to environmental stresses (Kinoshita and Seki, 2014). The question arises whether epigenetic memories of stress are inheritable via the reproductive lineage that hypothetically appears after stress exposure; however, it is possible to genetically alleviate some barriers against the inheritance of stress-induced transcriptional signatures (Iwasaki and Paszkowski, 2014). This situation occurs in the double mutant mom1 ddm1, which is compromised in the activity of the DECREASE IN DNA METHYLATION 1 (DDM1) SWI/SNF chromatin remodeler and the transcriptional silencer MORPHEUS’ MOLECULE 1 (MOM). How these factors act in preventing the transmission of somatically acquired transcriptome signatures, thought to arise from stress-induced epigenetic changes, needs further investigation. The situation is further complicated by the fact that some stresses may induce genomic changes via the activation of transposable elements (e.g. Ito et al., 2011), and by the observation that the meristem has mechanisms safeguarding transposon silencing (Baubec et al., 2014). There is currently no unequivocal evidence for a naturally occurring transgenerational inheritance of stress-induced epigenetic changes in the absence of genomic changes (Heard and Martienssen, 2014).
Intergenerational or parental effects should be distinguished from transgenerational epigenetic inheritance stricto sensu. The former are heritable changes influencing the epigenome of the offspring, in which maintenance of the triggering signal is necessary to ensure the inheritance of the epigenetic state. Transgenerational epigenetic inheritance sensu stricto creates true epialleles, stably inherited independently of the genetic or environmental inducer signal (reviewed in Heard and Martienssen, 2014; Paszkowski and Grossniklaus, 2011). The study of experimentally induced epialleles in descendant generations of parents transmitting randomly demethylated genomes, owing to met1 mutation, have revealed a particular role for the maternal reproductive lineage. The active state of the EVADE retrotransposon is transmitted only through the male germline, but is reset to inactive when transmitted through the maternal germline. Genetic analysis showed that this maternally mediated safeguarding requires a functional RdDM pathway in (female) sporophytic ovule tissues. This mechanism may operate in a non-cell autonomous manner on the reproductive lineage, reminiscent of the mode of action in the AGO9 pathway during female SMC formation (Reinders et al. 2013). Alternatively, reprogramming of the inactive state may take place in the female SMCs as a consequence of a massive, increased deposition of silencing mark H3K9me2 (She et al., 2013).
Similarly, the well-studied case of paramutation, a particular form of epialleles identified in maize, probably involves germline reprogramming (reviewed in Grimanelli and Roudier, 2013; Hollick, 2012). Paramutagenic alleles act in trans to stably convert naive homologous (paramutable) alleles into a paramutagenic state requiring RdDM-related mechanisms for transgenerational maintenance. Whereas the mechanisms operating epigenetic state switches at paramutable/paramutagenic alleles are not fully elucidated, genetic analyses indicate that paramutation occurs during reproduction, probably around meiosis (Coe, 1966; Hollick, 2012; Grimanelli and Roudier, 2013). This suggests a major role of the reproductive lineage in creating an epigenetic context or reprogramming to establish the paramutagenic potential. In Arabidopsis, a similar trans-acting epigenetic phenomenon has been described: differentially methylated regions in inter-ecotype hybrids potentially contribute to heterosis and inbreeding depression (Greaves et al., 2014). Paramutation mechanisms might also involve maternal cytoplasmic small RNAs as inherited templates, as shown recently for the first time in animals. In Drosophila, the stable trans-silencing of repetitive elements through more than 50 generations is mediated by maternal inheritance of cytoplasm carrying Piwi-interacting RNAs (piRNAs) homologous to the transgenes (de Vanssay et al., 2012). In flowering plants, no PIWI protein was found so far, but members of the plant ARGONAUTE family, expressed during female reproductive development, can fulfil a similar task (Vaucheret, 2008). Paramutation thus illustrates the interplay between nuclear chromatin mechanisms and cytoplasmic determinants in female gamete reprogramming.
Another carefully documented example in plants is the dynamics of epigenetic marks influencing the expression of the flowering repressor FLC and carrying vernalization memory through the generations (reviewed in Grimanelli and Roudier, 2013; Mozgova et al., 2015). During vernalization, low temperatures gradually induce FLC silencing in the somatic cells, mainly via H3K27me3 deposition at the FLC locus, a situation resulting in progressive flowering derepression until spring. The flowers producing the reproductive lineage do not express the flowering repressor FLC, yet the requirement of vernalization is reset at each generation. It is thought that the repressed epigenetic state of FLC is propagated in the gametophyte, but then reprogrammed (transcriptionally activated) in the embryo and maintained in late embryogenesis and in the adult plant (Choi et al., 2009). Mutation in ELF6, a H3K27me3 JumonJi-domain demethylase, impaired the reactivation of FLC in the embryo, leading to the inheritance of a partially vernalized state (Crevillen et al., 2014). Interestingly, ELF6 is expressed in young ovules (Crevillen et al., 2014), raising the possibility that the resetting of FLC epigenetic status may occur concomitantly with the massive loss of H3K27me3 observed in the SMCs and FMSs (She et al., 2013). In this scenario, FLC expression would be effective only at embryogenesis thanks to embryo-specific transcriptional activators. This model linking chromatin dynamics in the early reproductive lineage to a transcriptional status after fertilization deserves further attention.
Hence, it is clear from these examples that the understanding of transgenerational epigenetic inheritance largely relies on our knowledge of the successive epigenetic reprogramming phases during female gamete formation. These are likely to shape the maternally inherited chromatin required for the correct deletion and resetting of epigenetic memory through generations, although which of the pre-meiotic or gametic processes is crucial remains to be determined.
Speculations and Perspectives
Despite significant advances in our knowledge of chromatin biology and epigenetically mediated gene regulation in differentiated plant tissues, our understanding of those processes in reproductive cells is still very scarce, at the levels of both descriptive and functional analyses. Yet recent findings provided evidence for the idea that chromatin dynamics are likely to support a whole sequence of cellular reprogramming processes in the female reproductive lineage, including: (i) the switch from mitotic to meiotic fate during SMC differentiation; (ii) the establishment of a pluripotent megaspore after meiosis; (iii) the epigenetic differentiation of the gametes in the multicellular female gametophyte; and (iv) the establishment of a maternal chromatin environment compatible with zygote totipotency and seed development.
Plant SMCs are functionally equivalent to animal primordial germ cells (PGCs), as plant egg cells are to animal oocytes. At least in Arabidopsis large similarities in chromatin dynamics and transcriptional rewiring between SMCs and PGCs (Schmidt et al., 2011; She et al., 2013), and egg cells and oocytes (Ingouff et al., 2007; Pillot et al., 2010b; Wuest et al., 2010), were already stressed, and allow for speculating analogous and specific roles of chromatin dynamics between both kingdoms.
The SMCs, PGCs and gametes are highly specialized cells, but at the same time carry a unique potential towards totipotency in the zygote. In animals, this duality is achieved by highly dynamic epigenetic reprogramming in PGCs and gamete differentiation, enabling the establishment of heterogenous epigenetic states throughout the genome. These states poise developmental genes for expression during lineage specification while repressing the somatic programs typical of terminally differentiated cells (Hemberger et al., 2009). Thus, similarly in plants, a major role of chromatin dynamics in the female reproductive lineage is likely to provide a functional architecture for cellular plasticity. This is a very exciting hypothesis that promotes the concept of a direct mechanical link between chromatin dynamics and a canalized plasticity in genome expression, enabling the establishment of pluripotency or totipotency. The possibility that a loose or open nuclear architecture is progressively locked when embryonic cells progress towards their differentiated state has been proposed for animals (Meister et al., 2011) and plants (Costa and Shaw, 2007; Rosa et al., 2014). Moreover, recent studies in mouse early embryos clearly showed that the mobility of core histones in itself is inversely correlated with the transition from totipotency to pluripotency, and to lineage commitment (Boskovic et al., 2014). Impressively, super-resolution nanoscopy also revealed the topological differentiation of chromatin fibers between pluripotent and somatic cells with respect to nucleosome groups called ‘clutches’: whereas the pluripotent chromatin is enriched in small and dispersed clutches strongly associated with Pol II, and is depleted of somatic linker histones, the somatic chromatin possess large clutches enriched in linker H1 and additional heterochromatic features (Ricci et al., 2015). In light of these data, it is tempting to speculate a working model for chromatin dynamics in the SMCs and the egg influencing nucleosome mobility, which might provide a mechanistic support to restoring the cellular plasticity necessary for the somatic-to-reproductive transition and the priming for totipotency in the egg cell (Figure 4). Instrumental to the acquisition of chromatin mobility might be the replacement of somatic linker H1 histones by related, plant-specific HMGA proteins (Jerzmanowski and Kotlinski, 2011) in the SMCs and egg cells, which remains to be investigated.
Figure 4.
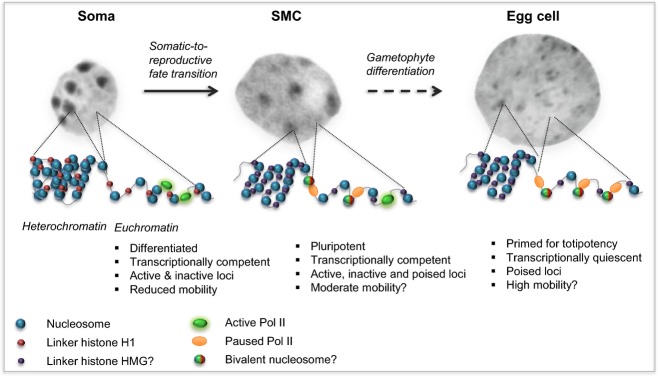
Speculative model for chromatin dynamics in the female reproductive lineage of flowering plants – towards a functional architecture for pluri/totipotency? The spore mother cell (SMC) and the gametes are highly specialized cells, but at the same time carry a unique potential towards totipotency in the zygote, a function implying highly dynamic chromatin architectures. The somatic-to-reproductive transition restores cellular plasticity in the SMC via global chromatin decondensation, loss of H1 and reduced histone acetylation levels, which altogether might promote the higher chromatin mobility favorable to pluripotency in the SMC. By contrast to the euchromatin of somatic cells, which harbors epigenetic and transcriptional differentiation of active and inactive loci, euchromatin in SMC harbors an epigenetic set-up that is favorable to transcriptional competence, yet shows reduced transcriptional activity. It is possible that this state might allow developmental loci to be poised, possibly involving the pausing of Pol II or bivalent domain nucleosomes, or both. The differentiation of the pluripotent gametophyte is associated with additional chromatin dynamics, particularly at the establishment of the egg cell fate. Chromatin dynamics during gametogenesis triggers the following: globally decondensed chromatin, devoid of somatic H1s; an epigenetically repressive set-up; and relative transcriptional quiescence, which is hypothesized to be necessary for totipotency in the zygote. By analogy to animal systems, a scenario involving poised developmental loci is proposed, possibly involving the pausing of Pol II and bivalent nucleosomes (where both repressive and activating marks are deposited), or both. The absence of somatic H1s in the SMC and egg cell might be functionally substituted by plant-specific HMGA proteins, which remain to be identified. The references related to these proposed scenarios are provided in the main text.
A functional specificity of the animal germline is the relative transcriptional quiescence leading to a global transcriptional repression of the somatic program, which is necessary for the acquisition of totipotency in the zygote (reviewed by Nakamura et al., 2010). Gamete maturation in yeast also involves a transcriptionally quiescent phase (Xu et al., 2012). Strikingly, the epigenetic and transcriptionally repressive chromatin state in the egg seems conserved in plants (see above). How does the germline chromatin accommodate both the transcriptional repression of the somatic program before fertilization and the priming of totipotency for the zygote? In mammals, developmental genes are poised via dual and antagonistic marking with the presence of repressive H3K27me3 and permissive H3K4me3, called bivalent marking, in correlation with paused Pol II at promoter regions. Upon lineage-specifying cues, these poised genes are rapidly primed for activation (reviewed by Vastenhouw and Schier, 2012; Voigt et al., 2013); however, in Drosophila, no bivalent states were detected and the control of Pol II occupancy and pausing is thought to be the main promoter-priming mechanism. Whether bivalent marking and/or RNA Pol II pausing in plants also serve the priming of developmental genes involved in the pluripotent development of the female gametophyte and in the acquisition of zygote totipotency remains an exciting, open question to investigate. In Arabidopsis, although the molecular components of the promoter-proximal Pol II pausing checkpoint are missing (Hajheidari et al., 2013), RNA Pol II pausing seems functional, and was shown to convey rapid transcriptional responses upon repeated drought stress (Ding et al., 2012). Whether bivalent chromatin states occur in Arabidopsis remains unsure, as the experiments suggesting these were based on mixed cell types (Berr et al., 2010; Roudier et al., 2011). Nevertheless, we propose a speculative model involving RNA Pol II pausing and/or bivalently marked nucleosomes in the egg cell that poise the developmental loci involved in totipotency (Figure 4).
The current perspectives are thus twofold. First, we need to elucidate further the mechanisms and developmental functions of chromatin dynamics in female reproductive development, particularly at the somatic-to-reproductive transition and at gamete differentiation. This objective requires elaborated genetic approaches to create spatially, and temporally, controlled perturbations of chromatin dynamics (for instance, interfering locally in the SMCs or FMSs upon histone turnover and histone modification). The availability of cell- and stage-specific promoters, as well as chemically inducible systems, should allow progress in this direction. Second, we need a radical improvement of our temporal and spatial resolution in the description of chromatin dynamics. Notably, although genetic and reporter gene analyses provided some evidence that enabled building a working model, a direct readout of DNA methylation dynamics in different sequence context and direct evidence of differential DNA methylation between the female gametes are currently lacking. The same is true for the dynamics of the effectors of small RNA pathways. Despite the existence of novel methods enabling cell type-specific isolation of nuclei for chromatin immunoprecipitation (Deal and Henikoff, 2011), elucidating the epigenome profile of the different cell types, from the SMC to the gametes, which are present in a limited number and are deeply embedded in ovule tissues, remains a major challenge. Collectively, gaining a precise functional understanding of plant chromatin dynamics in the female reproductive lineage requires technical and experimental innovations.
Acknowledgments
The authors are indebted to the two anonymous reviewers for their critical reading and constructive suggestions. CB and DA receive support from: the University of Zürich, the Swiss National Science Fondation (SNSF); the Institut de Recherche pour le Développement (IRD) and the Agence Nationale pour la Recherche (ANR), respectively. The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Figure S1. LHP1-GFP expression in Arabidopsis female gametophytes.
References
- Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 2013;76:729–741. doi: 10.1111/tpj.12336. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Baubec T, Finke A, Mittelsten Scheid O, Pecinka A. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 2014;15:446–452. doi: 10.1002/embr.201337915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Menard R, Meyer D, Fuchs J, Dong A, Shen WH. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22:3232–3248. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LT, Wente SR. From hypothesis to mechanism: uncovering nuclear pore complex links to gene expression. Mol. Cell. Biol. 2014;34:2114–2120. doi: 10.1128/MCB.01730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettoor AM, Givan SA, Cole RA, Coker CT, Unger-Wallace E, Vejlupkova Z, Vollbrecht E, Fowler JE, Evans MM. Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 2014;15:414. doi: 10.1186/s13059-014-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hyun Y, Kang MJ, et al. Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. 2009;57:918–931. doi: 10.1111/j.1365-313X.2008.03776.x. [DOI] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, et al. Arabidopsis meiotic crossover hotspots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 2013;45:1327–1336. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EH. The properties, origin, and mechanism of conversion-type inheritance at the B locus in maize. Genetics. 1966;53:1035–1063. doi: 10.1093/genetics/53.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature. 2006;439:493–496. doi: 10.1038/nature04269. [DOI] [PubMed] [Google Scholar]
- Costa S, Shaw P. ‘Open minded’ cells: how cells can change fate. Trends Cell Biol. 2007;17:101–106. doi: 10.1016/j.tcb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillen P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature. 2014;515:587–590. doi: 10.1038/nature13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro-De Leon G, Garcia-Aguilar M, Gillmor CS. Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature. 2014;514:624–627. doi: 10.1038/nature13620. [DOI] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- Duran-Figueroa N, Vielle-Calzada JP. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signal. Behav. 2010;5:1476–1479. doi: 10.4161/psb.5.11.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Aichinger E, Shu H, Wildhaber T, Alfarano P, Caflisch A, Gruissem W, Kohler C, Hennig L. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE. 2009;4:e5335. doi: 10.1371/journal.pone.0005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Feng X, Zilberman D, Dickinson H. A conversation across generations: soma-germ cell crosstalk in plants. Dev. Cell. 2013;24:215–225. doi: 10.1016/j.devcel.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Feng CM, Qiu Y, Van Buskirk EK, Yang EJ, Chen M. Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 2014;5:3027. doi: 10.1038/ncomms4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl Acad. Sci. USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl Acad. Sci. USA. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, ten Hoopen R, Tessadori F. Composition and formation of heterochromatin in Arabidopsis thaliana. Chromosome Res. 2006;14:71–82. doi: 10.1007/s10577-005-1022-5. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns–from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Jovtchev G, Schubert I. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008;16:891–898. doi: 10.1007/s10577-008-1252-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell. 2010;22:3249–3267. doi: 10.1105/tpc.109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim. Biophys. Acta. 2007;1769:276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES. Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc. Natl Acad. Sci. USA. 2014;111:2017–2022. doi: 10.1073/pnas.1323656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D, Roudier F. Epigenetics and development in plants: green light to convergent innovations. Curr. Top. Dev. Biol. 2013;104:189–222. doi: 10.1016/B978-0-12-416027-9.00006-1. [DOI] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Res. 2013;14:R129. doi: 10.1186/gb-2013-14-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Koncz C, Eick D. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013;18:633–643. doi: 10.1016/j.tplants.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat. Rev. Mol. Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FC. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell. 2012;24:4096–4109. doi: 10.1105/tpc.112.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Osman K, Jones GH, Franklin FCH. Factors underlying restricted crossover localization in barley meiosis. Annu. Rev. Genet. 2014;48:29–47. doi: 10.1146/annurev-genet-120213-092509. [DOI] [PubMed] [Google Scholar]
- Hollick JB. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 2012;15:536–543. doi: 10.1016/j.pbi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Houben A, Demidov D, Gernand D, Meister A, Leach CR, Schubert I. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 2003;33:967–973. doi: 10.1046/j.1365-313x.2003.01681.x. [DOI] [PubMed] [Google Scholar]
- Huanca-Mamani W, Garcia-Aguilar M, Leon-Martinez G, Grossniklaus U, Vielle-Calzada JP. CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2005;102:17231–17236. doi: 10.1073/pnas.0508186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang H, Xie X, Gao H, Guo G. Developmental changes in DNA methylation of pollen mother cells of David lily during meiotic prophase I. Mol. Biol. 2010;44:853–858. [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, Zalenskaya IA, Schultz RM, Meyer RG. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet. 2014;10:e1004317. doi: 10.1371/journal.pgen.1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 2007;17:1032–1037. doi: 10.1016/j.cub.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljic L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 2010;20:2137–2143. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Paszkowski J. Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl Acad. Sci. USA. 2014;111:8547–8552. doi: 10.1073/pnas.1402275111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janousek B, Zluvova J, Vyskot B. Histone H4 acetylation and DNA methylation dynamics during pollen development. Protoplasma. 2000;211:116–122. [Google Scholar]
- Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33:471–480. doi: 10.1046/j.1365-313x.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A, Kotlinski M. Conserved chromatin structural proteins – a source of variation enabling plant-specific adaptations? New Phytol. 2011;192:563–566. doi: 10.1111/j.1469-8137.2011.03918.x. [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U. Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 2007;8:R204. doi: 10.1186/gb-2007-8-10-r204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr. Biol. 2012;22:1825–1830. doi: 10.1016/j.cub.2012.07.061. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V. Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 2014;77:639–652. doi: 10.1111/tpj.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Egger RL, Zhang H, Walbot V. Unresolved issues in pre-meiotic anther development. Fron. Plant Sci. 2014;5:347. doi: 10.3389/fpls.2014.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014;55:1859–1863. doi: 10.1093/pcp/pcu125. [DOI] [PubMed] [Google Scholar]
- Kohler C, Hennig L. Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 2010;20:541–547. doi: 10.1016/j.gde.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Kohler C, Wolff P, Spillane C. Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 2012;63:331–352. doi: 10.1146/annurev-arplant-042811-105514. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Evolutionary genetics of life cycles. Annu. Rev. Ecol. Syst. 1997;28:391–435. [Google Scholar]
- Kubo T, Fujita M, Takahashi H, Nakazono M, Tsutsumi N, Kurata N. Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 2013;54:750–765. doi: 10.1093/pcp/pct029. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Lieber D, Lora J, Schrempp S, Lenhard M, Laux T. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr. Biol. 2011;21:1009–1017. doi: 10.1016/j.cub.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu Q, Yan Q, Shi L, Fang Y. Nucleolus-tethering system (NoTS) reveals that assembly of photobodies follows a self-organization model. Mol. Biol. Cell. 2014;25:1366–1373. doi: 10.1091/mbc.E13-09-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari P. An Introduction to the Embryology of Angiosperms. New York: McGraw-Hill: 1950. [Google Scholar]
- Mainiero S, Pawlowski WP. Meiotic chromosome structure and function in plants. Cytogenet. Genome Res. 2014;143:6–17. doi: 10.1159/000365260. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Kohler C. Mechanism of PHERES1 imprinting in Arabidopsis. J. Cell Sci. 2008;121:906–912. doi: 10.1242/jcs.023077. [DOI] [PubMed] [Google Scholar]
- Marques M, Laflamme L, Gervais AL, Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Curr. Opin. Genet. Dev. 2011;21:167–174. doi: 10.1016/j.gde.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl Acad. Sci. USA. 2012;109:5880–5889. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- Mozgova I, Köhler C, Hennig L. Keeping the gate closed: functions of the Polycomb repressive complex PRC2 in development. Plant J. 2015;83:121–132. doi: 10.1111/tpj.12828. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Shirae-Kurabayashi M, Hanyu-Nakamura K. Repression of early zygotic transcription in the germline. Curr. Opin. Cell Biol. 2010;22:709–714. doi: 10.1016/j.ceb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Naumann K, Fischer A, Hofmann I, et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Wang H, Li J, Holec S, Berger F. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol. Open. 2014;3:794–802. doi: 10.1242/bio.20148680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeley EJ, Podestà A, Jost J-P. Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proc. Natl Acad. Sci. USA. 1997;94:11721–11725. doi: 10.1073/pnas.94.21.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada J-P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GT, Frost JM, Park JS, Kim TH, Lee JS, Oh SA, Twell D, Brooks JS, Fischer RL, Choi Y. Nucleoporin MOS7/Nup88 is required for mitosis in gametogenesis and seed development in Arabidopsis. Proc. Natl Acad. Sci. USA. 2014;111:18393–18398. doi: 10.1073/pnas.1421911112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 2011;14:195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP. Chromosome organization and dynamics in plants. Curr. Opin. Plant Biol. 2010;13:640–645. doi: 10.1016/j.pbi.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113:258–269. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- Perrella G, Consiglio MF, Aiese-Cigliano R, Cremona G, Sanchez-Moran E, Barra L, Errico A, Bressan RA, Franklin FC, Conicella C. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J. 2010;62:796–806. doi: 10.1111/j.1365-313X.2010.04191.x. [DOI] [PubMed] [Google Scholar]
- Pillot M, Autran D, Leblanc O, Grimanelli D. A role for CHROMOMETHYLASE3 in mediating transposon and euchromatin silencing during egg cell reprogramming in Arabidopsis. Plant Signal. Behav. 2010a;5:1167–1170. doi: 10.4161/psb.5.10.11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M, Baroux C, Vazquez MA, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell. 2010b;22:307–320. doi: 10.1105/tpc.109.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhao L, Skaggs MI, et al. ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell. 2014;26:1612–1628. doi: 10.1105/tpc.113.120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U. Regulation and flexibility of genomic imprinting during seed development. Plant Cell. 2011;23:16–26. doi: 10.1105/tpc.110.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Mirouze M, Nicolet J, Paszkowski J. Parent-of-origin control of transgenerational retrotransposon proliferation in Arabidopsis. EMBO Rep. 2013;14:823–828. doi: 10.1038/embor.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, Garcia-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- Rosa S, Ntoukakis V, Ohmido N, Pendle A, Abranches R, Shaw P. Cell differentiation and development in arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell. 2014;26:4821–4833. doi: 10.1105/tpc.114.133793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 2011;9:e1001155. doi: 10.1371/journal.pbio.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Grossniklaus U. Analysis of plant germline development by high-throughput RNA profiling: technical advances and new insights. Plant J. 2012;70:18–29. doi: 10.1111/j.1365-313X.2012.04897.x. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Grossniklaus U. Plant germline formation: common concepts and developmental flexibility in sexual and asexual reproduction. Development. 2015;142:229–241. doi: 10.1242/dev.102103. [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Schubert I, Shaw P. Organization and dynamics of plant interphase chromosomes. Trends Plant Sci. 2011;16:273–281. doi: 10.1016/j.tplants.2011.02.002. [DOI] [PubMed] [Google Scholar]
- She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013;140:4008–4019. doi: 10.1242/dev.095034. [DOI] [PubMed] [Google Scholar]
- Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN. The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics. 1999;153:933–941. doi: 10.1093/genetics/153.2.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H, Nakamura M, Siretskiy A, Borghi L, Moraes I, Wildhaber T, Gruissem W, Hennig L. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol. 2014;15:R62. doi: 10.1186/gb-2014-15-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis MT, Rodriguez-Serrano M, Meijon M, Canal MJ, Cifuentes A, Risueno MC, Testillano PS. DNA methylation dynamics and MET1a-like gene expression changes during stress-induced pollen reprogramming to embryogenesis. J. Exp. Bot. 2012;63:6431–6444. doi: 10.1093/jxb/ers298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz P. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Otero S, Desvoyes B, Ramirez-Parra E, Jacobsen SE, Gutierrez C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2012;109:5370–5375. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 2007;120:1200–1208. doi: 10.1242/jcs.000026. [DOI] [PubMed] [Google Scholar]
- Tiang CL, He Y, Pawlowski WP. Chromosome organization and dynamics during interphase, mitosis, and meiosis in plants. Plant Physiol. 2012;158:26–34. doi: 10.1104/pp.111.187161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Koltunow AM. Traffic monitors at the cell periphery: the role of cell walls during early female reproductive cell differentiation in plants. Curr. Opin. Plant Biol. 2014;17:137–145. doi: 10.1016/j.pbi.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development. 2012;139:1399–1404. doi: 10.1242/dev.075390. [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl Acad. Sci. USA. 2006;103:13244–13249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25:246–256. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Vallejos RH, Christou P, Stoger E, Shaw P. Large-scale chromatin decondensation induced in a developmentally activated transgene locus. J. Cell Sci. 2005;118:1021–1031. doi: 10.1242/jcs.01685. [DOI] [PubMed] [Google Scholar]
- Weismann A. Das Keimplasma; eine Theorie der Vererbung. Jena: Fischer; 1892. [Google Scholar]
- Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H, Holec S, Alden K, Clarke ND, Jacques PE, Berger F. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenfuhrer J, von Mering C, Grossniklaus U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Xiao W, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 2006;142:1160–1168. doi: 10.1104/pp.106.088849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Soloveychik M, Ranger M, et al. Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation. Dev. Cell. 2012;23:1059–1071. doi: 10.1016/j.devcel.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 2012;8:e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HJ, Hogan P, Sundaresan V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 2005;139:1853–1869. doi: 10.1104/pp.105.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The arabidopsis nucleosome remodeler DDM1 Allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Pawlowski WP. Regulation of meiotic gene expression in plants. Fron. Plant Sci. 2014;5:413. doi: 10.3389/fpls.2014.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zubko E, Gentry M, Kunova A, Meyer P. De novo DNA methylation activity of methyltransferase 1 (MET1) partially restores body methylation in Arabidopsis thaliana. Plant J. 2012;71:1029–1037. doi: 10.1111/j.1365-313X.2012.05051.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. LHP1-GFP expression in Arabidopsis female gametophytes.