Abstract
Recent findings have shown that microRNAs play critical roles in the pathogenesis of diabetic nephropathy. miR-34c has been found to inhibit fibrosis and the epithelial-mesenchymal transition of kidney cells. However, the role of miR-34c in diabetic nephropathy has not been well studied. The current study was designed to investigate the role and potential underlying mechanism of miR-34c in regulating diabetic nephropathy. After treating podocytes with high glucose (HG) in vitro, we found that miR-34c was downregulated and that overexpression of miR-34c inhibited HG-induced podocyte apoptosis. The direct interaction between miR-34c and the 3’-untranslated region (UTR) of Notch1 and Jagged1 was validated by dual-luciferase reporter assay. Moreover, Notch1 and Jagged1 as putative targets of miR-34c were downregulated by miR-34c overexpression in HG-treated podocytes. Overexpression of miR-34c inhibited HG-induced Notch signaling pathway activation, as indicated by decreased expression of the Notch intracellular domain (NICD) and downstream genes including Hes1 and Hey1. Furthermore, miR-34c overexpression increased the expression of the anti-apoptotic gene Bcl-2, and decreased the expression of the pro-apoptotic protein Bax and cleaved Caspase-3. Additionally, the phosphorylation of p53 was also downregulated by miR-34c overexpression. Taken together, our findings suggest that miR-34c overexpression inhibits the Notch signaling pathway by targeting Notch1 and Jaggged1 in HG-treated podocytes, representing a novel and potential therapeutic target for the treatment of diabetic nephropathy.
Keywords: Diabetic nephropathy, podocytes, Notch, microRNAs, cell apoptosis
Introduction
Diabetic nephropathy, a severe and common complication of diabetes, has become the leading cause of end stage renal disease [1]. Diabetic nephropathy not only affects the quality of life of patients, but also endangers their lives [2]. Understanding the underlying mechanism of diabetic nephropathy pathogenesis could provide novel insights for the development of effective therapeutics for diabetic nephropathy. Podocytes, attached to the outer surface of the glomerular basement membrane, have been suggested to play an important role in the development of diabetic nephropathy [3,4]. High glucose (HG) has been found to contribute to podocyte injury, which is characterized by increased albumin filtration and decreased expression of podocin, nephrin and slit diaphragm-associated proteins [5,6]. It has been reported that HG induces podocyte apoptosis in vitro and in vivo [7]. Loss of podocytes has been suggested to be the early mechanism of the pathogenesis of diabetic nephropathy [7,8]. Therefore, targeting podocytes may provide new renal-protective drugs for diabetic nephropathy.
The initiation factors of diabetic nephropathy include glucose and lipid metabolism disorder, oxidative stress and inflammatory mediators that activate various signaling pathways [9-12] including the Notch pathway [13]. The Notch pathway is an important signal for cell differentiation and cell survival [14,15], and is known to play critical roles in kidney development [16]. Notch proteins, including Notch1-Notch4, are transmembrane receptors and bind many ligands including Delta-like (Dll) 1, Dll3, Dll4, Jagged1 and Jagged2 [17]. The activation of Notch results in the sequential proteolytic cleavage of the Notch receptor, which releases Notch intracellular domain (NICD) into the nucleus, which in turn activates downstream gene transcription including that of the Hes and Hey genes [18,19]. Nowadays, there is increasing evidence that the Notch signaling pathway is extensively involved in HG-induced podocyte apoptosis [20,21]. Therefore, targeting the Notch pathway may provide novel insights into the mechanism underlying diabetic nephropathy and possible targets for drug development.
MicroRNAs (miRNAs) with ~24 nucleotides are non-coding RNAs that regulate gene expression by targeting the 3’-untranslated region (UTR) of the target gene to inhibit protein translation [22-24]. In this way, miRNAs have been found to be involved in various cellular processes including cell survival, differentiation and apoptosis [22]. Compelling studies have suggested that miRNAs play an important role in podocytes [25-28]. miR-93 has been found to be decreased in diabetic mice and HG-treated podocytes [29]. miR-29c is upregulated in diabetic mice and HG-treated podocytes, where it contributes to Rho kinase activation and podocyte apoptosis by targeting Sprouty homolog 1 [30]. With the development of technology, miRNAs have become key targets for disease therapy. Thereby, anti-miRNAs have already been developed in clinical trials for diseases such as hepatitis C [31]. Given the tremendous role of miRNA in disease, targeting the key miRNAs in the pathogenesis of diabetic nephropathy is expected to provide novel and potentially therapeutic strategies.
Using bioinformatic algorithms, we determined that Notch1 and Jagged1, which are critical molecules in the Notch pathway [32], are the predicted target genes of miR-34c. Given the critical roles of the Notch pathway in diabetic nephropathy, we aimed to investigate the role of miR-34c in HG-induced podocyte apoptosis and the potential underlying mechanism. Here, we demonstrate that miR-34c plays an important role in HG-induced podocyte apoptosis by interacting with the 3’-UTR of Notch1 and Jagged1 to inhibit Notch signaling activation and abrogate apoptotic and p53 pathways.
Materials and methods
Cell culture and treatment
Mouse podocytes were purchased from the Cell Resource Center of Peking Union Medical College (Beijing, China) and conditionally immortalized using a temperature-sensitive SV40 large T-cell antigen (tsA58 Tag). To induce cell proliferation, podocytes were grown in Dulbecco’s modified eagle medium (DMEM)-F12 (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) containing 10 U/ml γ-interferon, at 33°C and 5% CO2 atmosphere (growth permissive conditions). Then, cells were cultured in fresh DMEM-F12 supplemented with 10% FBS without γ-interferon and maintained at 37°C and 5% CO2 atmosphere for 10-14 days to induce quiescence (growth restrictive conditions) [33]. After that, podocytes were cultured in serum-free medium containing 30 mM D-glucose (high glucose, HG) or 5 mM D-glucose (normal glucose, NG) for 24, 48 or 72 h. pre-miR-34c (Genepharma, Shanghai, China) was administered with lipofectamine 2000 as per the supplier’s instructions.
Apoptosis assay
Cell apoptosis was examined by annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. After treatment of pre-miR-34c for 48 h, the cultured podocytes were collected and washed three times with ice-cold phosphate-buffered saline (PBS). Then, the cells were trypsinized and centrifuged at 200 g for 5 min. The pellets were collected, washed with PBS, and resuspended in binding buffer. Annexin V stock solution was added and the cells were incubated for 30 min at 4°C then stained with PI for 5 min at room temperature in the dark. Finally, apoptotic podocytes were quantified by fluorescence-activated cell sorting scan (BD Biosciences, San Jose, CA, USA).
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) or a one-step primescript miRNA cDNA synthesis kit (Takara, Dalian, China). Complementary DNA was produced using M-MLV reverse transcriptase (Life Technologies, Carlsbad, CA, USA). Gene expression was detected by RT-qPCR as follows. A mix containing the cDNA template, corresponding primers and SYBR Green qPCR Master Mix (Thermo Fisher, Shanghai, China) was subjected to RT-qPCR quantification (94°C for 4 min; 94°C for 20 s, 55°C for 30 s, and 72°C for 20 s; 2 s for plate reading for 35 cycles; and melt-curve analysis from 65 to 95°C). The primers used were as follows: miR-34c (Forward: GCTGCTGTAGGCAGTGTAGTTAG, Reverse: CTCAACTGGTGTCGTGGAGTC); U6 SnRNA (Forward: CTCGCTTCGGCAGCACA, Reverse: AACGCTTCACGAATTTGCGT); Notch1 (Forward: GTGGATGACCTAGGCAAGTCG, Reverse: GTCTCCTCCTTGTTGTTCTGC); Jagged1 (Forward: AGAAGTCAGAGTTCAGAGGCGTCC, Reverse: AGTAGAAGGCTGTCACCAAGCCAAC; Hes1 (Forward: CACGACACCGGACAAACCA, Reverse: GCCGGGAGCTATCTTTCTTAAGTG; Hey1 (Forward: AAGACG GAGAGGCATCATCGAG, Reverse: CAGATCCCTGCTTCTCAAAGGCAC; β-actin (Forward: TTCCTTCTTGGGTATGGAAT, Reverse: GAGCAATGATCTTGATCTTC). β-actin and U6 SnRNA were used as internal references for quantification of the relative expression of mRNA and miRNA, respectively, using the 2-ΔΔCt method.
Western blot analysis
Cells were lysed in cell lysis buffer (Beyotime, Haimen, China) and the protein concentration was measured using the Bradford method. A total of 25 μg proteins were separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electrophoretically transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The membranes were blocked by 2.5% nonfat milk for 1 h at 37°C, and then incubated with primary antibodies at 4°C overnight. After incubating with horseradish peroxidase-conjugated secondary antibody (Boster Corporation, Wuhan, Hubei, China) for 1 h at room temperature, protein bands were visualized using an enhanced chemiluminescence detection system (Amersham, Little Chalfont, UK). The following primary antibodies were used: antibodies against Notch1, Hes1, p53, Bcl-2, Bax and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); antibodies against NCID were purchased from Abcam (Cambridge, UK); antibodies against Hey1 were purchased from Bioss (Beijing, China); antibodies against p-p53 and cleaved Caspase-3 were purchased from Cell Signaling Technology (Danvers, MA, USA).
Dual-luciferase reporter assay
Fragments from the 3’-UTR of Notch1 and Jagged1 containing the predicted binding sequences for miR-34c were amplified and sub-cloned into pGL3 luciferase promoter vector (Promega, Madison, WI, USA). The site-directed mutagenesis of miR-34c binding sites in the 3’-UTR of Notch1 and Jag1 was performed using a Site-Directed Mutagenesis Kit (Stratagene, Hangzhou, China). pGL3 vectors (0.1 μg) containing wild type or mutated 3’-UTR fragments were co-transfected with pre-miR-34c (50 nM) in human embryonic kidney 293 (HEK293) cells using lipofectamine 2000 and cultured for 48 h. After the cells were harvested and lysed, the luciferase activity was measured using the dual-luciferase reporter assay kit (Promega) as per the supplier’s instructions.
Statistical analysis
Data are presented as mean ± standard deviation (SD). Statistical analysis was performed using SPSS version 11.5 (SPSS Inc., Chicago, IL, USA) and statistical significance was calculated by one-way analysis of variance (ANOVA) among multiple groups, or Student t test between two groups. Differences were regarded statistically significant at P < 0.05.
Results
Expression of miR-34c is decreased in HG-treated podocytes
To gain an insight into the role of miR-34c in HG-treated podocytes, we firstly examined the expression profiles of miR-34c in podocytes post HG challenge via RT-qPCR. The results showed that the expression of miR-34c was significantly decreased in podocytes after stimulation with HG for 24 h, continuously decreased after 48 and 72 h (Figure 1). The data suggested a critical role for miR-34c in HG-treated podocytes.
Figure 1.
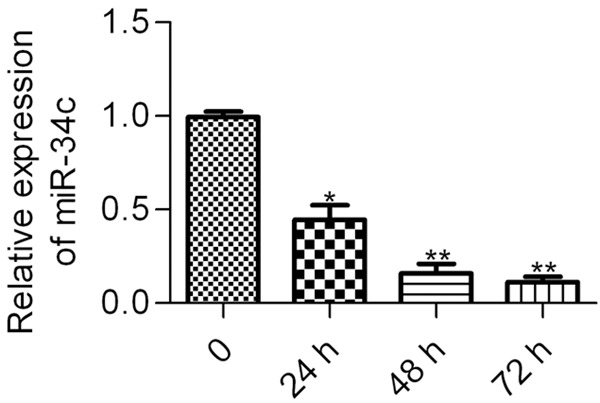
The expression of miR-34c in HG-treated podocytes. Podocytes were incubated with HG (30 nM) and the miR-34c expression was analyzed by RT-qPCR at the indicated times (0-72 h). Statistical analysis was calculated by one-way ANOVA. N = 3, *P < 0.05, **P < 0.01 vs. control (0 h).
Overexpression of miR-34c inhibits HG-induced cell apoptosis in podocytes
To investigate the contribution of miR-34c to HG-induced cell apoptosis in podocytes, the effect of miR-34c overexpression on cell apoptosis induced by HG was examined by flow cytometry. The results showed that HG-treated podocytes exhibited a significant increase in apoptotic cells as compared with normal glucose-treated podocytes, whereas pre-miR-34c treatment significantly decreased HG-induced cell apoptosis in podocytes (Figure 2A and 2B). These results imply that miR-34c is associated with HG-induced cell apoptosis in podocytes.
Figure 2.
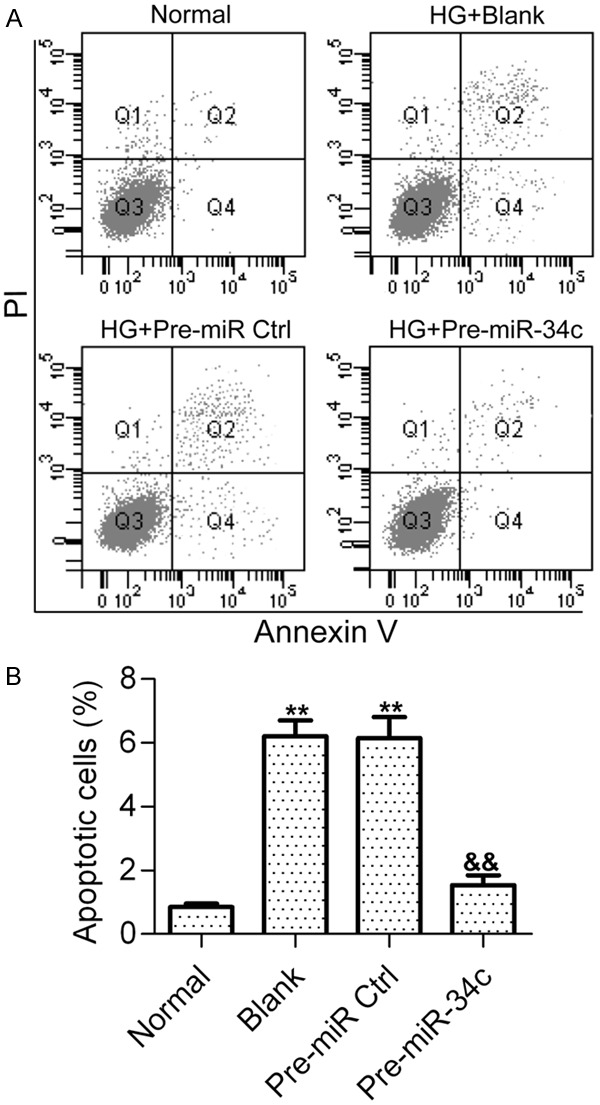
Effect of miR-34c on HG-induced podocyte apoptosis. A. Cell apoptosis was detected by flow cytometry after HG stimulation for 48 h. Podocytes treated with normal glucose (5 nM) was used as the control. Podocytes stimulated with HG were treated with PBS (Blank), control pre-miRNAs (pre-miR Ctrl) or pre-miR-34c. Podocytes were harvested and stained with Annexin V and PI before apoptotic cells were examined. B. Representative histograms show the fractions of apoptotic cells. Statistical analysis was calculated by one-way ANOVA. N = 3, **P < 0.01 vs. Normal, &&P < 0.01 vs. Blank or Pre-miR Ctrl.
miR-34c inhibits the expression of Notch1 and Jagged1 by targeting the 3’-UTR
To investigate the potential mechanism of miR-34c in regulating HG-induced podocyte apoptosis, we screened the putative targets of miR-34c using bioinformatic algorithms. Interestingly, we found that Notch1 and Jagged1, which are the core molecules in the Notch signaling pathway and play critical roles in HG-induced podocyte apoptosis [32], were the predicted targets of miR-34c (Figure 3A and 3B). To validate whether miR-34c directly targets the 3’-UTR region of Nocth1 and Jagged1 mRNA, a dual-luciferase reporter assay was performed. The results showed that co-transfection of pGL3-Notch1-3’-UTR with pre-miR-34c in HEK293 cells significantly decreased luciferase activity in comparison with the control group, whereas co-transfection of pre-miR-34c with pGL3-Notch1-Mut-3’-UTR containing mutations in the predicted consensus sequences for miR-134 had no apparent effect on luciferase activity (Figure 3C). Similar results were obtained using pGL3-Jagged1-3’-UTR and pGL3-Jagged1-Mut-3’-UTR with pre-miR-34c (Figure 3D).
Figure 3.
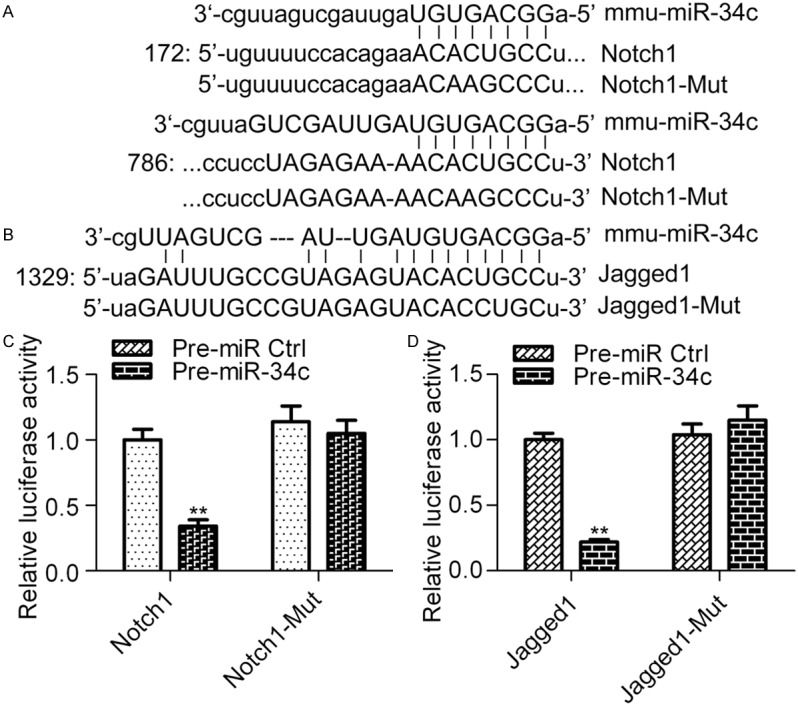
miR-34c directly targets the 3’-UTR of Notch1 and Jagged1. The predicted binding sequences of the 3’-UTR of Notch1 (A) and Jagged1 (B) mRNA with miR-134. (B) Examination of the interaction between miR-34c and the 3’-UTR of Notch1 (C) and Jagged1 (D) mRNA. Cells were transfected with pGL3 reporter plasmids containing either the wild type or mutant versions of the 3’-UTR of Notch1 or Jagged1 in the presence of pre-miR-34c and cultured for 48 h before being harvested for analysis. Pre-miR Ctrl was used as the control. Statistical analysis was calculated Student t test. N = 3, **P < 0.01 vs. Pre-miR Ctrl.
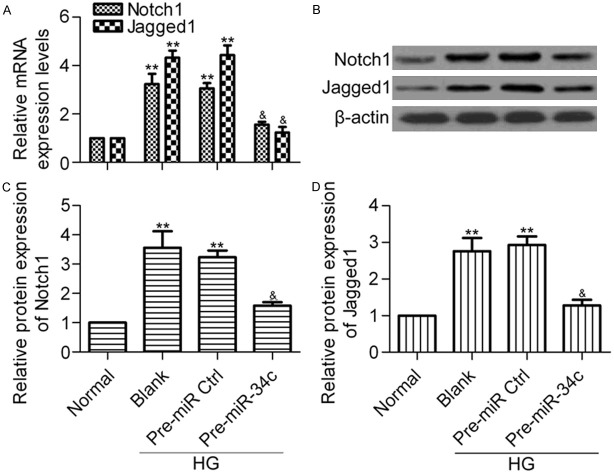
Next, to verify whether miR-34c regulates the expression of Notch1 and Jagged1 in podocytes, we treated cells with pre-miR-34c and examined the mRNA and protein expression of Notch1 and Jagged1 by RT-qPCR and western blot analysis, respectively. The results showed that overexpression of miR-34c significantly decreased the mRNA expression levels of Notch1 and Jagged1 (Figure 4A). Consistently, western blot results (Figure 4B) showed that the protein expression of Notch1 (Figure 4C) and Jagged1 (Figure 4D) was markedly decreased by miR-34c overexpression. These findings suggest that Notch1 and Jagged1 are direct targets of miR-34c.
Figure 4.
Effect of miR-34c on the expression of Notch1 and Jagged1. (A) RT-qPCR analysis of the mRNA expression of Notch1 and Jagged1 in different groups. Podocytes treated with pre-miR-34 were stimulated with HG for 48 h before being collected for analysis. N = 3, **P < 0.01 vs. Normal, &P < 0.05 vs. Blank or Pre-miR Ctrl. (B) Western blot analysis of Notch1 and Jagged1 protein expression levels in different groups with indicated antibodies. β-actin was used as the control. Protein levels of Notch1 (C) and Jagged1 (D) were quantified using Image-Pro Plus 6.0 software. Statistical analysis was calculated by one-way ANOVA. N = 3, **P < 0.01 vs. Normal; &P < 0.05 vs. Blank or Pre-miR Ctrl.
Overexpression of miR-34c suppresses the activation of Notch signaling pathway
To determine the effects of miR-34c on the Notch pathway, we measured the transcription levels of Hes1 and Hey1, which are downstream genes of the Notch pathway, via RT-qPCR. The results showed that the elevated mRNA expression levels of Hes1 and Hey1 induced by HG in podocytes were significantly decreased by miR-34c overexpression (Figure 5A). Furthermore, the protein expression of Notch intracellular domain (NICD), Hes1 and Hey1 was examined by western blot analysis (Figure 5B). Consistently, the protein expression levels of NICD, Hes1 and Hey1 were found to be markedly increased in HG-treated podocytes, but were reversed by miR-34c overexpression (Figure 5C). These results suggest that miR-34c is capable of inhibiting the Notch pathway.
Figure 5.
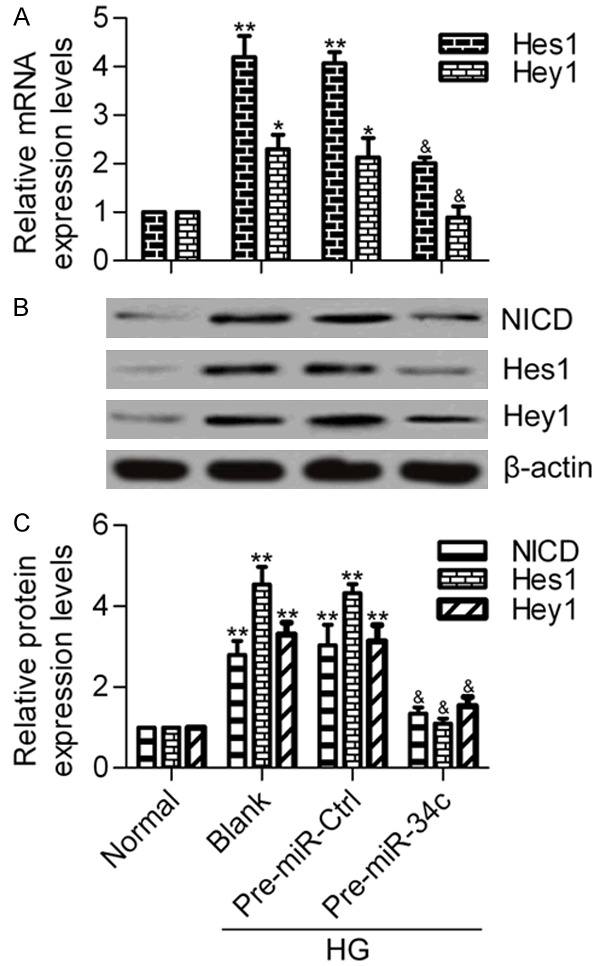
Effect of miR-34c on Notch pathway. A. RT-qPCR analysis of the mRNA expression levels of Hes1 and Hey1 in different treated groups. Podocytes treated with pre-miR-34 were stimulated with HG for 48 h before being collected for analysis. N = 3, *P < 0.05 and **P < 0.01 vs. Normal; &P < 0.05 and &&P < 0.01 vs. Blank or Pre-miR Ctrl. B. Western blot analysis of the protein expression levels of NICD, Hes1 and Hey1 in different treated groups. C. Quantitative protein levels were normalized to β-actin using Image-Pro Plus 6.0 software. Statistical analysis was calculated by one-way ANOVA. N = 3, **P < 0.01 vs. Normal; &P < 0.05 and &&P < 0.01vs. Blank or Pre-miR Ctrl.
Overexpression of miR-34c inhibits apoptotic and p53 pathways
To further delineate the molecular mechanism of miR-34c in HG-induced podocyte apoptosis, we detected the effect of miR-34c on Bcl-2, Bax, cleaved Caspase-3, phosphorylation level of p53 (p-p53), and total p53 protein levels in HG-treated podocytes. Western blot analysis revealed that HG stimulation markedly decreased the protein levels of Bcl-2, but significantly increased the protein levels of Bax and cleaved Caspase-3 (Figure 6A and 6B). Furthermore, overexpression of miR-34c significantly inhibited the p-p53 protein levels induced by HG in podocytes (Figure 6C and 6D). However, there was no difference in total p53 protein levels among the different groups. These results indicate that miR-34c inhibits the apoptotic and p53 pathways, which are activated by HG in podocytes.
Figure 6.
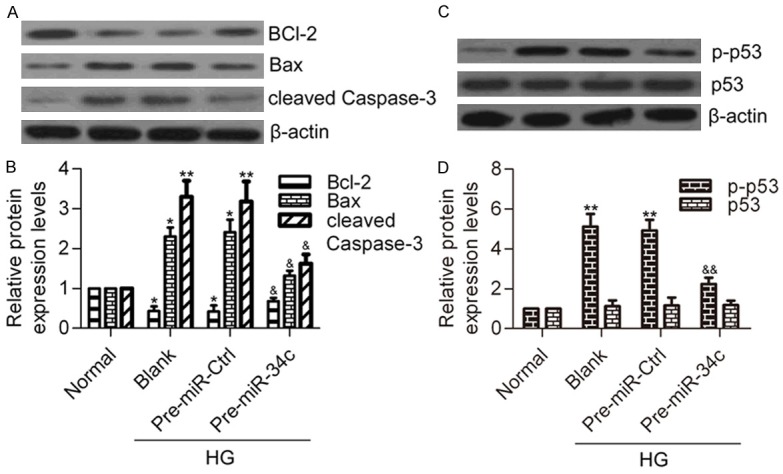
Effect of miR-34c on apoptotic and p53 pathways. A. Western blot analysis of Bcl-2, Bax and cleaved Casapse-3 protein levels in different groups. B. Quantitative protein levels of Bcl-2, Bax and cleaved Casapse-3 were normalized to β-actin using Image-Pro Plus 6.0 software. N=3, *P < 0.05, **P < 0.01 vs. Normal; &P < 0.05 vs. Blank or Pre-miR Ctrl. C. Western blot analysis of p-p53 and total p53 protein levels in different groups. D. Quantitative protein levels of p-p53 and p53. Statistical analysis was calculated by one-way ANOVA. N = 3, **P < 0.01 vs. Normal; &&P < 0.01 vs. Blank or Pre-miR Ctrl.
Discussion
It has recently been reported that miR-34c can inhibit kidney fibrosis and the epithelial-mesenchymal transition [34]. The results of the present study provide evidence that the expression of miR-34c is affected by HG treatment in podocytes and that overexpression of miR-34c inhibits Notch signaling and apoptotic pathways by targeting Notch1 and Jagged1 in HG-treated podocytes. These results suggest an important role of miR-34c in regulating podocyte apoptosis.
The role of the Notch signaling pathway in diabetic nephropathy has been widely studied in recent years. Previous studies found that the Notch ligand Jagged1 and its downstream effector, Hes1, were detected in renal biopsies and tubuli of kidneys from diabetic nephropathy patients, but not in healthy controls, implying an important role of Notch pathways in the pathogenesis of diabetic nephropathy [13]. Niranjan et al. reported that NCID was overexpressed in glomerular epithelial cells in diabetic nephropathy and that overexpression of NCID induced podocyte apoptosis in vitro and in vivo through activation of p53 [35]. Notch1 signaling was found to be significantly increased in HG-treated podocytes and inhibition of Notch1 decreased vascular endothelial growth factor expression and increased nephrin expression, and ameliorated HG-induced podocyte apoptosis [20]. More recently, Gao et al. reported that the expression of Notch1, NCID, jagged1, Hes1 and Hey1 was highly upregulated in HG-treated podocytes and that a chemical inhibitor or specific short hairpin RNA of Notch1 effectively inhibited HG-induced podocyte apoptosis by suppressing Bcl-2 and p53-dependent apoptotic pathways [32]. Notch1, jagged1 and Hes1 were upregulated in HG-treated glomerular mesangial cells, leading to transforming growth factor-beta signaling pathway activation, which was involved in the pathogenesis of diabetic nephropathy [36]. A recent study reported that coactivator-associated arginine methyltransferase 1 degradation induced by HG facilitated Notch1-mediated podocyte apoptosis [37]. These studies have indicated that modulation of Notch1 signaling may provide novel strategies for diabetic nephropathy therapy. Here, we have demonstrated that inhibition of the Notch signaling pathway by miR-34c overexpression inhibited HG-induced podocyte apoptosis. Our data further confirm the critical role of the Notch signaling pathway in HG-induced podocyte apoptosis, revealing that miR-34c is a novel inhibitor of the Notch signaling pathway.
We found that miR34c inhibited the Notch signaling pathway by directly interacting with the 3’UTR of Notch1 and Jagged1, which are critical for Notch signaling pathway activation. The interaction between miR34c and Notch signaling pathways has been reported in other cellular processes. Bae et al. found that miR-34c directly targeted Notch1, Notch2 and Jagged1 to regulate osteoclast differentiation and bone homeostasis [38]. miR-34c was reported to attenuate the epithelial-mesenchymal transition of kidney cells by downregulation of Jagged1 [34]. Here, we found that miR-34c directly targets the 3’UTR of Notch1 and Jagged1 by dual-luciferase reporter assay. Moreover, overexpression of miR-34c inhibited the expression of Notch1 and Jagged1 in HG-treated podocytes. The activation of the Notch signaling pathway indicated by increased expression of NCID, Hes1 and Hey1 was also blocked by miR-34c overexpression.
In the current study, we found that overexpression of miR-34c inhibited the apoptotic and p53 pathway. This finding was consistent with the reports of Gao et al., who demonstrated that inhibition of the Notch pathway apparently inhibited HG-induced podocyte apoptosis by downregulation of the Bcl-2 and p53 pathways [32]. In lung cancer cells, miR-34c was found to contribute to resistance to paclitaxel-apoptosis by inhibiting p53 [39]. However, there have also been studies reporting that miR-34c has a positive role in promoting cell apoptosis [40,41]. The apparent discrepancies between studies indicate that miR-34c may have different biological functions in different cells following different types of stimulation. Nonetheless, we have provided evidence that miR-34c attenuates HG-induced podocyte apoptosis by targeting Notch1 and Jagged1 to inhibit Notch signaling pathway activation. Given that the Notch pathway contributes to HG-induced podocyte apoptosis, miR-34c can be expected to be a novel and effective therapeutic target for the treatment of diabetic nephropathy.
Disclosure of conflict of interest
None.
Abbreviations
- HG
high glucose
- 3’-UTR
3’-untranslated region
- Dll
Delta-like
- NICD
Notch intracellular domain
- FITC
isothiocyanate
- PI
propidium iodide
- DMEM
Dulbecco’s modified eagle medium
- FBS
fetal bovine serum
References
- 1.Nakai S, Masakane I, Akiba T, Shigematsu T, Yamagata K, Watanabe Y, Iseki K, Itami N, Shinoda T, Morozumi K, Shoji T, Marubayashi S, Morita O, Kimata N, Suzuki K, Tsuchida K, Nakamoto H, Hamano T, Yamashita A, Wakai K, Wada A, Tsubakihara Y. Overview of regular dialysis treatment in Japan as of 31 December 2006. Ther Apher Dial. 2008;12:428–456. doi: 10.1111/j.1744-9987.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- 2.Yoon KH, Lee JH, Kim JW, Cho JH, Choi YH, Ko SH, Zimmet P, Son HY. Epidemic obesity and type 2 diabetes in Asia. Lancet. 2006;368:1681–1688. doi: 10.1016/S0140-6736(06)69703-1. [DOI] [PubMed] [Google Scholar]
- 3.Coimbra TM, Janssen U, Grone HJ, Ostendorf T, Kunter U, Schmidt H, Brabant G, Floege J. Early events leading to renal injury in obese Zucker (fatty) rats with type II diabetes. Kidney Int. 2000;57:167–182. doi: 10.1046/j.1523-1755.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 4.Siu B, Saha J, Smoyer WE, Sullivan KA, Brosius FC 3rd. Reduction in podocyte density as a pathologic feature in early diabetic nephropathy in rodents: prevention by lipoic acid treatment. BMC Nephrol. 2006;7:6. doi: 10.1186/1471-2369-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li CX, Xia M, Han WQ, Li XX, Zhang C, Boini KM, Liu XC, Li PL. Reversal by growth hormone of homocysteine-induced epithelial-to-mesenchymal transition through membrane raft-redox signaling in podocytes. Cell Physiol Biochem. 2011;27:691–702. doi: 10.1159/000330078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riediger F, Quack I, Qadri F, Hartleben B, Park JK, Potthoff SA, Sohn D, Sihn G, Rousselle A, Fokuhl V, Maschke U, Purfurst B, Schneider W, Rump LC, Luft FC, Dechend R, Bader M, Huber TB, Nguyen G, Muller DN. Prorenin receptor is essential for podocyte autophagy and survival. J Am Soc Nephrol. 2011;22:2193–2202. doi: 10.1681/ASN.2011020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Susztak K, Raff AC, Schiffer M, Bottinger EP. Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes. 2006;55:225–233. [PubMed] [Google Scholar]
- 8.Steffes MW, Schmidt D, McCrery R, Basgen JM. Glomerular cell number in normal subjects and in type 1 diabetic patients. Kidney Int. 2001;59:2104–2113. doi: 10.1046/j.1523-1755.2001.00725.x. [DOI] [PubMed] [Google Scholar]
- 9.Yuan H, Ren ZL, Hu FQ, Liang W, Ding GH. Renin induces apoptosis in podocytes through a receptor-mediated, angiotensin II-independent mechanism. Am J Med Sci. 2012;344:441–446. doi: 10.1097/MAJ.0b013e318245fdaa. [DOI] [PubMed] [Google Scholar]
- 10.Kim KM, Kim YS, Jung DH, Lee J, Kim JS. Increased glyoxalase I levels inhibit accumulation of oxidative stress and an advanced glycation end product in mouse mesangial cells cultured in high glucose. Exp Cell Res. 2012;318:152–159. doi: 10.1016/j.yexcr.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Lim JC, Lim SK, Park MJ, Kim GY, Han HJ, Park SH. Cannabinoid receptor 1 mediates high glucose-induced apoptosis via endoplasmic reticulum stress in primary cultured rat mesangial cells. Am J Physiol Renal Physiol. 2011;301:F179–88. doi: 10.1152/ajprenal.00032.2010. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Gui D, Chen Y, Mou L, Liu Y, Huang J. Astragaloside IV improves high glucose-induced podocyte adhesion dysfunction via alpha3beta1 integrin upregulation and integrin-linked kinase inhibition. Biochem Pharmacol. 2008;76:796–804. doi: 10.1016/j.bcp.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Walsh DW, Roxburgh SA, McGettigan P, Berthier CC, Higgins DG, Kretzler M, Cohen CD, Mezzano S, Brazil DP, Martin F. Co-regulation of Gremlin and Notch signalling in diabetic nephropathy. Biochim Biophys Acta. 2008;1:10–21. doi: 10.1016/j.bbadis.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Cook KM, Figg WD. Angiogenesis inhibitors: current strategies and future prospects. CA Cancer J Clin. 2010;60:222–243. doi: 10.3322/caac.20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji X, Wang Z, Geamanu A, Sarkar FH, Gupta SV. Inhibition of cell growth and induction of apoptosis in non-small cell lung cancer cells by delta-tocotrienol is associated with notch-1 down-regulation. J Cell Biochem. 2011;112:2773–2783. doi: 10.1002/jcb.23184. [DOI] [PubMed] [Google Scholar]
- 16.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, Das P, Golde TE, Osborne BA. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 2008;112:1813–1821. doi: 10.1182/blood-2008-03-144980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCright B. Notch signaling in kidney development. Curr Opin Nephrol Hypertens. 2003;12:5–10. doi: 10.1097/00041552-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Hayward P, Kalmar T, Arias AM. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 20.Lin CL, Wang FS, Hsu YC, Chen CN, Tseng MJ, Saleem MA, Chang PJ, Wang JY. Modulation of notch-1 signaling alleviates vascular endothelial growth factor-mediated diabetic nephropathy. Diabetes. 2010;59:1915–1925. doi: 10.2337/db09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonegio R, Susztak K. Notch signaling in diabetic nephropathy. Exp Cell Res. 2012;318:986–992. doi: 10.1016/j.yexcr.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 25.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80:719–730. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao F, Yao M, Shi Y, Hao J, Ren Y, Liu Q, Wang X, Duan H. Notch pathway is involved in high glucose-induced apoptosis in podocytes via Bcl-2 and p53 pathways. J Cell Biochem. 2013;114:1029–1038. doi: 10.1002/jcb.24442. [DOI] [PubMed] [Google Scholar]
- 33.Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, Xing CY, Ni L, Mathieson PW, Mundel P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 34.Morizane R, Fujii S, Monkawa T, Hiratsuka K, Yamaguchi S, Homma K, Itoh H. miR-34c attenuates epithelial-mesenchymal transition and kidney fibrosis with ureteral obstruction. Sci Rep. 2014;4:4578. doi: 10.1038/srep04578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niranjan T, Bielesz B, Gruenwald A, Ponda MP, Kopp JB, Thomas DB, Susztak K. The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med. 2008;14:290–298. doi: 10.1038/nm1731. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Gao C, Chen G, Li X, Li J, Wan Q, Xu Y. Notch Signaling Molecules Activate TGF-beta in Rat Mesangial Cells under High Glucose Conditions. J Diabetes Res. 2013;2013:979702. doi: 10.1155/2013/979702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, Lim S, Park M, Choi J, Kim J, Han H, Yoon K, Kim K, Lim J, Park S. Ubiquitination-dependent CARM1 degradation facilitates Notch1-mediated podocyte apoptosis in diabetic nephropathy. Cell Signal. 2014;26:1774–1782. doi: 10.1016/j.cellsig.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catuogno S, Cerchia L, Romano G, Pognonec P, Condorelli G, de Franciscis V. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene. 2013;32:341–351. doi: 10.1038/onc.2012.51. [DOI] [PubMed] [Google Scholar]
- 40.Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One. 2012;7:e33861. doi: 10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]