Abstract
Triple-negative breast carcinoma (TN) is a heterogeneous cancer type expressing EGFR in 75% of cases. MUC1 is a large type I sialylated glycoprotein comprising two subunits (α and β chains, also called respectively MUC1-VNTR and MUC1-CT), which was found to regulate EGFR activity through endocytic internalisation. Endocytosis and autophagy use the lysosome pathway involving NEU1. Recently, a molecular EGFR-MUC1-NEU1 complex was suggested to play a role in EGFR pathway. In the aim to understand the relationship between EGFR-MUC1-NEU1 complex and autophagy in breast carcinoma, we compared triple negative (TN) showing a high-EGFR expression with luminal (LUM) presenting low-EGFR level. We studied the expression of MUC1-VNTR, MUC1-CT and NEU1 in comparison with those of two molecular actors of autophagy, PI3K (p110β) and Beclin1. A total of 87 breast cancers were split in two groups following the immunohistochemical classification of breast carcinoma: 48 TN and 39 LUM. Our results showed that TN presented a high expression of EGFR and a low expression of MUC1-VNTR, MUC1-CT, NEU1, Beclin-1 and PI3Kp110β. Moreover, in TN, a positive statistical correlation was observed between Beclin-1 or PI3Kp110β and MUC1-VNTR or NEU1, but not with EGFR. In conclusion, our data suggest that autophagy is reduced in TN leading likely to the deregulation of EGFR-MUC1-NEU1 complex and its associated cellular pathways.
Keywords: Breast, carcinoma, EGFR, MUC1, NEU1, PI3K, beclin-1, autophagy
Introduction
Breast cancers are the most common cause of cancer mortality in women. Most of them are routinely treated following their estrogen receptors (ER), progesterone receptors (PR) and human epidermal growth factor receptor type 2 (HER2) expressions. According to this clinical approach, a biological classification has been recently proposed by Perou et al and adopted by the St Galen International Expert Consensus. Briefly, this classification proposes three main molecular subtypes: luminal (ER+PR+HER2-), overexpressed HER2 (ER-PR-HER2+) and triple negative (ER-PR-HER2-) carcinomas [1-3]. However, triple negative breast carcinoma (TN) corresponds to a heterogeneous cancer subtype leading to difficulties to assign an appropriate treatment [4]. Interestingly, about 75% of TN expressed high amount of type 1 epidermal growth factor receptor (EGFR). Unfortunately, the treatments by a monoclonal anti-EGFR alone (Cetuximab) or in combination with carboplatin, were associated with a low rate of clinical response suggesting a complex signalling pathway [5-7].
MUC1, or CD227, is a large trans-membrane O-glycosylated protein affiliated to the insoluble mucin family. Structurally, MUC1 is a heterodimer consisting of a large extracellular α-subunit containing 20 to 125 tandem repeats of 20 amino acids broadly glycosylated (MUC1-VNTR), and a β-subunit containing the transmembrane domain and a cytoplasmic tail (MUC1-CT) [8-10]. Many breast cancers and other epithelial cancers over-express MUC1 presenting severe alterations of their glycosylation pattern leading to the exposure of repetitive peptide core epitopes that may represent potential targets for immunotherapy [11-14]. Kawaguchi et al demonstrated that MUC1 glycosylation changes are correlated to the tumoral capacity to develop metastasis [15]. Among the glycosylation processes, sialylation is crucial for a variety of cellular functions such as cell adhesion signal recognition, and biological stability of glycoproteins. Sialylation of glycoproteins is regulated by two opposing enzymatic activities: sialyltransferases and sialidases [16,17]. It is interesting to mention that NEU1, a well-known lysosome sialidase, has been proposed to regulate EGFR and MUC1 signalling (ref Lillehoj et al). Moreover, NEU1 forms a complex with both EGFR and MUC1 [18]. The β-subunit part of MUC1, MUC-CT, is involved in several cellular signalling pathways that could potentially induce cancerous transformation by either growth/survival pathways induction or apoptosis inhibition [19,20]. Some authors demonstrated a colocalisation between MUC1-CT and EGFR both at the cell membrane and in the nucleus, involving internalisation of EGFR and activation of the EGFR-PI3K-AKT pathway [20-22].
The phosphoinositide 3-kinases (PI3K) constitute a family of lipid kinases that can be activated by extracellular stimuli. PI3K are involved in tumour cell survival, proliferation and differentiation. They are grouped into three classes of isoforms mainly based on their substrate specificity. The two ubiquitously expressed PI3K isoforms p110α and p110β play different roles in cellular signalling. The p110α isoform promotes the main response of EGFR stimulation, whereas p110-β seems to finely tune this response [23,24]. Importantly, p110β is also involved in the endocytosis of EGFR and/or to promote autophagy by activation of the Rab5-Vps34-Vps15-Beclin-1 complex [25,26]. Interestingly, MUC1 expression is associated with increased lysosomal turnover of the autophagic maker LC3-II by stimulation of the AMP-activated protein kinase (AMPK), therefore highlighting the involvement of MUC1 in the regulation of autophagy [21,27]. Autophagy is a cellular degradation pathway involving double-membrane vesicles and the lysosome machinery, including catabolic enzymes such as NEU1. Autophagy is activated upon cellular stress in order to maintain cell homeostasis. Autophagy plays a role in differentiation, aging, immunity and tumour suppression [28]. Intriguingly, autophagy is also associated with resistance to chemotherapy [29,30].
To understand the relationship between EGFR-MUC1-NEU1 complex and autophagy in breast carcinoma, we compared TN showing a high-EGFR expression, with LUM presenting low-EGFR level. We studied the expression of MUC1-VNTR, MUC1-CT and NEU1 in comparison with those of PI3K (p110β) and Beclin1.
Materials and methodology
Patient population
Between 2010 to 2013, archival paraffin embedded surgical material and clinical data of 48 triple negative breast carcinomas (TN, age = 61.1 ± 14.9 years) and 39 luminal carcinoma (LUM, age = 60.4 ± 12.4 years, P = ns) using as control group, were available for this study. All cases were classified following the immunohistochemical classification in mean of a preliminary immunohistochemical study confirmed by the tissue microarray (TMA) [1-3]. Among those, 19 patients presented lymph node metastasis (LUM = 15/39 (38.4%) vs. TN = 4/48 (10.4%), P = 0.0008) and 15 had haematogenous metastasis (mainly lung, liver and brain; LUM = 1/39 (2.5%) vs. TN = 14/48 (29.1%), P = 0.0007). Tumour recurrence was described in 13 patients (LUM = 2/39 (5.1%) vs. TN = 11/48 (22.9%), P = 0.02). No neo-adjuvant chemotherapy was performed. The mean of follow-up was 101.6 ± 60.4 weeks.
This study was made according to the approval of the local ethic committee, and all patients were informed and agreed to contribute to this study.
Histological procedures and Tissue Micro Array (TMA) construction
All surgical specimens were initially fixed in 4% buffered formaldehyde solution for 8 to 48 hours, then embedded in paraffin and cut into 4 µm thick slides. The slides were stained with a classical haematoxylin-eosin stain to perform the initial diagnosis. From these archival formol/paraffin blocs, we built a TMA receive paraffin block that could be used for all immunohistochemical slides. We used an automated TMA device (Minicore2, Mitogen UK) associated with a needle core of 0.6 mm diameter. We chose 3 distant core needle samples of each donor tumour paraffin block. The final TMA receive paraffin block was cut in serial slides. These slides were consecutively used for immunohistochemistry.
Immunohistochemical methods
Immunohistological staining was performed with a Dako Autostainer Link 48® immunostaing system (Dako Glostrub, Denmark). After dewaxing, antigenic retrieval were performed using citrate buffered (pH 6) or EDTA buffered (pH 9) antigenic retrieval solution at 99°C in a warm bath (EnVision Flex Target Retrieval solutions high and low pH, Dako). Endogen peroxydase were inhibiting with a hydrogen peroxide phosphate buffered solution (EnVision Flex Peroxydase Blocking Reagent, Dako). After the incubation of the primary antibodies, the immunological reaction was revealed by a polymer dextran coupled with secondary antibody and peroxydase for 15 min (EnVision Flex HRP, Dako) and diaminobenzidine for 10 minutes (EnVision DAB + chromogen, Dako). Counterstain was made with haematoxylin for 10 min (EnVision Flex haematoxylin, Dako). Negative controls were obtained using mouse IgG1 (Negative Control Mouse, Dako) diluted at 1:100, in place of primary antibodies. Primary antibodies, dilution and antigenic retrieval are described in Table 1.
Table 1.
Primary antibodies, dilution, antigenic retrieval, incubation times and abbreviations used in this study
| Antibodies | Clone | Abbreviation | Manufacture | Dilution | Retrievial | Incubation (minutes) |
|---|---|---|---|---|---|---|
| Beclin-1 | H-300 | Beclin-1 | Santa Cruz | 1:50 | Citrate, pH 6 | 60 |
| EGFR wild-type | DAK-H1-WT | EGFR | Dako | 1:200 | EDTA, pH 9 | 30 |
| α-Estrogen Receptor | SP1 | ER | Dako | RTU | EDTA, pH 9 | 20 |
| HER2 | c-erB-2 | HER2 | Dako | 1:800 | Citrate, pH 6 | 30 |
| MUC1 core glycoprotein | Ma552 | MUC1-VNTR | Novocastra | 1:50 | EDTA, pH 9 | 10 |
| MUC1-ter C | ARP41446 | MUC1-CT | Aviva System Biology | 1:400 | EDTA, pH 9 | 60 |
| Neuraminidase1 | NEU1 | ARP44186_T100 | Aviva System Biology | 1:1000 | EDTA, pH 9 | 60 |
| Pi3K p110 β | N/A | PI3K | Spring | 1:100 | Citrate, pH 6 | 30 |
| Progesterone Receptor | PgR636 | PR | Dako | RTU | EDTA, pH 9 | 20 |
Classification of breast cancers by immunohistochimestry
HER2 immunostaining were considered positive as described in the Guideline of College of American Pathologists and controlled by a FISH technique for all cases (HercepTest® Dako) [31]. ER and PR were subsequently scored using a score consisting to sum the intensity and proportion of the nuclear immunostaining. A result superior to 2 was considered as positive [32]. According the St Galen guideline [3] and the results of these immunostainings, all cases were classified following the immunohistochemical classification [1,2].
Immunostaining quantification
Staining results were evaluated by CG and CM, based on the intensity and percentage of staining tumour cells, with agreement reached. The parametric results were edited as a score by a multiplication of intensity (0 = none, 1 = weak, 2 = intermediated, 3 = strong) and the percentage of tumour cells (0 = none, 1 = 1%, 2 = between 1% to 10%, 3 = between 10% to 33%, 4 = between 33% to 66% and 5 = between 66 to 100%) [modified from 32].
Statistics
T-test and Spearman’s test were performed. A p value < 0.05 was considered significant. The WinSTAT® version 2012 (Fitch Software, Bad Krozinger, Germany) and Excel 2013 (Microsoft Corp., Redmond, Washington U.S.A.) programs were used for statistical analysis. The results were expressed in means and standard error.
Results
Characteristics of patients with TN and LUM breast carcinomas
In this retrospective study, we analysed data from 87 patients prognosticated either with triple negative breast carcinomas (TN, n = 48, age = 61.1 ± 14.9 years) or with luminal breast carcinoma (LUM, n = 39, age = 60.4 ± 12.4 years, P = ns) [1-3]. Among the whole population of breasts cancer patients, those diagnosed with lymph node metastasis was significantly higher in the LUM group compared to the TN group (15/39 (38.4%) vs. TN = 4/48 (10.4%), respectively, P = 0.0008). Conversely, the number of patients with haematogenous metastasis (mainly lung, liver and brain) was lower within the LUM group than in the TN group (LUM = 1/39 (2.5%) vs. TN = 14/48 (29.1%), P = 0.0007). Tumour recurrence was described in 13 patients (LUM = 2/39 (5.1%) vs. TN = 11/48 (22.9%), P = 0.02). No neo-adjuvant chemotherapy was performed. The mean of follow-up was 101.6 ± 60.4 weeks.
Morphological differences between TN and LUM
First, we studied the morphological cell distribution of EGFR, MUC1-VNTR, MUC1-CT and of NEU1 (Figure 1), PI3Kp110β and Beclin-1 (Figure 2) in the 48 TN in comparison with the 39 LUM. To that aim, we compared the immunohistological score obtained for each antibody in the TN and LUM groups (Table 2). Interestingly, all immunostainings presented a significant statistical difference between LUM and TN, suggesting an important biological difference between these 2 groups of breast tumours. Globally, TN showed a lower expression of MUC1-VNTR (P = 0.002), MUC1-CT (P < 0.0001), NEU1 (P = 0.03), PI3Kp110β (P < 0.0001) and Beclin-1 (P < 0.0001) as compared to LUM. A higher expression of EGFR (P < 0.0001) was observed in TN. Although TN breast cancers are well-known to highly express EGFR, in this study 14 TN were EGFR-negative and 2 LUM were EGFR-positive. However, no change within our data was observed if these cases were discarded EGFR is expressed both in the cytoplasm and the cell membrane.
Figure 1.
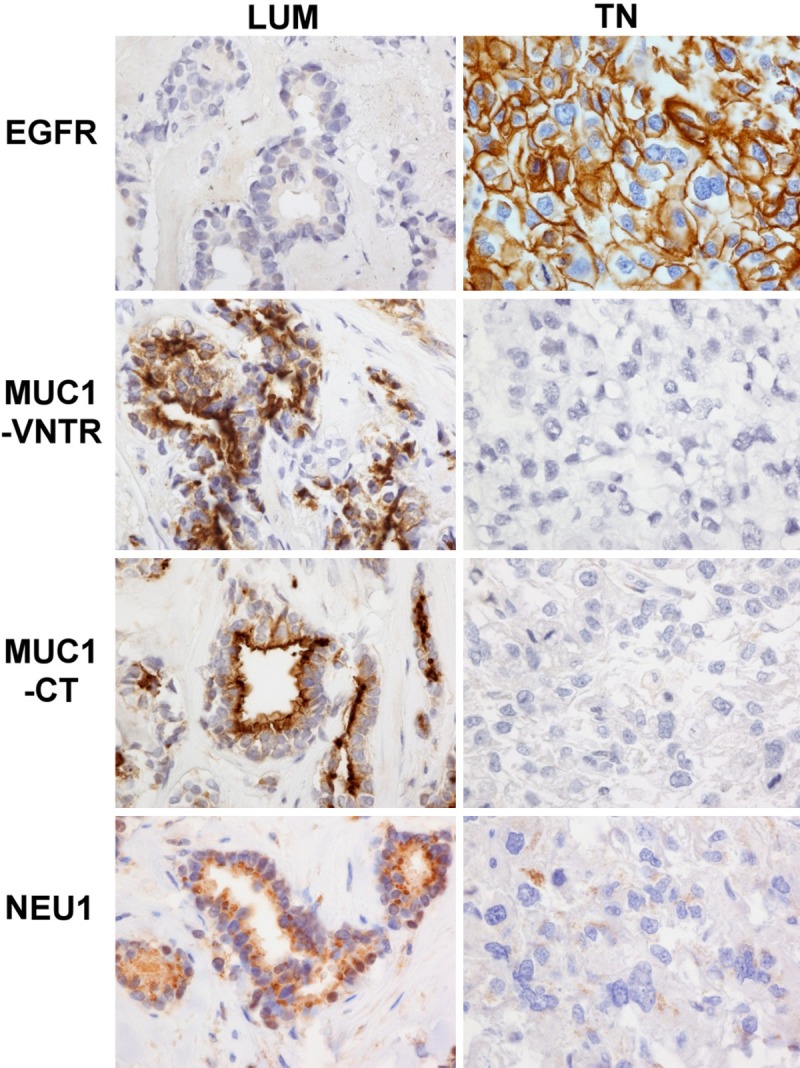
EGFR, MUC1-VNTR, MUC1-CT and NEU1 difference and morphological distribution between luminal (LUM) and triple-negative (TN) breast carcinoma. EGFR is negative in LUM and positive in only membrane in this TN case/MUC1-VNTR and MUC-CT are positive in cytoplasm of LUM and negative in TN/NEU1 is positive in cytoplasm of LUM and negative in TN. This figure illustrated observations described in Table 2: the low-expression of MUC1-VNTR, MUC1-CT, NEU1 and the high-expession of EGFR in TN, suggesting that the EGFR/MUC1/NEU1 molecular complex could be deregulated in breast cancers. Immunohistochemistry on the same LUM and TN cases. Magnification 400×.
Figure 2.
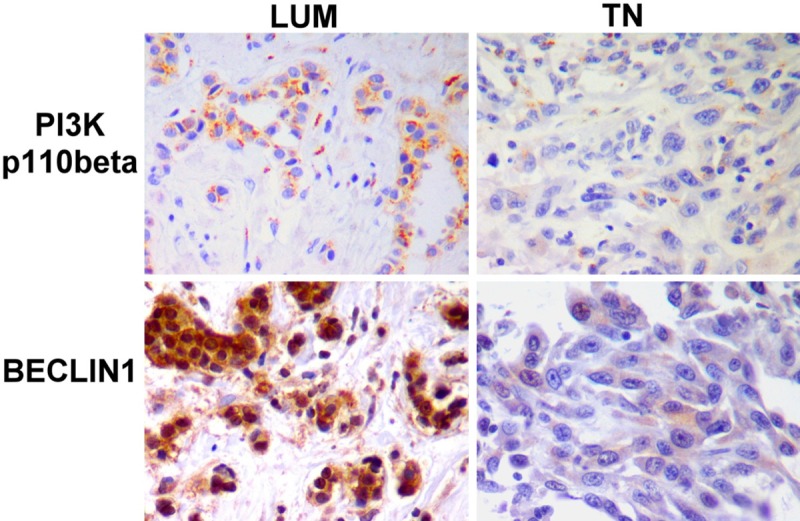
PI3Kp110β and Beclin-1 difference and morphological descriptions between luminal (LUM) and triple-negative (TN) breast carcinoma. PI3Kp110β and Beclin-1 are high-positive in LUM and low-positive in TN. This figure illustrated observations described in Table 2: the low-expression of Beclin-1 and PI3Kp110β in TN, suggesting a low action of autophagy. Immunohistochemistry on the same LUM and TN cases. Magnification 400×.
Table 2.
Biological difference between LUM and TN antigenic expressions. Results are expressed in mean and standard deviation of histological score
| LUM | TN | P | |
|---|---|---|---|
| N | 39 | 48 | |
| EGFR | 0.35 ±1.18 | 6.60 ± 5.04 | < 0.0001 |
| MUC1-VNTR | 10.71 ± 4.89 | 7.14 ± 5.76 | 0.002 |
| MUC1-CT | 9.53 ± 4.62 | 5.50 ± 4.18 | < 0.0001 |
| NEU1 | 8.05 ± 3.16 | 6.06 ± 5.01 | 0.03 |
| PI3Kp110β | 9.55 ±4.09 | 6.10 ± 3.96 | < 0.0001 |
| BECLIN1 | 9.65 ± 4.26 | 6.23± 3.77 | < 0.0001 |
Then, we investigated the morphological localisation of these molecules. As we previously described, MUC1-VNTR is expressed both at the cytoplasm membrane and in the cytoplasm [33]. MUC1-CT showed the same expression pattern. Importantly, we noted that MUC1-VNTR and MUC1-CT expression were not always observed at the cell membrane of each patient group, indicating that MUC1 epitopes are not always accessible for a target therapy using monoclonal antibodies. NEU1 and PI3Kp110β were localized mainly in the cytoplasm. Beclin-1 was observed either in the cytoplasm or in the nuclei.
Relative expression of the EGFR/MUC1/NEU1 complex molecules
Although MUC1 expression has been well illustrated in breast carcinoma, to date the comparison of MUC1-VNTR and MUC1-CT in TN and LUM is still not described in the literature. Here, we found a positive correlation between MUC1-VNTR and MUC1-CT within the TN group (P < 0.0001, r = 0.64) but not for the LUM group, suggesting that in TN, MUC1-VNTR and MUC1-CT were produced at the same rate (Figure 3). Previous studies suggested the possibility of a MUC1-EGFR-NEU1 molecular complex [18,34]. We then sought a relationship between the expressions of EGFR with each one of these two forms of MUC1 in the two groups of breast cancers. In contrast to the previous documented studies, we did not observe a positive correlation neither between EGFR and MUC1-VNTR, nor with MUC1-CT whatever the breast cancer group analysed (data not shown). Likewise, EGFR expression was not positively correlated to Neu1 expression. Furthermore, in the setting of the above-mentioned results in Figure 1, we also questioned the possibility of an inverse correlation between EGFR and MUC1 or NEU1. Though, no correlation was found between these molecules. Conversely, MUC1-VNTR was statistically and positively correlated to NEU1 expression in the TN group (P = 0.04; r = 0.25), but not in the LUM group (Figure 4). No correlation was observed with the intracellular MUC1 domain (MUC1-CT) (Figure 4). These results suggest that only an interaction between NEU1 and the extracellular domain of MUC1 may occur in the TN group.
Figure 3.
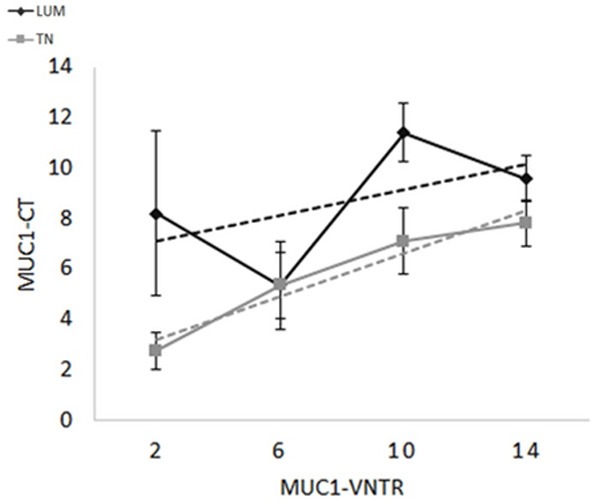
Histological score relationship between MUC1-VNTR and MUC1-CT in LUM (grey, P = ns) or TN (black, P < 0.0001). The linear positive correlation between MUC1-VNTR and MUC1-CT observed only in TN, suggests that these both antigens were produced in the same concentration. Consequently, MUC1 seems less modified in TN than in LUM.
Figure 4.
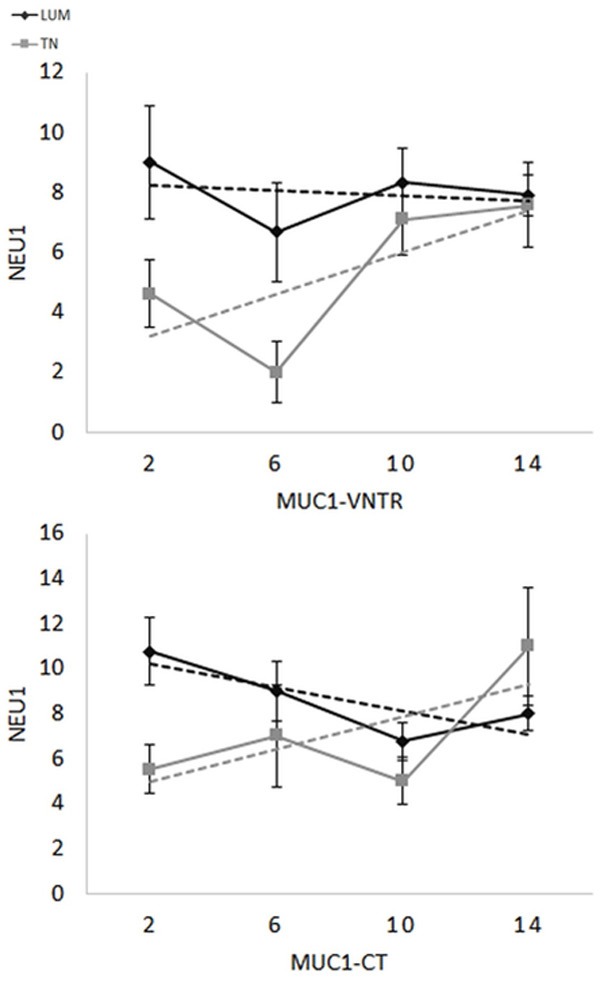
Histological score relationship between NEU1 and MUC1-VNTR in LUM (grey, p=ns) or TN (black, p=0.04) or MUC1-CT for LUM (grey, P = ns) or TN (black, P = ns). Correlations between MUC1-VNTR or MUC1-CT and NEU1 were only significant between MUC1-VNTR and NEU1 in TN (P = 0.04) suggesting that the extracellular chain of MUC1 is associated with NEU1 only in TN.
Autophagy and TN breast cancer
Autophagy has been involved in breast cancer [29,30]. We studied two different proteins involved in the autophagy pathway: the subunit PI3Kp110β and Beclin-1 [26-28]. Our results indicate a positive correlation between PI3Kp110β and Beclin-1 either in LUM (P = 0.001, r = 0.41) and TN (P = 0.002, r = 0.40), demonstrating the relationship between the two proteins. TN presented a low-level of both PI3Kp110β and Beclin-1 suggesting a decreasing of autophagy (Table 2). Moreover, NEU1 presented a positive correlation for PI3Kp110β and Beclin-1 in TN (respectively P = 0.0003, r =0.48 and P = 0.01, r = 0.31) (Figure 5). In the LUM group, NEU1 was positively correlated to PI3Kp110β (P = 0.04, r = 0.29) but not with beclin-1 (Figure 5). These observations pointed out the implication of NEU1 in the autophagy through the lysosomal machinery. MUC1-VNTR also showed positive correlations in TN for both PI3Kp110β (P = 0.009, r = 0.32) and Beclin 1 (P = 0.01, r = 0.32). MUC1-CT was only correlated with PI3Kp110β in the TN group (P = 0.04) but not with Beclin-1 (Figure 6), suggesting that MUC1-VNTR was the main MUC1 subunit involved in the autophagy process in TN (Figure 7). This relationship was less obvious in LUM. We concluded that autophagy is reduced in TN involving likely MUC1-VNTR and NEU1.
Figure 5.
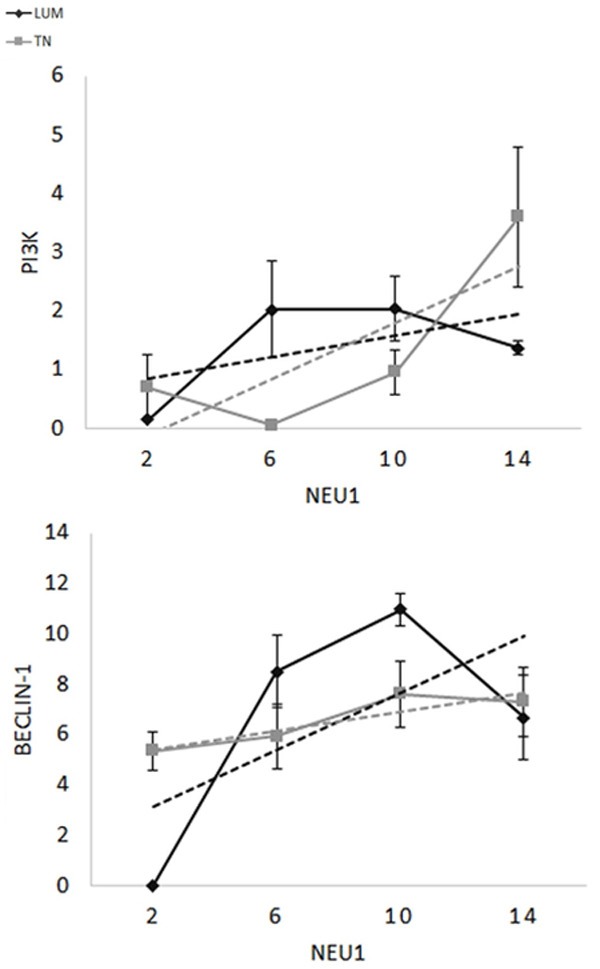
Histological score relationship between NEU1 and PI3Kp110β for LUM (grey, P = 0.04) or TN (black, P = 0.0003) or Beclin1 for LUM (grey, P = ns) or TN (black, P = 0.01). NEU1 presented a positive correlation with the two antigens of autophagy: PI3K (p110β) (for TN P = 0.01 and for LUM P = 0.04) and Beclin-1 (For Tn P = 0.0003 and for LUM P = ns). This suggests that the lysosomial enzyme NEU1 is involved in the autophagy pathway.
Figure 6.
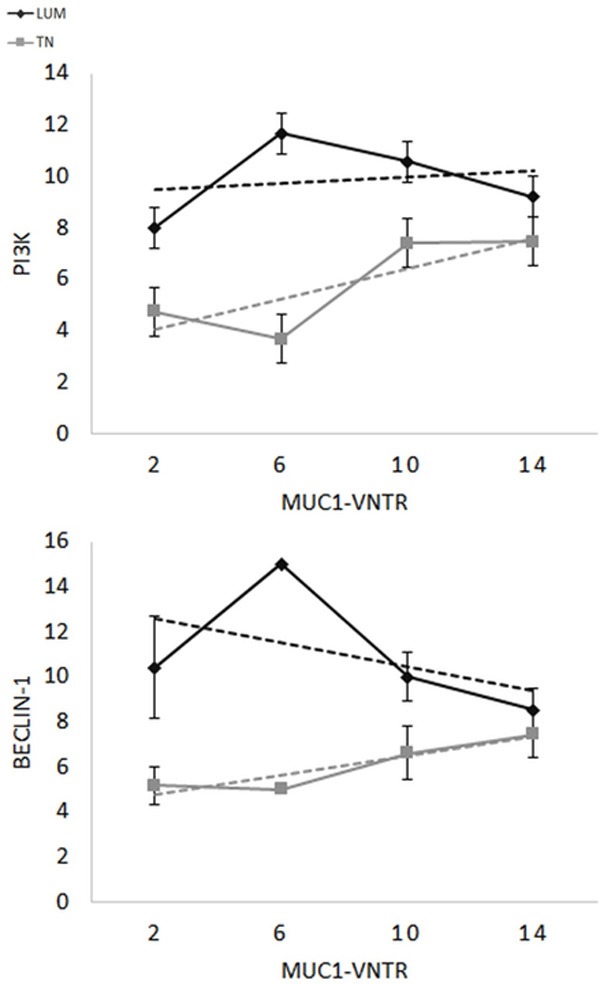
Histological score relationship between MUC1-VNTR and PI3Kp110β for LUM (grey, P = 0.04) or TN (black, P = 0.009) or Beclin-1 for LUM (grey, P = 0.01) or TN (black, P = 0.01). Only MUC1-VNTR is correlated with two antigens of autophagy (Beclin-1 and PI3Kp110β) (P = …) in TN and in LUM. This suggests that the extracellular part of MUC1 could be played a role in the autophagy of breast carcinoma.
Figure 7.
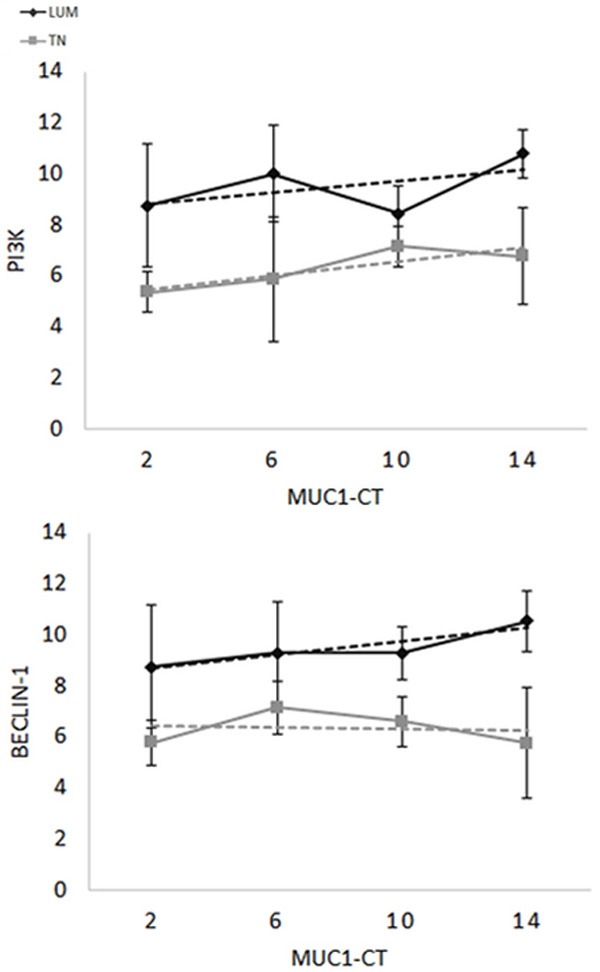
Histological score realtionship between MUC1-CT and PI3Kp110β for LUM (grey, P = ns) or TN (black, P = 0.04) or Beclin-1 for LUM (grey, P = ns) or TN (black, P = ns). Only MUC1-CT is only correlated PI3K (p110β) in TN (P = 0.04). This suggests that the intracellular part of MUC1 is less involved in the autophagy pathway than the extracellular part of MUC1. As illustrated in the Figure 2, MUC1 is less modified by the MUC1-VNTR splicing in TN, suggesting that an independent action of the extracellular MUC1-VNTR and the intracellular MUC1-CT.
Discussion
Recently on routine histological material, we have demonstrated that MUC1 protein was associated with the tumour aggressive biological behaviour of breast carcinoma [33]. Several authors also illustrated the secretion of MUC1 by breast cancer cells [35-37]. Most of them were mainly interested by the extracellular α-subunit of the MUC1 (MUC1-VNTR) because in tumour cells, the antigenic sides of the α-subunit protein core are specifically denuded by an aberrant lack of glycosylation. The core protein is then more exposed and constitutes a potential target for immunotherapy [38]. Deepening the knowledge of breast carcinomas, we here showed an important heterogeneity, both in the quantitative and the qualitative expression of MUC1 in luminal and triple negative breast carcinomas. In agreement with our data, Siroy et al had already showed that 67% of early stage basal-like triple negative breast cancers strongly expressed MUC1, 27% showed a weak secretion and 6% were negative [37]. Our study also supports a previous report on MUC1-VNTR expression in a small group of patients (10 cases) [33]. Such variability is of clinical interest. Indeed, we here showed that both MUC1 subunits (MUC1-VNTR and MUC1-CT), and thereof epitopes exposure, are less expressed in TN than in LUM breast cancers.
Lillehoj et al suggested the possibility of a MUC1-EGFR-NEU1 molecular complex [18]. Consequently, we looked at the association between MUC1, EGFR and/or NEU1 in the two groups of breast cancers. However, we did not find any correlation between EGFR and MUC1-VNTR, MUC1-CT or NEU1. We even found that while TN was expressed a high level of EGFR, both MUC1-VNTR and MUC1-CT were down-regulated in TN as compared to LUM breast cancer. These results are discordant with those of Neeraja Dharmanarj et al who recently demonstrated a statistical correlation between MUC1 and EGFR and concluded that the activation of EGFR stimulates MUC1 expression in multiple cellular contexts [39], but are in setting with those of others authors who showed that EGFR stimulation promotes the cleavage α-subunit MUC1 [20,40,41]. Such discrepancies highlight the importance to define in situ the expression of these molecules according to the type of cancer. In vitro study on breast cell lines demonstrated that MUC1 and EGFR are associated in a molecular complex in breast cancers and that MUC1 inhibits EGFR down-regulation and endocytosis [34]. Based on our in situ results, we then hypothesize that EGFR overexpression observed in TN is the result of EGFR accumulation in the cytoplasm or cell membrane rather than EGFR-overproduction.
Our results demonstrate that the expression of MUC1-VNTR and MUC1-CT were only correlated in TN, suggesting that the MUC1 is not cleaved yet, and therefore that MUC1 endocytosis is reduced in TN. Accordingly, Wreschner et al demonstrated that full length MUC1 is modified following a limited proteolysis event of its extracellular part (MUC1-VNTR) by the recycling of MUC1 by endocytosis [34,38]. Subsequently, Crose et al showed that MUC1-CT constitutes a better indicator of MUC1 production than MUC1-VNTR because it does not depend on the MUC1 proteolysis [42]. Therefore, the positive correlation between MUC1-VNTR and MUC1-CT observed in TN, is the indirect reflect of the lack of MUC1 recycling. Reduced recycling of MUC1 in TN therefore bound to a reduced level of MUC1 glycosylation supports the fact that MUC1 epitopes could be better recognized in this type of breast cancer. Furthermore, because MUC1-CT is not or less altered or glycosylated, it constitutes a better indicator of the primary secretion of the MUC1 [42]. This also supports our above mentioned hypothesis that EGFR accumulates in TN.
The high level of EGFR and our hypothesis that MUC1 is not cleaved suggest that EGFR-MUC1 pathway is deregulated in TN. The membrane associated PI3K plays an important role in the EGFR intracytoplasmic signalling. Using conditional gene knockout mice deficient in the class IA PI3K p110α or p110β catalytic subunit, Dou et al demonstrated that p110β subunit promotes autophagy by activation of the complex Rab5-Vps34-Vps15-Beclin-1, independently of its kinase activity [26]. This pathway seems to be independent of EGFR stimulating pathway which is associated with the cascade of PI3K p110α/AKT/mTOR, well-known as an inhibitor of autophagy [23,43]. Our observation of the low expression of both Beclin-1 and PI3Kp110β confirms that the autophagy pathway is reduced in TN breast cancer. In these cancers, the positive correlation between MUC1-VNTR with both PI3K p110β and Beclin-1, strongly advocates for a link between MUC1-VNTR and autophagy [45]. Then, our in situ results support a previous in vitro study showing that MUC1 promotes autophagy in human tumour cells in response to glucose deprivation [27].
Autophagy is an adaptive phenomena widely used by tumour cells using the lysosomial machinery [44]. Debnath et al pointed out the important role of autophagy in breast carcinogenesis. Indeed, reduced autophagy can promote tumour development by genomic instability. We found that EGFR was highly expressed in TN. Interestingly, in a series of 107 TN, Tilch et al did not identify any mutation of the EGFR gene suggesting that EGFR protein is physiologically normal [45]. In the setting of a reduced level of autophagy in TN breast cancer, it is worth to note that IL17A has been described to attenuate the autophagy process by regulation of PI3K [46]. Recently, in 3 of our patients presenting a TN breast cancer, Cochaud et al showed a high production of IL17A, supporting the implication of IL17A as inhibitor of autophagy in TN [47]. Furthermore, EGFR activation in TN surely plays a role in Beclin-1 phosphorylation and, consequently on autophagy suppression. Indeed, Wei et al demonstrated that this mechanism could contribute to tumour progression and chemoresistance in lung carcinoma [48]. Another interesting regulator of autophagy is the oncoprotein p53 which is often mutated in TN. Of note, genetically altered p53 was also demonstrated to inhibit autophagy [49-51]. Interestingly, we also observed a high level of p53 in TN (data not show).
Tumour cells are also able to activate autophagy in diverse conditions such as hypoxia, extracellular matrix fragmentation or other metabolic modifications [52]. The recycling of MUC1 through the cytoplasm and the cellular membrane and its cleavage and release in the extracellular matrix are well documented in the literature [34,38]. Although NEU1 has been recently suggested to be associated with the EGFR-MUC1 membrane complex, we found that NEU1 is less expressed in TN than LUM. Nevertheless, in TN NEU1 was positively correlated with the extracellular MUC1 domain or MUC1-VNTR (P = 0.04, r = 0.25) but not with the intracellular MUC1 domain (MUC1-CT). Interestingly, NEU1 is a lysosome enzyme that is able to de-sialylate several macromolecules such as MUC1 or EGFR. NEU1 is known as a modulator of cell receptors, and has been involved in endocytosis and MUC1 regulation [18]. It can activate phagocytosis in macrophages and dendritic cells through de-sialylation of surface receptors [53,54]. NEU1 is also involved in processing extracellular matrix fragmentation signals such as elastin peptides [55]. Gilmour et al showed that matrix metalloproteinase-9 and NEU1 form a complex with EGFR on the cell surface [56]. These observations suggest that the extracellular matrix could play an important role in the engagement of the molecular complex EGFR-NEU1-MUC1 and its associated intracellular signals.
To conclude, we demonstrated that autophagy is reduced in TN breast cancers leading likely to deregulation of the EGFR-MUC1-NEU1 complex and associated cellular pathways. Nevertheless, further studies will be needed to show the co-localizations of EGFR-MUC1-NEU1 and the close regulations of these molecular actors in different type of breast cancers.
Disclosure of conflict of interest
None.
Abbreviations
- TN
triple-negative breast carcinoma
- LUM
luminal type of breast carcinoma
- MUC1-VNTR
α-subunit of MUC1
- MUC1-CT
β-subunit of MUC1
- TMA
tissue micro-array
- NEU1
neuraminidase-1
- EGFR
human epidermal growth factor receptor type 1
- HER2
human epidermal growth factor receptor type 2
- ER
Estrogen receptors
- PR
progesterone receptors
- CK
cytokeratin
- PI3K
phosphoinositide 3-kinases
References
- 1.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 2.Strehl JD, Wachter DL, Fasching PA, Beckmann MW, Hartmann A. Invasive Breast Cancer: Recognition of Molecular Subtypes. Breast Care (Basel) 2011;6:258–264. doi: 10.1159/000331339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ Panel members. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–23. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivenbark AG, O’Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol. 2013;183:1113–24. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, Wolff AC, Hobday TJ, Ivanova A, Chiu WK, Ferraro M, Burrows E, Bernard PS, Hoadley KA, Perou CM, Winer EP. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J. Clin. Oncol. 2012;30:2615–23. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baselga J, Gómez P, Greil R, Braga S, Climent MA, Wardley AM, Kaufman B, Stemmer SM, Pêgo A, Chan A, Goeminne JC, Graas MP, Kennedy MJ, Ciruelos Gil EM, Schneeweiss A, Zubel A, Groos J, Melezínková H, Awada A. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J. Clin. Oncol. 2013;31:2586–92. doi: 10.1200/JCO.2012.46.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–50. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baruch A, Hartmann M, Zrihan-Licht S, Greenstein S, Burstein M, Keydar I, Weiss M, Smorodinsky N, Wreschner DH. Preferential expression of novel MUC1 tumor antigen isoforms in human epithelial tumors and their tumor-potentiating function. Int J Cancer. 1997;71:741–9. doi: 10.1002/(sici)1097-0215(19970529)71:5<741::aid-ijc9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Joshi MD, Ahmad R, Yin L, Raina D, Rajabi H, Bubley G, Kharbanda S, Kufe D. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther. 2009;8:3056–65. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubinstein DB, Karmely M, Ziv R, Benhar I, Leitner O, Baron S, Katz BZ, reschner DH. MUC1/X protein immunization enhances cDNA immunization in generating anti-MUC1 alpha/beta junction antibodies that target malignant cells. Cancer Res. 2006;66:11247–53. doi: 10.1158/0008-5472.CAN-06-1486. [DOI] [PubMed] [Google Scholar]
- 11.Senapati S, Das S, Batra SK. Mucin-interacting proteins: from function to therapeutics. Trends Biochem Sci. 2010;35:236–45. doi: 10.1016/j.tibs.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coveler AL, Bates NE, Disis ML. Progress in the development of a therapeutic vaccine for breast cancer. Breast Cancer (Dove Med Press) 2010;2:25–36. doi: 10.2147/bctt.s6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limacher JM, Quoix E. TG4010: A therapeutic vaccine against MUC1 expressing tumors. Oncoimmunology. 2012;1:791–792. doi: 10.4161/onci.19863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillai K, Pourgholami MH, Chua TC, Morris DL. MUC1 as a Potential Target in Anticancer Therapies. Am J Clin Oncol. 2013;38:108–18. doi: 10.1097/COC.0b013e31828f5a07. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi T, Takazawa H, Imai S, Morimoto J, Watanabe T. Lack of polymorphism in MUC1 tandem repeats in cancer cells is related to breast cancer progression in Japanese women. Breast Cancer Res Treat. 2005;92:223–30. doi: 10.1007/s10549-005-2469-y. [DOI] [PubMed] [Google Scholar]
- 16.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83:727–37. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 17.Monti E, Preti A, Venerando B, Borsani G. Recent development in mammalian sialidase molecular biology. Neurochem Res. 2002;27:649–63. doi: 10.1023/a:1020276000901. [DOI] [PubMed] [Google Scholar]
- 18.Lillehoj EP, Hyun SW, Feng C, Zhang L, Liu A, Guang W, Nguyen C, Luzina IG, Atamas SP, Passaniti A, Twaddell WS, Puché AC, Wang LX, Cross AS, Goldblum SE. NEU1 sialidase expressed in human airway epithelia regulates epidermal growthfactor receptor (EGFR) and MUC1 protein signaling. J Biol Chem. 2012;287:8214–31. doi: 10.1074/jbc.M111.292888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–12. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 21.Bitler BG, Goverdhan A, Schroeder JA. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J Cell Sci. 2010;123:1716–23. doi: 10.1242/jcs.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–81. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia S, Roberts TM, Zhao JJ. Should individual PI3 kinase isoforms be targeted in cancer? Curr Opin Cell Biol. 2009;21:199–208. doi: 10.1016/j.ceb.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, Iglehart JD, Roberts TM, Muller WJ, Zhao JJ. The p110α and p110β isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–86. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dbouk HA, Backer JM. Novel approaches to inhibitor design for the p110β phosphoinositide 3-kinase. Trends Pharmacol Sci. 2013;34:149–53. doi: 10.1016/j.tips.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, Lin RZ, Zong WX. Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50:29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein promotes autophagy in a survival response to glucose deprivation. Int J Oncol. 2009;34:1691–9. doi: 10.3892/ijo_00000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–48. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon JH, Ahn SG, Lee BH, Jung SH, Oh SH. Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA-PKcs and PARP-1. Biochem Pharmacol. 2012;83:747–57. doi: 10.1016/j.bcp.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 30.Ajabnoor GM, Crook T, Coley HM. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis. 2012;3:e260. doi: 10.1038/cddis.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF American Society of Clinical Oncology; College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 32.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- 33.Garbar C, Mascaux C, Curé H, Bensussan A. Muc1/Cd227 immunohistochemistry in routine practice is a useful biomarker in breast cancers. J Immunoassay Immunochem. 2013;34:232–45. doi: 10.1080/15321819.2012.699491. [DOI] [PubMed] [Google Scholar]
- 34.Wreschner DH, McGuckin MA, Williams SJ, Baruch A, Yoeli M, Ziv R, Okun L, Zaretsky J, Smorodinsky N, Keydar I, Neophytou P, Stacey M, Lin HH, Gordon S. Generation of ligand-receptor alliances by “SEA” module-mediated cleavage of membrane-associated mucin proteins. Protein Sci. 2002;11:698–706. doi: 10.1110/ps.16502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay P, Chakraborty S, Ponnusamy MP, Lakshmanan I, Jain M, Batra SK. Mucins in the pathogenesis of breast cancer: implications in diagnosis, prognosis and therapy. Biochim Biophys Acta. 2011;1815:224–40. doi: 10.1016/j.bbcan.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Vegt B, de Roos MA, Peterse JL, Patriarca C, Hilkens J, de Bock GH, Wesseling J. The expression pattern of MUC1 (EMA) is related to tumour characteristics and clinical outcome of invasive ductal breast carcinoma. Histopathology. 2007;51:322–35. doi: 10.1111/j.1365-2559.2007.02757.x. [DOI] [PubMed] [Google Scholar]
- 37.Siroy A, Abdul-Karim FW, Miedler J, Fong N, Fu P, Gilmore H, Baar J. MUC1 is expressed at high frequency in early-stage basal-like triple-negative breast cancer. Hum Pathol. 2013;44:2159–66. doi: 10.1016/j.humpath.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litvinov SV, Hilkens J. The epithelial sialomucin, episialin, is sialylated during recycling. J Biol Chem. 1993;268:21364–71. [PubMed] [Google Scholar]
- 39.Neeraja Dharmaraj, Engel BJ, Carson DD. Activated EGFR stimulates MUC1 expression in human uterine and pancreatic cancer cell lines. J Cell Biochem. 2013;114:2314–22. doi: 10.1002/jcb.24580. [DOI] [PubMed] [Google Scholar]
- 40.Lau SK, Shields DJ, Murphy EA, Desgrosellier JS, Anand S, Huang M, Kato S, Lim ST, Weis SM, Stupack DG, Schlaepfer DD, Cheresh DA. EGFR-mediated carcinoma cell metastasis mediated by integrin αvβ5 depends on activation of c-Src and cleavage of MUC1. PLoS One. 2012;7:e36753. doi: 10.1371/journal.pone.0036753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pochampalli MR, Bitler BG, Schroeder JA. Transforming growth factor alpha dependent cancer progression is modulated by Muc1. Cancer Res. 2007;67:6591–8. doi: 10.1158/0008-5472.CAN-06-4518. [DOI] [PubMed] [Google Scholar]
- 42.Croce MV, Isla-Larrain MT, Rua CE, Rabassa ME, Gendler SJ, Segal-Eiras A. Patterns of MUC1 tissue expression defined by an anti-MUC1 cytoplasmic tail monoclonal antibody in breast cancer. J Histochem Cytochem. 2003;51:781–8. doi: 10.1177/002215540305100609. [DOI] [PubMed] [Google Scholar]
- 43.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barth JM, Köhler K. How to take autophagy and endocytosis up a notch. Biomed Res Int. 2014;2014:960803. doi: 10.1155/2014/960803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol Biol Cell. 2000;11:819–31. doi: 10.1091/mbc.11.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilch E, Seidens T, Cocciardi S, Reid LE, Byrne D, Simpson PT, Vargas AC, Cummings MC, Fox SB, Lakhani SR, Chenevix Trench G. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat. 2014;143:385–92. doi: 10.1007/s10549-013-2798-1. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Mi S, Li Z, Hua F, Hu ZW. Interleukin 17A inhibits autophagy through activation of PIK3CA to interrupt the GSK3B-mediated degradation of BCL2 in lung epithelial cells. Autophagy. 2013;9:730–42. doi: 10.4161/auto.24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cochaud S, Giustiniani J, Thomas C, Laprevotte E, Garbar C, Savoye AM, Curé H, Mascaux C, Alberici G, Bonnefoy N, Eliaou JF, Bensussan A, Bastid J. IL-17A is produced by breast cancer TILs and promotes chemoresistance and proliferation through ERK1/2. Sci Rep. 2013;3:3456. doi: 10.1038/srep03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, Grishin NV, Peyton M, Minna J, Bhagat G, Levine B. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–84. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363–9. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–61. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 53.Debnath J. The multifaceted roles of autophagy in tumors-implications for breast cancer. J Mammary Gland Biol Neoplasia. 2011;16:173–87. doi: 10.1007/s10911-011-9223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seyrantepe V, Iannello A, Liang F, Kanshin E, Jayanth P, Samarani S, Szewczuk MR, Ahmad A, Pshezhetsky AV. Regulation of phagocytosis in macrophages by neuraminidase 1. J Biol Chem. 2010;285:206–15. doi: 10.1074/jbc.M109.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pshezhetsky AV, Hinek A. Where catabolism meets signalling: neuraminidase 1 as a modulator of cell receptors. Glycoconj J. 2011;28:441–52. doi: 10.1007/s10719-011-9350-5. [DOI] [PubMed] [Google Scholar]
- 56.Duca L, Blanchevoye C, Cantarelli B, Ghoneim C, Dedieu S, Delacoux F, Hornebeck W, Hinek A, Martiny L, Debelle L. The elastin receptor complex transduces signals through the catalytic activity of its Neu-1 subunit. J Biol Chem. 2007;282:12484–91. doi: 10.1074/jbc.M609505200. [DOI] [PubMed] [Google Scholar]
- 57.Gilmour AM, Abdulkhalek S, Cheng TS, Alghamdi F, Jayanth P, O’Shea LK, Geen O, Arvizu LA, Szewczuk MR. A novel epidermal growth factor receptor-signaling platform and its targeted translation in pancreatic cancer. Cell Signal. 2013;25:2587–603. doi: 10.1016/j.cellsig.2013.08.008. [DOI] [PubMed] [Google Scholar]


