Abstract
Background: Aberrant expression of miR-378 has been observed in various malignancies including acute myeloid leukemia (AML). However, the mechanism regulating of miR-378 expression remains unknown. This study was aimed to investigate miR-378 methylation and to explore its clinical significance in AML. Methods: Methylation status of miR-378 5’-flanking region was investigated by real-time quantitative methylation-specific PCR (RQ-MSP) and bisulfite-sequencing PCR (BSP). The expression of miR-378 was evaluated by real-time quantitative PCR (RQ-PCR). The correlation between expression of miR-378 and 5’-flanking region methylation was analyzed using 5-aza-2’-deoxycytidine (5-aza-dC) treatment. Results: miR-378 5’-flanking region was significantly hypomethylated in AML patients compared to controls (median 0.109 vs. 0.058) (P=0.048). miR-378 expression was correlated with miR-378 5’-flanking region in leukemic cell line treated with 5-aza-dC, but not in AML patients. The level of miR-378 hypomethylation significantly increased in M2 subtype compared to other subtypes. Moreover, patients with t(8;21) harbored the highest level of miR-378 hypomethylation. However, there was no significant difference in overall survival between patients with high and low miR-378 hypomethylation. The association of miR-378 expression with methylation was not observed in AML patients, but miR-378 expression in THP-1 line was increased while methylation status of miR-378 5-flanking region was decreased after 5-aza-dC treatment. Conclusions: Our findings suggest that miR-378 is reactivated by demethylation after 5-aza-dC treatment. 5’-flanking region of miR-378 is hypomethylated in AML especially in those with t(8;21).
Keywords: miR-378, hypomethylation, acute myeloid leukemia, 5-aza-dC
Introduction
MicroRNAs (miRNA) are a group of small noncoding RNAs that regulate gene expression by inducing degradation or repressing translation of targeted mRNAs [1,2]. It is well-known that miRNAs play pivotal roles in regulating cellular activities, including development, proliferation, differentiation, and apoptosis. Since the first direct link between miRNAs and cancer was made in 2002 when the loss of miR-15 and miR-16 was identified in chronic lymphocytic leukemia with del (13q14) [3], the association between alteration of miRNAs with cancer development and progression has been clear [4,5]. The mechanisms involved in the control of miRNA expression include mutation, deletion, amplification, loss of heterozygosity, and epigenetic mechanisms [6].
Aberrant DNA methylation linked to silencing of individual miRNA genes has been found in acute myeloid leukemia (AML) [6]. Several miRNAs, such as miR-212, miR-3151, miR-181, and miR-29b, have been identified relevant with the outcome of AML [7-10]. However, the prognostic value of more miRNA abnormalities in AML is yet to be determined. Our previous study revealed the overexpression of miR-378 in AML [11]. We identified that the 5’-flanking region of miR-378 gene, located on chromosome 5q32, is embedded in a large CpG island containing 212 CpG binucleotides. It remains unclear whether miR-378 overexpression is regulated by methylation. Moreover, the incidence of aberrant methylation of miR-378 in AML is also unknown. In this study, we analyzed the methylation pattern of miR-378 5’-flanking region in AML patients using a real-time methylation-specific PCR (RQ-MSP) and evaluated the effect of methylation on patient outcome.
Results
CpG islands in the miR-378 5’-flanking region
We focused on DNA methylation in the miR-378 5’-flanking region. Three CpG islands were predicted spanning bp -1775 to -612 (Figure 1). The University of California and Santa Cruz Genome Browser (UCSC Genome Browser) (http://genome.ucsc.edu/) and CpG Island Searcher (http://www.cpgislands. com/) also confirmed the presence of CpG islands (data not shown).
Figure 1.
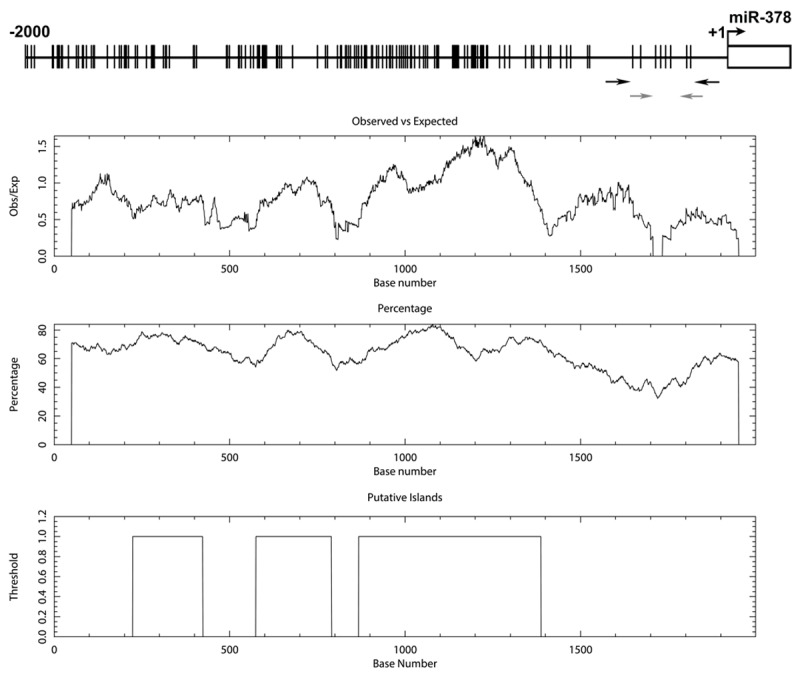
Bioinformatics analysis of the miR-378 5’-flanking region on chromosome 5. The vertical lines on the top horizontal line indicate the cytosine resides of CpGs. Numbers in the top panel represent nucleotide positions from miR-378. The rectangle indicates the position of the sequence encoding mature miR-378. The black arrows indicate the locations of primers used for bisulfite sequencing analysis, and the gray arrows indicate the locations of primers used for RQ-MSP analysis. Second panel represents the distribution of observed/expected ratios of CpG dinucleotides; third panel plots the GC content as a percentage of the total; bottom panel represents the putative 3 CpG islands within the -2.0 kb of analyzed sequence.
Hypomethylation of miR-378 5’-flanking region in AML
The 5’-flanking region of miR-378 was significantly hypomethylated in AML patients (median 0.109, range 0-20.258) compared to controls (median 0.058, range 0-1.000) (P=0.048). The representative electrophoresis results of RQ-MSP products were shown in Figure 2. In order to confirm the results of RQ-MSP, we evaluated the methylation density of 7-CpGs in miR-378 5’-flanking region in two normal controls, three miR-378-hypomethylated and three miR-378-methylated AML samples according to RQ-MSP. BSP results of normal controls presented that miR-378 5’-flanking region were strong methylated in normal BMNCs. The results of RQ-MSP and bisulfite-sequencing were significantly correlated in AML samples (R=-0.829, P=0.042) (Figure 3).
Figure 2.
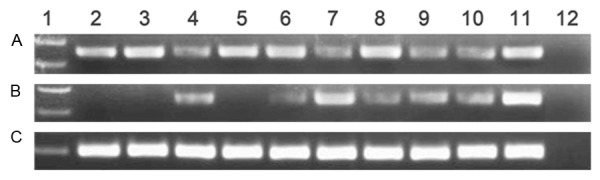
Electrophoresis results of RQ-MSP products in AML patients. A: miR-378 methylation; B: miR-378 unmethylation; C: ALU. 1: Gene RulerTM 100bp DNA ladder; 2-3: normal controls; 4-10: AML samples; 11: cloned plasmid; 12: negative control.
Figure 3.
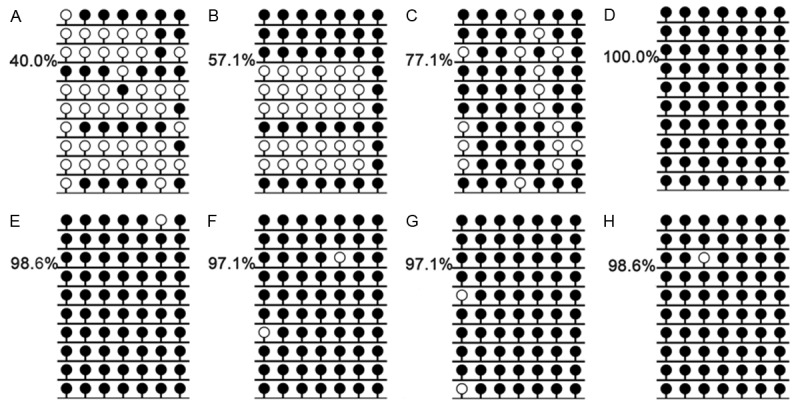
Methylation density of miR-378 5’-flanking region identified in two normal controls, three hypomethylated and three methylated AML patients by bisulfite sequencing. Black lollipop: methylated CpG dinucleotide; Blank lollipop: unmethylated CpG dinucleotide. A-C: three hypomethylated samples, A: M2, B: M4a, C: M5b; D-F: three methylated samples, D: M3, E: M1, F: M5b; G-H: normal controls. The results of RQ-MSP and bisulfite-sequencing were significantly negative correlated (R=-0.829, P=0.042).
There was no correlation between miR-378 hypomethylation with gender, age, and blood parameters. Among FAB subtypes, the level of miR-378 hypomethylation significantly increased in M2 subtype compared to other subtypes (Table 2; Figure 4).
Table 2.
Correlation between miR-378 methylation and patients’ parameters
| Parameter | mir-378 hypomethylation | ||
|---|---|---|---|
|
|
|||
| No. of cases | Median (range) | P-value | |
| Gender | |||
| Male | 80 | 0.148 (0-20.259) | 0.101 |
| Female | 50 | 0.045 (0-4.273) | |
| Age | |||
| ≥60 years | 46 | 0.133 (0-20.259) | 0.806 |
| <60 years | 84 | 0.085 (0-4.273) | |
| WBC | 130 | 17.5 (0.5-528.0) | 0.429 |
| FAB | |||
| M1 | 9 | 0.035 (0-2.719) | 0.126 |
| M2 | 51 | 0.183 (0-4.273) | |
| M3 | 21 | 0.031 (0-1.503) | |
| M4 | 26 | 0.204 (0-20.259) | |
| M5 | 18 | 0.082 (0-2.739) | |
| M6 | 5 | 0.012 (0-0.877) | |
| WHO | |||
| AML with t(8;21) | 12 | 1.611 (0-4.273) | 0.024 |
| APL with t(15;17) | 21 | 0.031 (0-1.503) | |
| AML with 11q23 | 4 | 0.397 (0-2.739) | |
| AML without maturation | 6 | 0.029 (0-0.885) | |
| AML with maturation | 41 | 0.137 (0-1.307) | |
| Acute myelomonocytic leukemia | 26 | 0.204 (0-20.259) | |
| Acute monoblastic and monocytic leukemia | 15 | 0.067 (0-2.635) | |
| Acute erythroid leukemia | 5 | 0.012 (0-0.877) | |
| Karyotype classification | |||
| Favorable | 33 | 0.083 (0-4.273) | 0.117 |
| Intermediate | 75 | 0.125 (20.259) | |
| Poor | 16 | 0.397 (0-2.739) | |
| No data | 6 | 0 (0-1.192) | |
| Karyotype | |||
| normal | 58 | 0.133 (0-20.259) | 0.004 |
| t(8;21) | 12 | 1.611 (0-4.273) | |
| t(15;17) | 21 | 0.031 (0-1.503) | |
| 11q23 | 4 | 0.397 (0-2.739) | |
| complex | 11 | 0.293 (0-1529) | |
| others | 18 | 0.076 (0-1.022) | |
| No data | 6 | 0 (0-0.129) | |
| C/EBPA mutant | |||
| + | 16 | 0.046 (0-2.719) | 0.541 |
| - | 113 | 0.115 (0-20.259) | |
| NPM1 mutant | |||
| + | 16 | 0.034 (0-20.259) | 0.363 |
| - | 113 | 0.115 (0-4.273) | |
| FLT3 ITD mutant | |||
| + | 17 | 0.022 (0-20.259) | 0.075 |
| - | 112 | 0.127 (0-4.273) | |
| C-KIT mutant | |||
| + | 5 | 0.026 (0-0.463) | 0.357 |
| - | 124 | 0.109 (0-20.259) | |
| N/K-RAS mutant | |||
| + | 14 | 0.314 (0-2.635) | 0.182 |
| - | 115 | 0.087 (0-20.259) | |
| DNMT3A mutant | |||
| + | 13 | 0.045 (0-1.216) | 0.376 |
| - | 116 | 0.120 (0-20.259) | |
| IDH1/2 mutant | |||
| + | 8 | 0.214 (0-0.932) | 0.582 |
| - | 121 | 0.095 (0-20.259) | |
| CR | |||
| + | 41 | 0.148 (0-20.259) | 0.301 |
| - | 53 | 0.136 (0-3.593) | |
WBC, white blood cells; FAB, French-American-British classification; AML, acute myeloid leukemia;CR, complete remission; *, percentage was equal to the number of mutated patients divided by total cases in each group.
Figure 4.
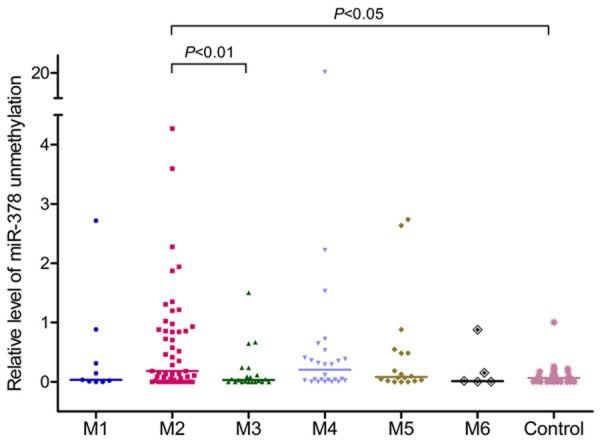
Relative levels of miR-378 5’-flanking hypomethylation in AML with different FAB subtypes.
Association of miR-378 methylation with karyotypes
Patients with t(8; 21) harbored the highest level of miR-378 hypomethylation compared to other karyotypes (Table 2; Figure 5). Due to miR-378 is located on chromosome 5q32, the level miR-378 hypomethylation was analyzed in patients with isolated -5/5q- or with concomitant other aberrations (n=5, median 0.708, range 0.144-0.877), which was significant higher than that in controls (P=0.0045, Figure 6). However, There was no difference in miR-378 hypomethylation between patients with and without -5/5q- (P=-0.124).
Figure 5.
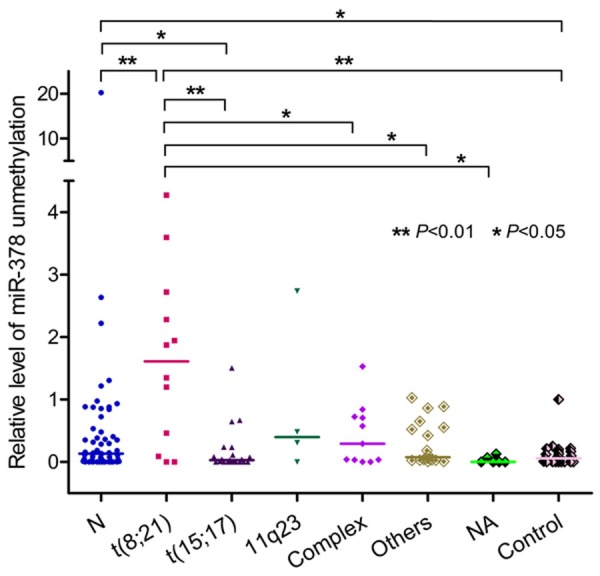
Relative levels of miR-378 5’-flanking hypomethylation in AML with different karyotypes.
Figure 6.

Relative levels of miR-378 5’-flanking hypomethylation in AML with and without -5/5q-.
Association between miR-378 hypomethylation and prognosis
There were 93 patients with available survival data. This cohort of AML patients was divided into two groups according to the median level of miR-378 hypomethylation. The patients with high level of miR-378 hypomethylation had similar overall survival (OS) as those with low level of miR-378 hypomethylation (median 7 months vs. 8 months, P=0.558, Figure 7). Multivariate analysis identified age and karyotypic risk but not miR-378 hypomethylation as independent prognostic factors (data not shown).
Figure 7.
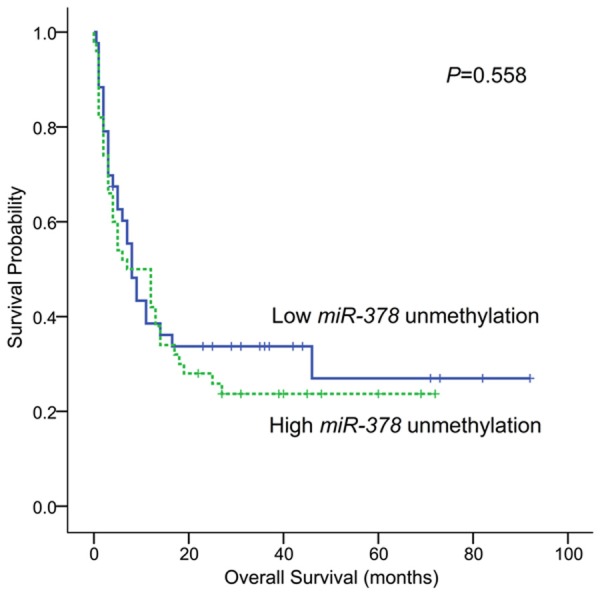
Overall survivals of AML patients.
Association of miR-378 methylation with miR-378 expression
The level of miR-378 expression was examined in 56 AML patients with available mRNA. No correlation was observed between the level of miR-378 expression and miR-378 unmethylation (R=0.018, P=0.898).
To determine whether miR-378 expression could be regulated by methylation, THP-1 cells were treated by 5-aza-dC. Obviously, miR-378 expression was significantly up-regulated after 5-aza-dC treatment (Figure 8A). The level of unmethylated miR-378 5’-flanking region was increased in a dose-dependent manner in THP-1 cells treated with 5-aza-dC (Figure 8B and 8C).
Figure 8.
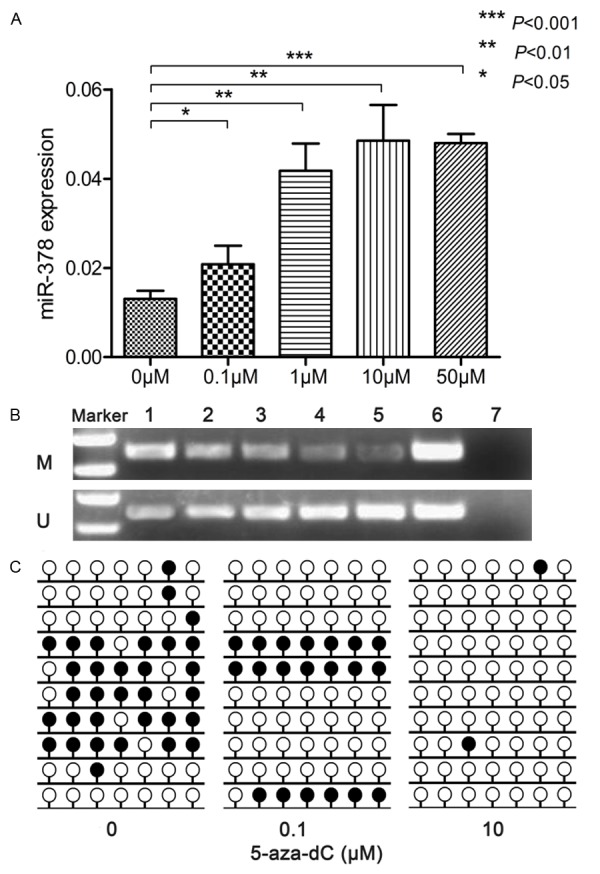
Methylation density of miR-378 5’-flanking region in THP-1 cells treated with 5-aza-dC. A: miR-378 expression; B: RQ-MSP results, 1-5: THP-1 treated with 5-aza-dC (0 μM, 0.1 μM, 1 μM, 10 μM, and 50 μM, respectively), 6: recombinant methylated or unmethylated miR-378 5’-flanking region plasmids, 7: ddH2O; C: BSP results. black lollipop: methylated CpG dinucleotide; blank lollipop: unmethylated CpG dinucleotide; methylation percentages at 7 CpG sites of miR-378 5’-flanking region in THP-1 treated with 0 μM, 0.1 μM and 10 μM of 5-aza-dC were 45.7%, 28.6% and 2.9%, respectively.
Discussion
Our previous study observed the overexpression of miR-378 in AML [11]. However, the mechanism underlying miR-378 upregulation remains unclear. In this study, we identified the large islands in the miR-378 5’-flanking region. Further study on primary samples confirmed the aberrant hypomethylation of miR-378 5’-flanking region in AML. However, miR-378 expression was not associated with the pattern of miR-378 methylation in AML patients. The treatment of THP-1 cell line with demethylating reagent 5-aza-dC indicated that miR-378 could be reactivated by demethylation. These results suggest that miR-378 expression may be regulated by other molecular mechanisms besides methylation in leukemia.
Interestingly, although the association of miR-378 expression with methylation was not observed in our cohort of AML patients, this study identified the prevalence of miR-378 hypomethylation in the FAB-M2 subtype, in accordance with our previous findings that the incidence of miR-378 overexpression was highest in the M2 subtype [11]. Further analysis disclosed the high level of miR-378 hypomethylation was associated with the t(8;21) chromosomal aberration, which also confirmed our previous observations [11]. Although miR-378 gene is located on chromosome 5q32, a commonly deleted region in myeloid malignancies, the incidence of miR-378 hypomethylation in this study or miR-378 overexpression reported previously was independent on -5/5q-. Further study is needed to determine the mechanism underlying miR-378 aberrations in the leukemogenesis involved in t(8;21) translocation.
The role of miR-378 in tumorigenesis remains controversial. Several studies indicated the oncogenic function of miR-378 due to enhancing cell survival, reducing apoptosis, accelerating tumor growth, angiogenesis and promoting cell migration and invasion in solid cancers including glioblastoma, non-small cell lung cancer (NSCLC), breast cancer, and nasopharyngeal carcinoma [20-25], whereas other studies suggest that miR-378 functions as a tumor suppressor in gastric and colorectal cancer [26-29]. The occurrence of miR-378 upregulation and miR-378 hypomethylation suggests the potential oncogenic role of miR-378 in AML.
The impact of miR-378 aberration on prognosis has been increasingly explored in cancers. In ovarian cancer, high miR-378 expression was associated with shorter survival after bevacizumab and chemotherapy [30]. Moreover, high miR-378 expression was associated with brain metastasis in NSCLC [21]. However, low miR-378 expression had significantly poorer overall survival in colorectal cancer [28]. The prognostic value of miR-378 expression was not observed in nasopharyngeal carcinoma and renal cell carcinoma [31,32]. In addition, a genetic variant in pri-miR-378 was associated with increased miR-378 expression and with a better survival in hepatocellular carcinoma [33]. Inconsistent with our previous report in which miR-378 overexpression conferred an unfavorable effect to outcome in AML [11], the present study did not observe the association of survival with miR-378 hypomethylation. Combined with the functional studies in cancers, these results indicate that miR-378 expression is regulated by various mechanisms that play different roles in tumorigenesis and clinical outcome, depending on the tissue- or time- context.
In conclusion, this is the first study on miR-378 methylation which discloses that miR-378 is reactivated by demethylation after 5-aza-dC treatment. 5’-flanking region of miR-378 is hypomethylated in AML especially in those with t(8;21), but is unlikely to provide helpful prognostic information in AML patients.
Materials and methods
Patients and samples
130 patients with primary AML presented at the Affiliated People’s Hospital of Jiangsu University were selected for this investigation based on the availability of stored leukemic cells. The diagnosis and classification of AML patients were made according to the revised French-American-British (FAB) classification and the 2008 World Health Organization criteria [12,13]. Karyotypes were analyzed by conventional R-banding method. Karyotype risk was classified according to reported previously [14]. Treatment protocol was described previously [15]. 25 healthy donors were collected as controls. Written informed consent was obtained from all patients and normal controls. The study was approved by the Ethics Committee of the Affiliated People’ Hospital of Jiangsu University. Bone marrow mononuclear cells (BMNCs) were separated by density gradient centrifugation using Ficoll solution and washed twice with PBS.
5-aza-2’-deoxycytidine treatment
The leukemic cell line THP-1 was plated at a density of 1×106/ml and was cultured in 5 ml RPMI 1640 medium at 37°C in a humidified atmosphere containing 5% CO2. 5-aza-2’-deoxycytidine (5-aza-dC) (Sigma-Aldrich, Steinheim, USA) diluted in dimethyl sulfoxide (DMSO) was added in four flasks of THP-1 cells once a day at the same time at different final concentrations of 0.1 μM, 1 μM, 10 μM and 50 μM as experimental group. THP-1 cells without treatment were used as the control. All cells were cultured until harvested for extraction of RNA and DNA.
Real-time quantitative PCR
Real time quantitative PCR (RQ-PCR) was performed to detect miR-378 expression as reported previously [11].
Survey of CpG island in mir-378 5’-flanking region
The 5’ sequences flanking the region of genomic DNA (gDNA) encoding miR-378 on chromosome 5 [strand (+), nucleotides (nt) 149110392-149112413] was surveyed for the presence of CpG islands using cpgplot software (http://emboss.bioinformatics.nl/cgi-bin/emboss/cpgplot). The criteria used were as follows: island size >100 bp, GC percent >50.0%, and ratio of observed (Obs) CpG sites to expected (Exp) CpG sites >0.6).
DNA isolation and bisulfite modification
Genomic DNA was obtained from BMNCs using DNA Purification Kit (Gentra, Minneapolis, MN, USA). 1 μg of genomic DNA was sodium bisulphite-modified as described in manufacturer instruction using the CpGenomeTM DNA Modification kit (Chemicon, Ternecula, CA, USA). Modified DNA was used immediately or stored at -80°C at once until analyzed.
Methylation-specific PCR (MSP) and real-time quantitative MSP (RQ-MSP)
RQ-MSP was performed using unmethylation-specific primers (Table 1) and ALU repetitive sequence on Step One Plus (Applied Biosystems, CA, USA). ALU repetitive sequence was used as reference sequence. A final volume of 20 µL of mixture containing 0.4 μM of primers, 10 μM SYBR Premix Ex Taq II, 0.4 μL 50×ROX (Takara, Japan) and 20 ng of modified DNA composed the reaction system. Amplification was carried out at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 62°C for 30 s and 72°C for 30 s, and a fluorescence collection step at 75°C for 30 s, followed by a melting program at 95°C for 15 s, 60°C for 60 s, 95°C for15 s, and 60°C for 15 s. Distilled water without DNA was used as negative control as well as recombined methylated and unmethylated miR-378 plasmids were positive controls for each set of PCR. The normalized ratio (Nunmethylation-miR-378) was calculated in relation to the reference ALU sequence [16] and was used to assess the degree of methylation of miR-378 5’-flanking region in samples. The standard curves were established using recombined methylated and unmethylated miR-378 plasmids as well as ALU plasmids from 1×108 copies/μl to 10 copies/μl before determining the cutoff level of specific fluorescence for the three corresponding sequences in samples. Nunmethylation-miR-378 was calculated according to the following formula. Nunmethylation-miR-378 = (Eunmethylation-miR-378)ΔCTunmethylation-miR-378 (control-sample)÷(EALU)ΔCTALU (control-sample). CT was the cycle number when the fluorescent signal intensity of detected gene in each reaction reaches to the setting threshold. E was the amplification efficiency of each PCR reaction and was computed as E=10(-1/slope). DNA methylation status of the miR-378 5’-flanking region was also determined by MSP using the primers listed in Table 1. MSP products were analyzed on 2% agarose gels and visualized under UV illumination after staining with ethidium bromide.
Table 1.
The sequences of primers used in RQ-MSP and BSP
| Forward (5’→3’) | Reverse (5’→3’) | Product (bp) | |
|---|---|---|---|
| RQ-MSP | |||
| M | GGATGAGTTTTGAGTCGTTT | CCATACAAACCGCTCACTCC | 129 |
| U | TAGGATGAGTTTTGAGTTGT | ATAATCCCATACAAACCACT | 137 |
| BSP | AGGATTTTTTGGTGATTTTTG | TCACCCTCTAACTACATAATCCC | 200 |
Bisulfite-sequencing PCR (BSP)
To further determine the comprehensive methylation status of the miR-378 5’-flanking region in AML patients, bisulfite-modified DNA sequencing PCR was performed in two normal controls, three miR-378-hypomethylated and three miR-378-methylated AML samples in accordance with the result of RQ-MSP. Each BSP reaction included 25 μL of mixture containing 10×PCR buffer (0.25 mM KCl), 6.25 μM of dNTP Mixture, 0.5 μM of primers, 0.75 U of Hot start DNA polymerase (Takara, Tokyo, Japan), and 20 ng of modified DNA. PCR conditions were 98°C for 10 s, 40 cycles for 10 s at 98°C, 30 s at 56°C, and 30 s at 72°C followed by a final 7 min extension step at 72°C. The reaction was performed on iCycler Thermal Cycler (Eppendorf, Hamburg, Germany). PCR products were cloned into pMD®. 19-T Vector (Takara, Dalian, China). 10 independent colonies of each sample were sequenced.
Mutations analysis
NPM1, C-KIT, IDH1, IDH2, N/K-RAS and DNMT3A mutations were detected by high-resolution melting analysis (HRMA) as reported previously [11,18,19]. Briefly, genomic DNA samples were amplified using gene-specific primers. Mutation scanning was performed for PCR products using HRMA with the LightScannerTM platform (Idaho, Salt Lake City, Utah). All positive samples were verified by direct DNA sequencing to confirm the results of HRMA. FLT3 internal tandem duplication (ITD) and C/EBPA mutations were detected using direct DNA sequencing [11].
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software package (SPSS, Chicago, IL, USA). Pearson chi-square analysis and Fisher exact test were carried out to compare the difference of categorical variables between patients group. Mann-Whitney’s U-test was carried out to compare the difference of continuous variables between two patient groups. The correlation between the frequency of miR-378 5’-flanking methylation and the clinical and hematologic parameters was analyzed with Spearman’s rank correlation. Survival was analyzed according to the Kaplan-Meier method and differences in the distribution were evaluated by means of the log-rank test. A P-value of less than 0.05 was considered statistically significant.
Acknowledgements
This study was supported by the National Natural Science foundation of China (81270630, 81172592), 333 Project of Jiangsu Province (BRA2013136), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), Science and Technology Infrastructure Program of Zhenjiang (SS2012003), Social Development Foundation of Zhenjiang (SH2013042, SH2013082, SH2014044), Key Medical Talent Program of Zhenjiang City, and Jiangsu Government Scholarship for Overseas Studies.
Disclosure of conflict of interest
None.
References
- 1.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–5. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 6.Agirre X, Martínez-Climent JÁ, Odero MD, Prósper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. 2012;26:395–403. doi: 10.1038/leu.2011.344. [DOI] [PubMed] [Google Scholar]
- 7.Sun SM, Rockova V, Bullinger L, Dijkstra MK, Döhner H, Löwenberg B, Jongen-Lavrencic M. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. 2013;27:100–6. doi: 10.1038/leu.2012.158. [DOI] [PubMed] [Google Scholar]
- 8.Eisfeld AK, Marcucci G, Maharry K, Schwind S, Radmacher MD, Nicolet D, Becker H, Mrózek K, Whitman SP, Metzeler KH, Mendler JH, Wu YZ, Liyanarachchi S, Patel R, Baer MR, Powell BL, Carter TH, Moore JO, Kolitz JE, Wetzler M, Caligiuri MA, Larson RA, Tanner SM, de la Chapelle A, Bloomfield CD. miR-3151 interplays with its host gene BAALC and independently affects outcome of patients with cytogenetically normal acute myeloid leukemia. Blood. 2012;120:249–58. doi: 10.1182/blood-2012-02-408492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, Dohner K, Neilly MB, Price C, Lussier YA, Zhang Y, Larson RA, Le Beau MM, Caligiuri MA, Bullinger L, Valk PJ, Delwel R, Lowenberg B, Liu PP, Marcucci G, Bloomfield CD, Rowley JD, Chen J. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–24. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, Liu S, Havelange V, Becker H, Schaaf L, Mickle J, Devine H, Kefauver C, Devine SM, Chan KK, Heerema NA, Bloomfield CD, Grever MR, Byrd JC, Villalona-Calero M, Croce CM, Marcucci G. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107:7473–8. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian J, Lin J, Qian W, Ma JC, Qian SX, Li Y, Yang J, Li JY, Wang CZ, Chai HY, Chen XX, Deng ZQ. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leuk Res. 2013;37:765–8. doi: 10.1016/j.leukres.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukaemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–5. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 13.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon France: IARC Press; 2008. [Google Scholar]
- 14.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Lin J, Yang J, Qian J, Qian W, Yao DM, Deng ZQ, Liu Q, Chen XX, Xie D, An C, Tang CY. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37:1642–7. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Qian J, Zhu ZH, Lin J, Ming Yao DM, Li Y, Yang J, Wang CZ. Hypomethylation of PRAME promoter is associated with poor prognosis in myelodysplastic syndrome. Br J Haematol. 2011;154:153–5. doi: 10.1111/j.1365-2141.2011.08585.x. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91:519–25. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Qian J, Sun A, Lin J, Xiao G, Yin J, Chen S, Wu D. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46:579–83. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–5. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, Wang SF. MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol. 2012;29:1673–80. doi: 10.1007/s12032-011-0083-x. [DOI] [PubMed] [Google Scholar]
- 22.Skrzypek K, Tertil M, Golda S, Ciesla M, Weglarczyk K, Collet G, Guichard A, Kozakowska M, Boczkowski J, Was H, Gil T, Kuzdzal J, Muchova L, Vitek L, Loboda A, Jozkowicz A, Kieda C, Dulak J. Interplay between heme oxygenase-1 and miR-378 affects non-small cell lung carcinoma growth, vascularization, and metastasis. Antioxid Redox Signal. 2013;19:644–60. doi: 10.1089/ars.2013.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Z, Du WW, Fang L, Shan SW, Qian J, Lin J, Qian W, Ma J, Rutnam ZJ, Yang BB. The intermediate filament vimentin mediates microRNA miR-378 function in cellular self-renewal by regulating the expression of the Sox2 transcription factor. J Biol Chem. 2013;288:319–31. doi: 10.1074/jbc.M112.418830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, Giguère V. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang L, Li G, Chen HH, Li XP. MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int J Oncol. 2014;44:1215–22. doi: 10.3892/ijo.2014.2283. [DOI] [PubMed] [Google Scholar]
- 26.Fei B, Wu H. MiR-378 inhibits progression of human gastric cancer MGC-803 cells by targeting MAPK1 in vitro. Oncol Res. 2012;20:557–64. doi: 10.3727/096504013X13775486749254. [DOI] [PubMed] [Google Scholar]
- 27.Deng H, Guo Y, Song H, Xiao B, Sun W, Liu Z, Yu X, Xia T, Cui L, Guo J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene. 2013;518:351–9. doi: 10.1016/j.gene.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 28.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Dai S, Zhen T, Shi H, Zhang F, Yang Y, Kang L, Liang Y, Han A. Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer. 2014;50:1207–21. doi: 10.1016/j.ejca.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Chan JK, Kiet TK, Blansit K, Ramasubbaiah R, Hilton JF, Kapp DS, Matei D. MiR-378 as a biomarker for response to anti-angiogenic treatment in ovarian cancer. Gynecol Oncol. 2014;133:568–74. doi: 10.1016/j.ygyno.2014.03.564. [DOI] [PubMed] [Google Scholar]
- 31.Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW, Ahn WS. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Devel Ther. 2014;8:293–314. doi: 10.2147/DDDT.S51969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Luo HN, Tian WD, Lu J, Li G, Wang L, Zhang B, Liang BJ, Peng XH, Lin SX, Peng Y, Li XP. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol Ther. 2013;14:1133–42. doi: 10.4161/cbt.26170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An J, Liu J, Liu L, Liu Y, Pan Y, Huang M, Qi F, Wen J, Xie K, Ma H, Shen H, Hu Z. A Genetic Variant in Primary miR-378 Is Associated with Risk and Prognosis of Hepatocellular Carcinoma in a Chinese Population. PLoS One. 2014;9:e93707. doi: 10.1371/journal.pone.0093707. [DOI] [PMC free article] [PubMed] [Google Scholar]


