Abstract
Hepatitis E virus (HEV) infection is a cause of hepatitis in humans worldwide and has been associated with a mortality rate of up to 30% in pregnant women. Recently, persistent and chronic HEV infections have been recognized as a serious clinical problem, especially in immunocompromised individuals. To date, there are no FDA-approved HEV-specific antiviral drugs. In this study, we evaluated antisense peptide-conjugated morpholino oligomers (PPMO) designed against HEV genomic sequences as potential HEV-specific antiviral compounds. Two genetically-distinct strains of human HEV, genotype 1 Sar55 and genotype 3 Kernow-C1, isolated from patients with acute and chronic hepatitis, respectively, were used to evaluate inhibition of viral replication by PPMO in liver cells. The anti-HEV PPMO produced a significant reduction in the levels of HEV RNA and capsid protein, indicating effective inhibition of HEV replication. PPMO HP1, which targets a highly conserved sequence in the start site region of ORF1, was also effective against the genotype 3 Kernow-C1 strain in stably-infected HepG2/C3A liver cells. The antiviral activity observed was specific, dose-responsive and potent, suggesting that further exploration of PPMO HP1 as a potential HEV-specific antiviral agent is warranted.
Keywords: hepatitis E virus, morpholino oligomers, antisense, PPMO, antiviral
1. INTRODUCTION
Hepatitis E virus (HEV) is a single-stranded positive-sense RNA virus in the family Hepeviridae (Emerson et al., 2004). HEV is an etiologic agent of acute hepatitis in humans, and is endemic to various tropical and subtropical regions of the world. In pregnant women, infection with HEV genotype 1 can lead to fulminant hepatitis having a mortality rate of up to 30% (Jameel, 1999; Kumar et al., 2013). Hepatitis E is now recognized as a zoonotic disease, and strains of HEV from pig, chicken, mongoose, rabbit, rat, ferret, bat, fish and deer have been genetically characterized (Haqshenas et al., 2001; Li et al., 2005; Meng, 2011; Meng et al., 1997). Over the past several years, chronic and persistent symptomatic HEV infections have been reported in increasing numbers of immunocompromised individuals, including organ transplant recipients and leukemia, lymphoma and HIV/AIDS patients, in industrialized countries (Kamar et al., 2014a).
The HEV genome is approximately 7.2 kb in length and consists of three open reading frames (ORFs) (Tam et al., 1991). ORF1 encodes all the putative nonstructural proteins involved in HEV replication. ORF2 encodes the capsid protein, the major structural protein in the HEV virion. ORF3 encodes a multi-functional phosphoprotein that is essential for establishing HEV infection in macaques and pigs (Graff et al., 2005; Huang et al., 2007). HEV strains are highly diverse in sequence and those strains infecting humans are classified into four major genotypes in the genus Orthohepevirus (Lu et al., 2006; Smith et al., 2014). HEV genotype 1 and 2 are restricted to humans and have no known animal reservoir, whereas genotype 3 and 4 infect several animal species in addition to humans and are known to be zoonotic (Ahmad et al., 2011; Meng, 2010).
Although it has been over two decades since the sequence of a full-length HEV genome was first published (Tam et al., 1991), there are as yet no FDA-approved HEV-specific drugs. Off-label use of ribavirin and pegylated interferon for treatment of acute and chronic hepatitis E patients has been reported (Gerolami et al., 2011; Kamar et al., 2010; Mallet et al., 2010; Wedemeyer et al., 2012), but there are side effects and efficacy concerns with respect to those options (Kamar et al., 2014b; Pischke et al., 2013). Ribavirin belongs to the FDA Pregnancy Risk Category X and is not recommended for use in pregnant women. Interferon cannot be used in most transplant patients. Thus, there is a pressing need for the development of a HEV-specific antiviral therapeutic agent, especially for treating severe infections in pregnant women and chronic infections in immunocompromised patients.
In this study, phosphorodiamidate morpholino oligomers (PMO) were tested for their ability to inhibit HEV replication in liver cells. PMO are nuclease-resistant single-stranded DNA analogs containing a backbone of morpholine rings and phosphorodiamidate linkages (Summerton, 1999). PMO bind to mRNA by Watson–Crick base pairing and can interfere with translation through steric blockade of the AUG-translation start site region. Conjugation of PMO to an arginine-rich cell penetrating peptide, yielding peptide-conjugated PMO (PPMO), facilitates delivery into cells (Abes et al., 2006; Summerton, 1999). PPMO are water soluble and enter cells readily. This study demonstrates that several anti-HEV PPMO display potent inhibition of HEV genotype 1 replication. Notably, PPMO HP1, which targets the start site region of HEV ORF1, also effectively inhibited replication of genotype 3 Kernow-C1 strain in liver cells.
2. MATERIALS AND METHODS
2.1. PPMO design and synthesis
Based on previous studies targeting viral RNAs with PPMO (Stein, 2008), the PPMO for this study were designed to target genomic sequence of the HEV Sar55 strain (GenBank Accession # AF444002). PPMO HP1 and HP2 are complementary to a sequence in the 5’end region of genomic and subgenomic RNA, respectively (Fig. 1 and Table 1). HP3U is complementary to a sequence in the terminal region of the 3′ UTR. HPN3 is the reverse complement of HP1 and was intended to interfere with the synthesis of positive-sense genomic RNA. A nonsense-sequence PPMO CP1 (Zhang et al., 2007), having little agreement with HEV or human mRNA sequences, was used as a negative control PPMO. CP1 with fluorescein conjugated at its 3’end (CP1-F) was used in the PPMO uptake assay. PPMO were synthesized with an arginine-rich cell-penetrating peptide (P7) conjugated at the 5’end at AVI BioPharma Inc (Corvallis, OR) as previously described (Abes et al., 2006).
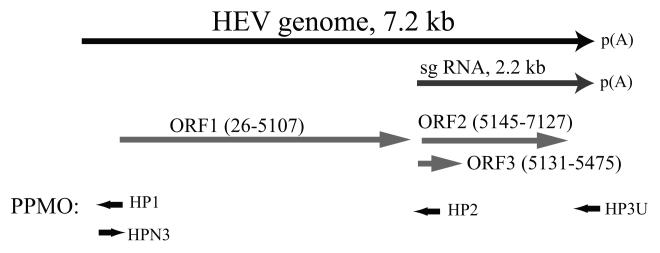
Fig.1. Schematic illustration of HEV genome, subgenomic RNA (sg RNA), ORFs, and PPMO target locations.
The numbers after ORFs indicate the nucleotide position of the beginning and end of the open reading frames (drawn in light gray arrows) within the HEV Sar55 genome (GenBank Accession # AF444002). The black arrows shown in the PPMO indicate their 5′ to 3′ orientation relative to the positive-sense HEV RNA genome.
Table 1.
PPMO sequences and their target sites in HEVa
| Name | PPMO sequence (5′ to 3′) | Target site in HEV genome (position)b |
|---|---|---|
| HP1 | GGGCCTCCATGGCATCGACC | Start site region of ORF1 (18-37) |
| HP2 | CATGGGCGCAGCAAAAGACA | Start site region of ORF2 (5116-5135) |
| HP3U | GCGCGAAACGCAGAAAAGAG | Terminal region of 3′ UTR (7169-7188) |
| HPN3 | GGTCGATGCCATGGAGGCCC | 3′ terminal region of negative sense RNAc |
| CP1 | GATATACACAACACCCAATT | None |
2.2. Cell-free translation
PPMO target sequences were cloned upstream of the luciferase gene in reporter vector pCiNeoLucr as previously described (Zhang et al., 2007). Briefly, oligomers of 30-nt in length containing the target sequence for PPMO HP1, HP2, and HP3U were each cloned upstream of luciferase coding sequence in pCiNeoLucr vector. The in vitro transcription and translation reactions were carried out as previously described (Zhang et al., 2008). Luminescence signal was measured with VICTOR3™ Multilabel Counter (Perkin-Elmer Life and Analytical Sciences, Wellesley, MA).
2.3. Cells, viruses and transfections
S10-3 cells, a subclone of Huh-7 hepatoma cells (Graff et al., 2006), and hepatoma cells HepG2/C3A (ATCC CRL-10741) were maintained in DMEM medium supplemented with 10% fetal bovine serum.
The PPMO uptake assay was performed in uninfected S10-3 cells. Approximately 1.2×105 cells were seeded per well into 12-well plates, producing confluent monolayers in 24 h. PPMO CP1-F was added to the cell supernatant at a final concentration of 8 μM and the cells further incubated at 37°C for 4 h. The medium was removed and the cells rinsed with PBS, before observation with fluorescence microscopy to assess PPMO uptake efficiency.
Transfection of S10-3 cells with HEV RNA in vitro transcribed from pSK-E2 (an infectious cDNA clone of HEV Sar55 strain) or pSK-E2-Luc (containing luciferase reporter) was performed using DMRIE-C reagent (Invitrogen, Grand Island, NY) as previously described (Nan et al., 2014a; Nan et al., 2014b). For PPMO treatment of the S10-3 cells in 12-well plates, cell culture supernatant was discarded 5 hours after RNA transfection and the cells were rinsed twice with Opti-MEM. PPMO suspended in 0.5 mL Opti-MEM was then added to the cell monolayers. Four hours after PPMO treatment, the PPMO solution was removed and 1 mL DMEM with 10% FBS was added to each well. The cells were then further incubated at 34.5°C for 7 days prior to further analysis for viral protein, RNA or luciferase signal. Luciferase activity from pSK-E2-Luc was determined by using the Bright-Glo™ Luciferase Assay System (Promega, Madison, WI).
The HEV genotype 3 Kernow-C1 strain p6 was used to infect HepG2/C3A cells at a multiplicity of infection (MOI) of 1 (Shukla et al., 2011). Kernow-C1 replication does not cause cytopathic effect and infected HepG2/C3A cells were passaged five times to produce stably infected cells. Immunofluorescence assay (IFA) with chimpanzee anti-HEV antibody (a gift from Suzanne Emerson at the National Institutes of Health) was conducted to confirm virus replication. Subsequently, the Kernow-C1-infected cells were seeded into 12-well plates. PPMO was then added to the HepG2/C3A cells in fresh medium once every two days for 6 days (3 treatments total). The cells were maintained at 37°C and harvested for analysis one day after the final PPMO treatment.
2.4. Cell viability assay
Viability of S10-3 cells after PPMO treatment was determined with CellTiter-Glo® Luminescent Cell Viability Assay (Promega). S10-3 cells were treated with the PPMO as described above for seven days, and viability measured according to manufacturer’s instructions.
2.5. IFA
IFA and confocal fluorescence microscopy were carried out as reported previously with chimpanzee antibody against the HEV capsid protein (Nan et al., 2014b).
2.6. Western blot analysis
Cells were lysed in Laemmli sample buffer. Total protein was subjected to sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting as described previously (Patel et al., 2010; Zhang et al., 2007). An anti-HEV ORF2 monoclonal antibody (clone 1E6, EMD Millipore, Billerica, MA) was used at dilution of 1:1000 (Riddell et al., 2000). The Quantity One Program (Version 4.6) and a ChemiDoc XRS imaging system (Bio-Rad Laboratories, Hercules, CA) were used for digital signal acquisition and densitometry analyses. β-tubulin was also detected as a protein load control.
2.7. Reverse transcription and real-time PCR (RT-qPCR)
RNA isolation, reverse transcription and real-time PCR were performed as previously described (Nan et al., 2012; Nan et al., 2014b). For the detection of HEV-specific RNA, HEV specific reverse primer (Sar55-R3, CAGAATCCACGCAGACCTTA) was used in reverse transcription. Primers Sar55-F3 (TGAGTTTGATTCCACCCAGA) and Sar55-R3 were used for real-time PCR (SYBR Green-based) on Sar55 cDNA. For absolute quantification of HEV RNA, the pSK-E2 (Sar55) plasmid served as the template to establish a standard curve.
2.8. Statistical analysis
Student t-test was used to assess the statistical significance in the difference between the level of luciferase signal or HEV RNA copies of groups of cells receiving the indicated PPMO treatments compared to no treatment. A two tailed P-value of less than 0.05 was considered significant.
3. RESULTS
3.1. PPMO inhibit target mRNA translation in cell-free luciferase reporter assay
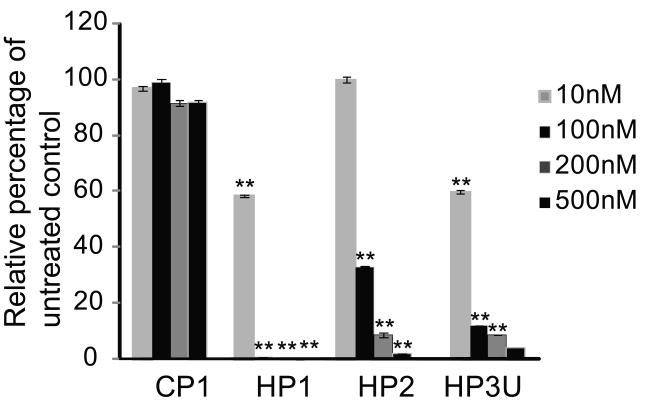
To validate binding of the PPMO to their respective target sequences, each PPMO was tested against RNA containing the PPMO target region upstream of and in frame with luciferase coding sequence. PPMO were added at various concentrations to cell-free translation reactions containing in vitro transcribed RNA from each reporter plasmid. Compared with CP1, each HEV-targeted PPMO reduced luciferase signal significantly (Fig. 2). PPMO HP1 produced a 99% reduction at 100 nM (Fig. 2). Similarly, PPMO HP2, and HP3U reduced luciferase expression by around 90% at 200 nM (Fig. 2). All the PPMO behaved in a dose-dependent manner with HP1 producing the most potent inhibition.
Fig. 2. Cell-free luciferase reporter assay.
PPMO were added to in vitro translation reactions containing RNA transcribed from reporter constructs that include PPMO target sequences upstream of and in-frame with firefly luciferase coding sequence. Reactions with a construct containing HP1 target sequence and treated with PPMO CP1 were used as a negative control. Luciferase activity in the presence of the various PPMO is graphed as the relative percentage of untreated control reactions, set as 100%. The average of three tests is shown and the error bars represent standard errors among the experiments. ** indicates significant differences from CP1 at corresponding concentrations (P < 0.01).
3.2. PPMO inhibit HEV capsid production
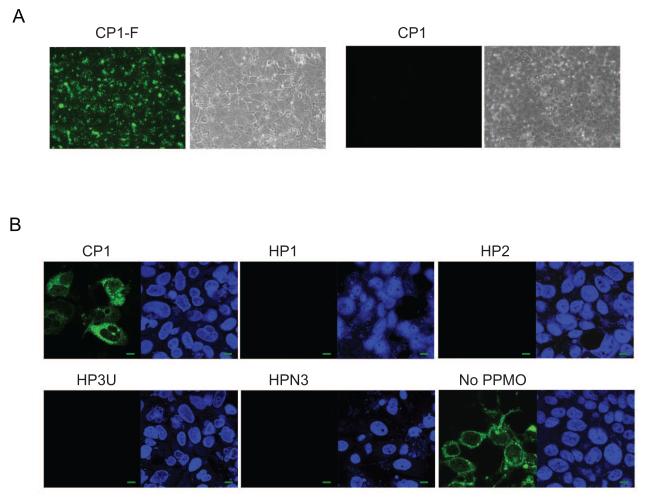
We next conducted a PPMO uptake assay in uninfected S10-3 cells with PPMO CP1-F. Highly efficient uptake of the CP1-F was observed, as indicated by the presence of green fluorescence signal present in all cells (Fig. 3A). Having established that PPMO enter S10-3 cells effectively, we next tested whether the PPMO designed were able to inhibit HEV replication. S10-3 cells were transfected with full-length Sar55 RNA, then treated with 16 μM PPMO, a dose chosen based on previous studies (Zhang et al., 2006; Zhang et al., 2007). PPMO HP1, HP2, HP3U and HPN3 produced marked reduction of capsid protein expression, indicating inhibition of HEV replication, while CP1 had minimal effect (Fig. 3B). The results indicate that the four HEV-targeted PPMO generated specific inhibition of HEV replication.
Fig. 3. PPMO enter S10-3 liver cells and inhibit HEV replication.
A. PPMO uptake assay in S10-3 cells. Fluorescein-conjugated CP1 (CP1-F) or CP1 without fluorescein conjugation were added to S10-3 cells and incubated for 4 h before fluorescence microscopy. Green fluorescence indicates uptake of PPMO. Bright field illuminations are shown next to the fluorescence images. B. Immunofluorescence assay of S10-3 cells infected with HEV. Cells were transfected with Sar55 RNA transcribed from pSK-E2, treated with indicated PPMO (16 μM) 5 hours later, and fixed for IFA at 7 days post-transfection. In each panel, the left image shows IFA using HEV-specific antibody, and the right image shows the same field with cell nuclei stained by DAPI. A green scale bar in the lower right corner represents 10 μm.
We also tested whether the PPMO produced cytotoxicity to S10-3 cells, as an impact on cell viability could produce non-specific inhibition of viral replication. When uninfected cells were treated with 16 μM PPMO HP1 under the same conditions as the antiviral assays above, no impact on cell viability was observed (data not shown).
3.3. PPMO treatment generates dose-dependent inhibition of HEV replication
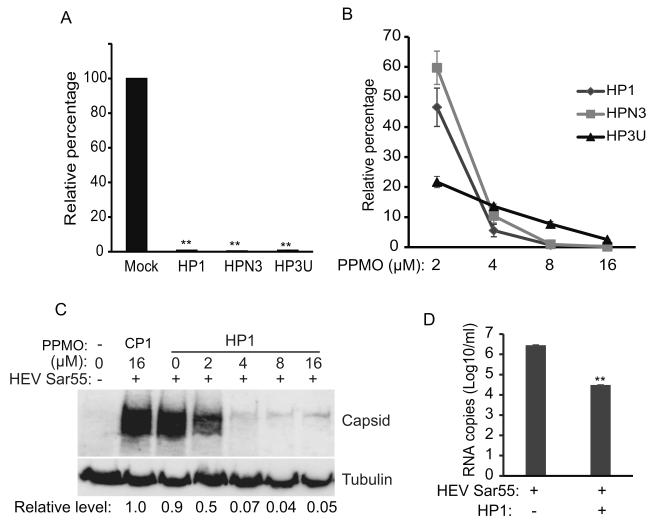
Next, an HEV replicon system containing a luciferase reporter (pSK-E2-Luc) was used to further verify the antiviral effect of selected PPMO. In the pSK-E2-Luc replicon, insertion of luciferase coding sequence into HEV ORF2/3 region disrupts ORF2 and ORF3 expression but provides a quantitative means to measure translation of subgenomic viral RNA (Graff et al., 2006). PPMO HP2 was not included in further testing due to the presumed disruption of its binding site by the luciferase insertion. Cells were transiently transfected with pSK-E2-Luc and treated with 16 μM PPMO. Luciferase yields in cells treated with HP1, HPN3 and HP3U were reduced by 99.1%, 99.4% and 98.9%, respectively, compared to mock-treated cells (Fig. 4A).
Fig. 4. Dose-dependent inhibition of HEV replication by PPMO.
A. Luciferase assay of S10-3 cells transfected with Sar55 RNA from HEV replicon pSK-E2-Luc. Cells were transfected with the viral RNA, treated with 16 μM PPMO 5 h later, and harvested for luciferase assay at 7 days post-transfection. Relative percentages of luciferase activity are shown in comparison with mock-treated S10-3 cells. Error bars represent standard errors among three repeat experiments. ** indicates significant difference from the mock-treated cells (P < 0.01). B. Dose-dependent inhibition of HEV replication by PPMO, using the same experimental scheme as in A above. C. Treatment of S10-3 cells with PPMO HP1 inhibits HEV capsid protein production in a dose-dependent manner. Cells were transfected with HEV RNA from pSK-E2, treated with PPMO HP1 5 h later and harvested 7 days later for Western blotting. A representative blot of three independent experiments is shown. D. HEV RNA levels present in supernatant of S10-3 cells treated with PPMO. Cells were transfected with Sar55 RNA and treated 8 μM PPMO HP1 5 h later. The cell culture supernatant was harvested 7 days post-transfection and RNA detected by RT-qPCR. The result shows average of four repeat treatments.
Further evaluations showed that PPMO HP1, HPN3 and HP3U generated dose-dependent reductions of luciferase expression (Fig. 4B). Luciferase expression in the cells treated with HP1 at 2, 4, and 8 μM was reduced by 53.4%, 94.4%, and 99.7%, respectively, compared to that of the mock-treated control. PPMO HPN3 reduced luciferase expression by 40.3%, 89.6% and 99.03%, when used at 2, 4, and 8 μM, respectively. PPMO HP3U at 2, 4, and 8 μM reduced the luciferase expression by 78.3%, 86.4% and 92.3%, respectively.
Of the three PPMO tested above in two cell-based systems, HP1 produced more potent inhibition of HEV replication than HPN3. In addition, sequence analysis indicated that the HP1 target sequence is more conserved than that of HP3U. Thus, HP1 was selected for further evaluation in S10-3 cells, where inhibition of virus replication was evaluated by Western blot detection of the HEV capsid protein. Cells receiving HP1 treatment at 2, 4, and 8 μM had relative capsid protein at 0.5, 0.07 and 0.04-fold, respectively, of cells treated with CP1, as indicated by densitometry analysis of the Western blots (Fig. 4C).
We next tested if PPMO treatment reduced the level of HEV RNA production. Cells were transfected with Sar55 RNA and, based on the above results, treated with 8 μM HP1. HEV RNA present in the supernatant of cell cultures was detected by RT-qPCR at seven days post transfection. The HP1 treatment led to significant reduction of HEV RNA from 2.8 × 106 copies to less than 3.1 × 104 copies per mL (Fig. 4D). The results were consistent with those of capsid protein detection and the luciferase reporter assay (pSK-E2-Luc) described above.
3.4. PPMO HP1 inhibits replication of HEV genotype 3 Kernow-C1
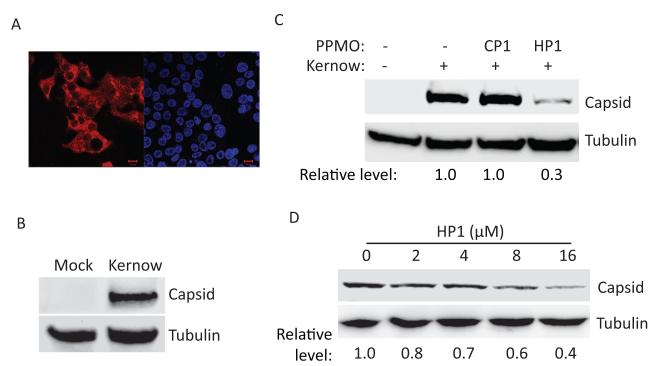
Kernow-C1, a genotype 3 HEV strain, has been successfully adapted to propagate in HepG2/C3A cells (Shukla et al., 2011). Since Kernow-C1 replication does not cause cytopathic effect, we established HepG2/C3A cells stably infected with the Kernow-C1 virus by multiple rounds of passaging. Active replication of HEV Kernow-C1 in HepG2/C3A cells was confirmed by both IFA and Western blotting (Fig. 5A and B). Sequence alignment revealed that the target sequence of PPMO HP1 in the genotype 1 Sar55 and genotype 3 Kernow-C1 is 100% conserved, while there are 4 nt mismatches between Kernow-C1 and Sar55 strains at the HP3U target site. We therefore tested PPMO HP1 in Kernow-infected HepG2/C3A cells and found that it reduced capsid protein level to 0.3-fold of untreated cells (Fig. 5C). More thorough evaluation showed dose-dependent inhibition of Kernow-C1capsid protein expression by HP1 (Fig. 5D).
Fig. 5.
Inhibition of HEV Kernow-C1 virus replication in HepG2/C3A cells. A. IFA of Kernow-C1-infected HepG2/C3A cells. The left image shows HEV-positive cells, and the right image shows the same field of cells stained with DAPI. A red scale bar in the lower right corner represents 10 μm. B. Western blotting detection of HEV capsid protein in Kernow-C1-infected HepG2/C3A cells. A representative blot of three independent experiments is shown. C. PPMO-mediated inhibition of Kernow-C1 virus replication. Cells were treated with 16 μM PPMO in fresh medium every two days for six days, then harvested one day after the final treatment. Relative levels of HEV capsid protein production in PPMO-treated cells are shown below the images in comparison with non-treated cells. D. Dose-dependent inhibition of Kernow-C1 capsid production by HP1. The cells were treated with PPMO HP1, as in C above.
Taken together, the data from experiments using two HEV genotypes and three different cell-based systems showed PPMO HP1 to be an effective inhibitor of HEV replication.
4. DISCUSSION
Our results demonstrate that PPMO targeting HEV RNA can inhibit HEV replication effectively. Inhibition of HEV replication in cells was demonstrated by reductions in both viral RNA and capsid protein levels. PPMO HP1, which targets the initiation region of ORF1, effectively inhibited the replication of the genotype 1 Sar55 strain as well as established infections of genotype 3 Kernow-C1 strain. The HP1 target site is perfectly conserved between HEV genotypes 1-3, and has one base of variance with type 4 (data not shown). The conserved nature of its target site and overall efficacy of PPMO HP1 in this study suggest it may be an HEV-specific inhibitor with antiviral activity across multiple HEV genotypes.
PPMO HPN3 and HP3U were able to inhibit the Sar55 replication in a dose-dependent manner. The target sites of HP3U and HPN3 are in the terminal region of the 3′ ends of HEV genomic plus-strand and replicative-intermediate minus-strand, respectively, where the HEV RNA-dependent RNA polymerase (RdRp) presumably associates during RNA synthesis. We speculate that those two PPMO may obstruct access of the RdRp to the respective RNA, thereby interfering with viral RNA synthesis.
Antisense PMO are currently in clinical trials, including a treatment for Duchenne muscular dystrophy in humans (Anthony et al., 2012; Mendell et al., 2013). PPMO have also been used in a clinical trial, albeit in an ex-vivo model (Moulton, 2013). PPMO have been documented as effective against numerous types of viral infections of the liver in experimental animal models. Importantly, upon systemic administration, PPMO distribute to liver tissue, remain pharmacologically viable, and have been effective at reducing viral titers (Amantana et al., 2007; Burrer et al., 2007; Paessler et al., 2008). These qualities, along with the efficacy against HEV replication in cultured cells that we observed in this study, suggest PPMO should be considered for further development as an inhibitor of HEV infections. Further evaluation and development of anti-HEV PPMO will require in vivo investigation, and the pig model of infection with genotype 3 HEV appears to be suitable for such study (Meng et al., 1998).
In summary, our results indicate that PPMO can be effective antiviral compounds against HEV infection. PPMO HP1 has potent activity against strains of HEV from two different genotypes, including an established infection of HepG2/C3A cells with the genotype 3 Kernow-C1 strain. The results suggest that HP1 is a promising candidate for further development as a HEV-specific antiviral compound.
Highlights.
Peptide-conjugated morpholino oligomers (PPMO) were designed against hepatitis E virus (HEV).
Four PPMOs are effective in inhibition of replication of a genotype 1 HEV strain.
PPMO HP1 also inhibits HEV Kernow-C1, a genotype 3 HEV strain, in stably infected cells.
HP1 may be a good candidate to be further explored as a HEV-specific antiviral compound.
ACKNOWLEDGEMENT
We thank Suzanne Emerson at the National Institutes of Health for generously providing the S10-3 cells, pSK-E2, pSK-E2-Luc, Kernow-C1 virus, and chimpanzee antibody, and the Chemistry Group at AVI BioPharma for their expert production of PPMO. This work was supported by NIH grant 1R21AI068881.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL, Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J Control Release. 2006;116:304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Holla RP, Jameel S. Molecular virology of hepatitis E virus. Virus Res. 2011;161:47–58. doi: 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amantana A, Moulton HM, Cate ML, Reddy MT, Whitehead T, Hassinger JN, Youngblood DS, Iversen PL. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjug Chem. 2007;18:1325–1331. doi: 10.1021/bc070060v. [DOI] [PubMed] [Google Scholar]
- Anthony K, Feng L, Arechavala-Gomeza V, Guglieri M, Straub V, Bushby K, Cirak S, Morgan J, Muntoni F. Exon skipping quantification by quantitative reverse-transcription polymerase chain reaction in Duchenne muscular dystrophy patients treated with the antisense oligomer eteplirsen. Human gene therapy methods. 2012;23:336–345. doi: 10.1089/hgtb.2012.117. [DOI] [PubMed] [Google Scholar]
- Burrer R, Neuman BW, Ting JP, Stein DA, Moulton HM, Iversen PL, Kuhn P, Buchmeier MJ. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J Virol. 2007;81:5637–5648. doi: 10.1128/JVI.02360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S, Anderson D, Arankalle VA, Meng X-J, Purdy M, Schlauder GG, Tsarev S. Hepevirus. In: Fauquest CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy, VIIIth report of the ICTV. Elseiver/Academic Press; London, UK: 2004. [Google Scholar]
- Gerolami R, Borentain P, Raissouni F, Motte A, Solas C, Colson P. Treatment of severe acute hepatitis E by ribavirin. J Clin Virol. 2011;52:60–62. doi: 10.1016/j.jcv.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Graff J, Nguyen H, Yu C, Elkins WR, St Claire M, Purcell RH, Emerson SU. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J Virol. 2005;79:6680–6689. doi: 10.1128/JVI.79.11.6680-6689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Torian U, Nguyen H, Emerson SU. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol. 2006;80:5919–5926. doi: 10.1128/JVI.00046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J Gen Virol. 2001;82:2449–2462. doi: 10.1099/0022-1317-82-10-2449. [DOI] [PubMed] [Google Scholar]
- Huang YW, Opriessnig T, Halbur PG, Meng XJ. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol. 2007;81:3018–3026. doi: 10.1128/JVI.02259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameel S. Molecular biology and pathogenesis of hepatitis E virus. Expert Rev Mol Med. 1999;1999:1–16. doi: 10.1017/S1462399499001271. [DOI] [PubMed] [Google Scholar]
- Kamar N, Dalton HR, Abravanel F, Izopet J. Hepatitis E virus infection. Clinical microbiology reviews. 2014a;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N, Izopet J, Tripon S, Bismuth M, Hillaire S, Dumortier J, Radenne S, Coilly A, Garrigue V, D’Alteroche L, Buchler M, Couzi L, Lebray P, Dharancy S, Minello A, Hourmant M, Roque-Afonso AM, Abravanel F, Pol S, Rostaing L, Mallet V. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med. 2014b;370:1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- Kamar N, Rostaing L, Abravanel F, Garrouste C, Lhomme S, Esposito L, Basse G, Cointault O, Ribes D, Nogier MB, Alric L, Peron JM, Izopet J. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis e virus infection. Gastroenterology. 2010;139:1612–1618. doi: 10.1053/j.gastro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Kumar S, Subhadra S, Singh B, Panda BK. Hepatitis E virus: the current scenario. Int J Infect Dis. 2013;17:e228–233. doi: 10.1016/j.ijid.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Li TC, Chijiwa K, Sera N, Ishibashi T, Etoh Y, Shinohara Y, Kurata Y, Ishida M, Sakamoto S, Takeda N, Miyamura T. Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis. 2005;11:1958–1960. doi: 10.3201/eid1112.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- Mallet V, Nicand E, Sultanik P, Chakvetadze C, Tesse S, Thervet E, Mouthon L, Sogni P, Pol S. Brief communication: case reports of ribavirin treatment for chronic hepatitis E. Ann Intern Med. 2010;153:85–89. doi: 10.7326/0003-4819-153-2-201007200-00257. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, Alfano L, Gomez AM, Lewis S, Kota J, Malik V, Shontz K, Walker CM, Flanigan KM, Corridore M, Kean JR, Allen HD, Shilling C, Melia KR, Sazani P, Saoud JB, Kaye EM. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- Meng XJ. Recent advances in Hepatitis E virus. J Viral Hepat. 2010;17:153–161. doi: 10.1111/j.1365-2893.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011;161:23–30. doi: 10.1016/j.virusres.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Mushahwar IK, Purcell RH, Emerson SU. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72:9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton HM. In vivo delivery of morpholino oligos by cell-penetrating peptides. Curr Pharm Des. 2013;19:2963–2969. doi: 10.2174/1381612811319160010. [DOI] [PubMed] [Google Scholar]
- Nan Y, Ma Z, Wang R, Yu Y, Kannan H, Fredericksen B, Zhang YJ. Enhancement of Interferon Induction by ORF3 Product of Hepatitis E Virus. J Virol. 2014a;88:8696–8705. doi: 10.1128/JVI.01228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Y, Wang R, Shen M, Faaberg KS, Samal SK, Zhang YJ. Induction of type I interferons by a novel porcine reproductive and respiratory syndrome virus isolate. Virology. 2012;432:261–270. doi: 10.1016/j.virol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. Hepatitis E Virus Inhibits Type I Interferon Induction by ORF1 Product. J Virol. 2014b;88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paessler S, Rijnbrand R, Stein DA, Ni H, Yun NE, Dziuba N, Borisevich V, Seregin A, Ma Y, Blouch R, Iversen PL, Zacks MA. Inhibition of alphavirus infection in cell culture and in mice with antisense morpholino oligomers. Virology. 2008;376:357–370. doi: 10.1016/j.virol.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Nan Y, Shen M, Ritthipichai K, Zhu X, Zhang YJ. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol. 2010;84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke S, Hardtke S, Bode U, Birkner S, Chatzikyrkou C, Kauffmann W, Bara CL, Gottlieb J, Wenzel J, Manns MP, Wedemeyer H. Ribavirin treatment of acute and chronic hepatitis E: a single-centre experience. Liver Int. 2013;33:722–726. doi: 10.1111/liv.12114. [DOI] [PubMed] [Google Scholar]
- Riddell MA, Li F, Anderson DA. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J Virol. 2000;74:8011–8017. doi: 10.1128/jvi.74.17.8011-8017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A. 2011;108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. Consensus proposals for classification of the family Hepeviridae. J Gen Virol. 2014;95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DA. Inhibition of RNA Virus Infections with Peptide-Conjugated Morpholino Oligomers. Current Pharmaceutical Design. 2008;14:2619–2634. doi: 10.2174/138161208786071290. [DOI] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemeyer H, Pischke S, Manns MP. Pathogenesis and treatment of hepatitis e virus infection. Gastroenterology. 2012;142:1388–1397. e1381. doi: 10.1053/j.gastro.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Bonaparte RS, Patel D, Stein DA, Iversen PL. Blockade of viral interleukin-6 expression of Kaposi’s sarcoma-associated herpesvirus. Molecular cancer therapeutics. 2008;7:712–720. doi: 10.1158/1535-7163.MCT-07-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Stein DA, Fan SM, Wang KY, Kroeker AD, Meng XJ, Iversen PL, Matson DO. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet Microbiol. 2006;117:117–129. doi: 10.1016/j.vetmic.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Wang KY, Stein DA, Patel D, Watkins R, Moulton HM, Iversen PL, Matson DO. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res. 2007;73:12–23. doi: 10.1016/j.antiviral.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]