Abstract
HIV-1 incidence has been increasing more rapidly in the Middle East and North Africa than in any other global region. Despite this trend, HIV epidemiology in the region remains poorly defined. We conducted an analysis of 3284 publicly available HIV-1 sequences from 15 countries in the Middle East and North Africa to better characterize the regional epidemic. A phylogenetic tree based on the reverse transcriptase gene revealed a complex mosaic of diverse HIV subtypes and circulating recombinant forms across the region.
Since 2001, HIV incidence has fallen by 50% in large parts of the developing world [1]. During the same time period, however, countries in the Middle East and North Africa (MENA) have experienced a 52% rise in the number of new HIV infections and near-tripling of AIDS-related deaths [2]. Although regional prevalence remains relatively low (0.1%), HIV incidence has increased more rapidly in the MENA countries than in any other region in the past decade. In 2012 alone, an estimated 260 000 people were living with HIV in the 23 countries that comprise the MENA region, 12% of whom were infected that year [3]. The regional HIV epidemic is currently concentrated in injection drug users, female sex workers, prisoners, and MSM [4]. Certain pockets within the region, however, harbor the threat of a generalized epidemic, as the virus spreads from the most-at-risk to the general population. HIV transmission in Iran, which has the largest number of estimated cases in the MENA region, has historically been transmitted via injection drug use, accounting for more than eight in 10 transmission events in 2001. In 2010, however, only 54% of new HIV infections could be directly attributed to injection drug use, whereas heterosexual transmission accounted for 34% of new cases, up from 5% a decade earlier [5]. Despite these alarming trends, the regional epidemic remains poorly described, HIV disease is still highly stigmatized, and antiretroviral therapy is only accessible to a relatively small proportion in need of treatment.
A key feature of the MENA epidemic is the paucity of available data, as nearly half of the countries in the region have limited or no formal HIV surveillance systems. In 2011, only eight of the 14 countries that reported new HIV cases had data on the total number of cases living with HIV [6]. Still, there are growing databases that can provide key information on HIV epidemiology in the MENA region. Since the start of the epidemic in the 1980s, HIV sequences from cases identified in the MENA countries have been sporadically published, providing a mosaic overview of the molecular epidemiology of HIV in the region. To date, however, no aggregate analysis of these sequences has been undertaken.
To better characterize the MENA HIV-1 epidemic, we performed a phylogenetic analysis of 3284 publicly available sequences from 15 countries in the region (data available as of 24 September 2014 in the Los Alamos HIV Sequence Database, http://www.hiv.lanl.gov). The MENA region was defined as the 23 countries in the WHO Eastern Mediterranean Region [7] and Algeria. No sequences were available from Bahrain, Iraq, Jordan, Oman, the Palestinian Territories, Qatar, Syria, or the United Arab Emirates. After selecting HIV-1 segments with the greatest depth of sequences available, alignments corresponding to the gag (HXB2 Gag AA32–156; 111 sequences; nine countries), protease (HXB2 Pro AA1–40; 618 sequences; 13 countries), reverse transcriptase (HXB2 RT AA39–220; 536 sequences; 12 countries), and envelope (HXB2 Env AA285–383; 339 sequences; 13 countries) genes were created based on nonhypermutated and independent sequences. These alignments included 111 (gag) to 618 sequences (protease), representing at least 9 (gag) and up to 13 countries (protease, env). Sequences were primarily from 2004 onward, although some sequences were older (e.g. 1989 for Somalia). Maximum likelihood phylogenetic trees were constructed based on nucleotide sequences, the General Time Reversible nucleotide substitution matrix, invariant sites, and four gamma-distributed rate categories using PhyML [8,9]. Phylogenetic trees were rooted with reference sequences for SIVcpz (Fig. 1; Supplementary Figs 1–3), rooted with SIVcpz and HIV-1 groups M, N, O, or unrooted (data not shown). The results did not change according to use of reference sequences.
Fig. 1.
Maximum likelihood phylogenetic tree based on 536 reverse transcriptase nucleotide sequences sampled between 1989 and 2012 in 12 countries from the Middle East and North Africa (MENA) region.
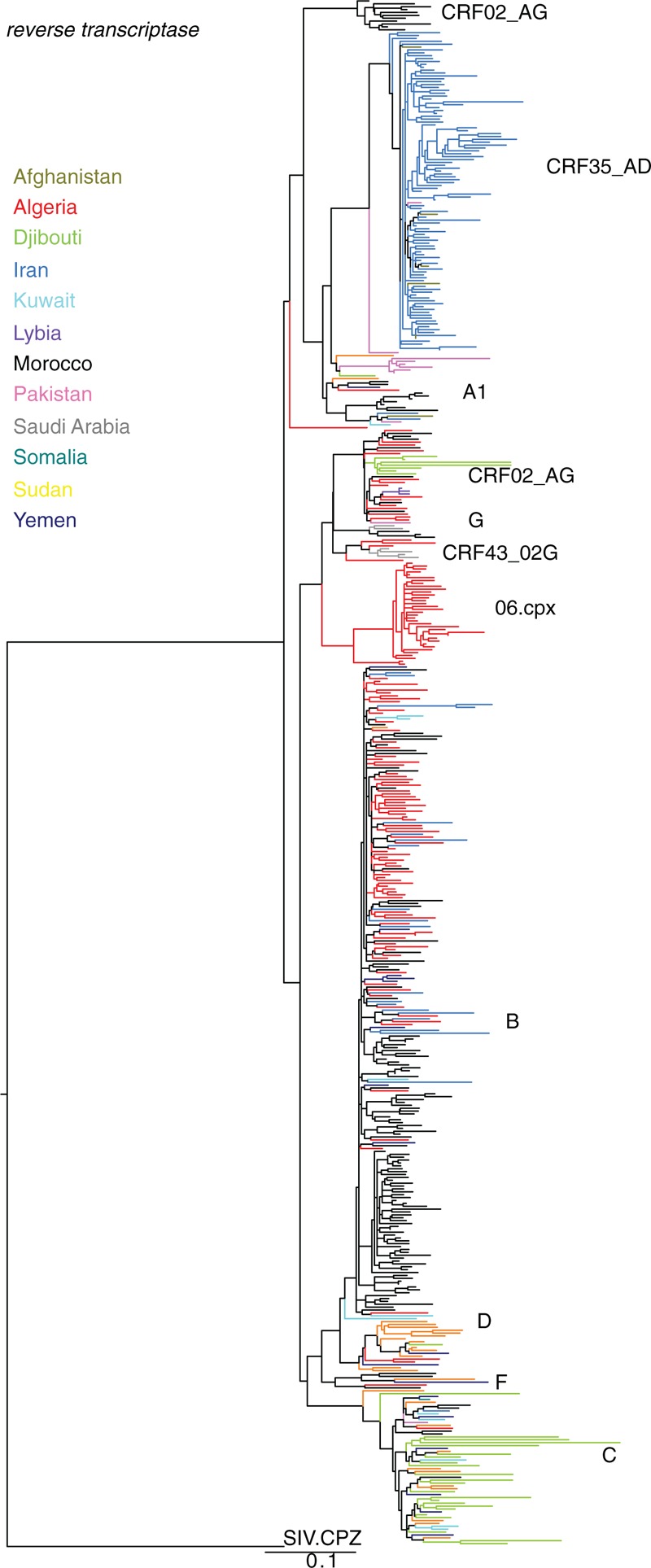
The tree is rooted on an Simian Immunodeficiency Virus (SIV) strain isolated from a chimpanzee (CPZ) in 1985 (GenBank accession number of the SIV.CPZ sequence: AF103818).
A phylogenetic tree based on the reverse transcriptase gene (Fig. 1) was selected over gag, protease, or env as it contained a strong phylogenetic signal, represented a broad geographic distribution, and contained a large number of available sequences (n = 536), including 98 unpublished sequences from Djibouti (n = 1), Iran (n = 75), Kuwait (n = 11), and Pakistan (n = 11) (Supplementary Table 1). The defining feature of this tree is its complexity, as multiple subtypes and circulating recombinant forms (CRFs) are co-circulating across countries in the MENA region. Several subtypes/CRFs have been circulating within each country except in Libya. Reflecting the subtype diversity, the mean pairwise diversity among all reverse transcriptase nucleotide sequences from each country ranged from 1.39% (Libya) to 13.85% (Djibouti), with an average of 8.12% (Supplementary Table 2). In Morocco, at least 17 separate subtypes/CRFs have been published. HIV subtypes were not restricted to specific countries, but could be found in several neighboring countries. For example, subtype B was found in 10 countries (six countries if only considering the reverse transcriptase data). Conversely, a large cluster of 112 CRF35_AD sequences was almost entirely restricted to Iran, with a few sequences (n = 6) in neighboring Afghanistan. When analyzing the data by subtype/CRF for each country, we found that the data were sparse for a number of subtypes/countries, with sometimes just single sequences for certain subtypes/CRFs, reflecting the known limited sampling in the region. Yet, the relatively large diversity estimates for some subtypes/CRFs illustrate that these local epidemics were neither recent nor narrow (Supplementary Tables 3–6). The wide array of HIV-1 subtypes found in each country tended to reflect known exchange routes between countries. For example, subtype B and CRF02_AG – typical in West Europe and West Africa, respectively [10] – were observed in North African countries.
Given the wide distribution of subtypes sampled, the current phylogenetic analysis suggests a diverse regional epidemic with a high potential for generating new HIV-1 recombinant forms. This analysis also suggests that HIV-1 has been introduced multiple times in several countries. The broad array of subtypes/CRFs indicates that the MENA epidemic is more complex than in many other regions of the world, where one subtype typically predominates. In North America and southern Africa, for example, the main circulating subtypes represent more than 95% of all HIV-1 infections in that region [10]. The complexity of the MENA epidemic has implications for prevention strategies, as the development of an effective vaccine would need to be tailored to the multiple circulating subtypes, the distribution of which has still not been mapped out in detail. Although the current study represents the most comprehensive analysis of unique HIV-1 sequences in the MENA region to date, sequences from only 1% of the estimated HIV infections in the region have been published. These findings, therefore, highlight the need for improved reporting of incident HIV infections and more comprehensive sequence sampling to better define the epidemiologic patterns of this expanding regional epidemic.
Acknowledgements
We thank Dr Nelson Michael, Jerome Kim, and Merlin Robb for their comments on the manuscript.
The study was supported by an Interagency Agreement between the National Institute of Allergy and Infectious Diseases and the US Army Medical Research and Material Command (Y1-AI-2642–12) and by a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the US Department of Defense (W81XWH-07-2-0067).
Conflicts of interest
Competing interests: The opinions expressed herein are those of the authors and should not be construed as official or representing the views of the US Department of Defense or the Department of the Army.
Supplementary Material
References
- 1.JUNPoHAU. Global report: UNAIDS report on the global AIDS epidemic. Geneva: UNAIDS; 2013. http://www.unaids.org/en/resources/campaigns/globalreport2013/globalreport [Google Scholar]
- 2.Hamidreza Setayes FR-F, Feki SE, Ashford LS. HIV and AIDS in the Middle East and North Africa. Population Reference Bureau; 2014. http://www.prb.org/Publications/Reports/2014/middle-east-hiv-aids.aspx [Google Scholar]
- 3.UNAIDS. Report on the global AIDS epidemic; 2013. http://www.unaids.org/sites/default/files/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/UNAIDS_Global_Report_2013_en.pdf [Google Scholar]
- 4.Mumtaz GR, Riedner G, Abu-Raddad LJ. The emerging face of the HIV epidemic in the Middle East and North Africa. Curr Opin HIV AIDS 2014; 9:183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islamic Republic of Iran National AIDS Committee Secretariat MoHaME. Islamic Republic of Iran Country Report on Monitoring of the United Nations General Assembly Special Session on HIV and AIDS. 2012. http://www.unaids.org/sites/default/files/en/dataanalysis/knowyourresponse/countryprogressreports/2012countries/IRIran%20AIDS%20Progress%20Report%202012%20English%20final1_1.pdf [Google Scholar]
- 6.Bozicevic I, Riedner G, Haghdoost A. HIV case reporting in the countries of North Africa and the Middle East. J Int AIDS Soc 2014; 17:18962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mediterranean WHOROftE. 2014. http://www.emro.who.int/countries.html [Google Scholar]
- 8.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, et al. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 2010; 48:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696–704. [DOI] [PubMed] [Google Scholar]
- 10.Hemelaar J, Gouws E, Ghys PD, Osmanov S, Isolation W-UNfH. Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011; 25:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


