Abstract
MicroRNA plays an important role in tumor proliferation and cell cycle. In this study, we suggested the level of miR-302a was increasing in the human ovarian cancer cells compared to the normal cells. We aimed to explore the role of miR-302a downregulation in human ovarian cancer cells. Functional studies demonstrate over expression of miR-302a could significant suppress ovarian cancer cells proliferation and promote the cell cycle progress. In vitro reporter assay suggested SDC1 is a direct target gene of miR-302a. Furthermore, the expressions of miR-302a in ovarian cancer cells were inversely corrected with that of SDC1. Upregulation of SDC1 could rescue the effect of over expressed miR-302a in the ovarian cancer cells. These findings provide evidence that miR-302a plays a key role in inhibition of the ovarian cancer cells proliferation, and enhancing the cells’ apoptosis through targeting SDC1, and strongly suggest that exogenous miR-302a may have therapeutic value in treating ovarian cancer.
Keywords: miRNA, miR-302a, SDC1, ovarian cancer
Introduction
Ovarian cancer (OC) is the most leading cause of gynecologic cancer-related deaths in Europe and North America, and the burden of this devastating cancer is expected to increase further in the coming years. Due to the difficulty of effectively diagnosing OC at its early stage, and optimal debulking with platinum- based chemotherapy remains the cornerstone of management. Unfortunately, despite frequent initial responses to chemotherapy, these tumors almost invariably relapse. Therefore, elucidating the molecular mechanisms involved in OC is essential for developing cancer prevention strategies and possible guiding disease management in the clinic [1].
In this several years, a class of novel non-coding RNAs called MicroRNAs (MiRNAs) had been discovered in plants and animals. MicroRNAs (miRNAs) include 18-26 nucleotides, which post-transcriptionally regulate gene expression in multicellular organisms by affecting both the stability and translation of mRNAs. In the process of tumor formation, the abnormal expression or the loss of the dynamic balance between oncogenes and tumor suppressor genes, leads tumorigenesis and development of cancer. MiRNA, as an important regulation factor of gene expression, also involved in tumor formation and progression. Considerable evidence showed critical functions for miRNAs in diverse biological processes, such as proliferation [2-10], apoptosis [11-18], angiogenesis [19-25], cell differentiation [26-28], adhesion and metastasis [29] of tumor cells. Therefore, down-regulation of certain miRNA expression may cause a variety of imbalance, including many kinds of cancers. Previous studies have confirmed cancer-specific miRNAs in many types of cancers, involved breast cancer [30], lung cancer [31], hepatocellular carcinoma [32] and so on.
The miR-302-367 cluster consists of four highly-homologous miRNA members, which are transcribed together as a noncoding RNA cluster containing mir-302b, mir-302c, mir-302a, mir-302d, and mir-367 in a 5’-to-3’ direction [33]. Most studies of this cluster have focused on the maintenance of stemness [34-38] and the ability of the cluster to reprogram somatic cells into induced pluripotent stem cells (iPSCs) [39-42]. To date, miR-302s have been proven to post-transcriptionally regulate CCND1 and CDK4, therefore affecting cell cycle progression. Other studies have demonstrated the tumor suppressive activity of miR-302 in human pluripotent stem cell by both the CCNE-CDK2 and CCND-CDK4/6 pathways in G1-S cell cycle transition [43]. In contrast, a recent study demonstrated that the miR-302-367 cluster compromised the maintenance of glioma-initiating cells (GiCs), strongly inhibited the clonogenicity of GiCs and promoted the loss of stem-like proteins, including OCT4 and NANOG, as well as the down-regulation of SOX1 and SHH [44,45]. Consistent with these findings, another report demonstrated the importance of the miR-302-367 cluster in cell differentiation by showing that the cluster controls mes-endodermal fate specification [46]. The subclass comprised of miR-302a-d (miR-302s) has also been shown to inhibit tumorigenicity and induce apoptosis in various tumors and cancer cells, including MCF7 breast cancer cells, HepG2 hepatocellular carcinoma cells, and Tera-2 embryonal teratocarcinoma cells [43]. So, the relationship between miR-320a and cancers needs to be investigated further.
In this research, we found that miR-302a was frequently down-regulated ovarian cancer cells. Next, we analyzed the miR-302a targets by bioinformatics software, and found that miR-302a can target SDC1. Further, in vitro experiments proved that the re-expression of miR-302a inhibited OC cells proliferation dramatically, and arrested the OC cell cycle at the G1/S phase. The dual-luciferase reporter assays further demonstrated that SDC1 was a novel target of miR-302a. The over expression of SDC1 led to rescue the effect of apoptosis related to miR-302a, such, strongly suggesting that miR-302a suppresses the growth of OC cells by targeting SDC1.
Materials and methods
Bioinformatics analysis
MicroRNA target prediction was performed using TargetScan v6.2 25, Microcosm v5.0 26, and miRanda 27, followed by expression correlation between miRNA-mRNA pairs. Matinspector (http://www.genomatix.de) and GeneGo (http://www.genego.com/metacore.php) were used for searching the transcription factor binding sites as well as analyzing the miRNA-mRNA network, respectively. MatInspector is a software tool that utilizes a large library of matrix descriptions for transcription factor binding sites (TFBS) to locate matches in DNA sequences. By introducing a matrix family concept, optimized thresholds, and comparative analysis, the program produces concise results that avoid redundant and false-positive matches. It assigns a quality rating to matches and thus allows quality-based filtering and selection of matches. GeneGo’s MetaCore™ is an integrated “knowledge-based” platform for pathway analysis of OMICs data and gene lists. MetaCore™ is based on a proprietary manually crated database of human protein-protein, protein-DNA, and protein compound interactions, as well as metabolic and signaling pathways for human, mouse, and rat, supported by proprietary ontologies and controlled vocabulary.
Cell culture, transfection
OVCAR3 and SKOV3 cells were cultured in RPMI 1640 (GIBCO) and McCoy’ s 5A Media, with 10% heat-inactivated fetal bovine serum, 100 IU penicillin/ml, 0.1 mg streptomycin/ml in a humidified 5% (v/v) atmosphere of CO2 at 37°C. Transfected with Lipofectamine 2000 Reagent (Invitrogen) followed the manufacturer’s protocol.
Fluorescent reporter assay
The Luciferase expression vector pGL3/Luciferase was constructed. The fragment of SDC1 3’UTR wild-type or mutant was cloned into pGL3/Luciferase at the same sites. Cells were transfected with miR-302a or control vector in 48-well plates, and with the reporter vector SDC1-WT or SDC1-MUT. The intensities of luciferase fluorescence were detected with Fluorescence Spectrophotometer F-4500 (HITACHI).
Quantitative RT-PCR
To detect the relative level of transcript, real-time RT-PCR was performed. Briefly, a cDNA library was generated through reverse transcription using M-MLV reverse transcriptase (Promega) with 2 μg of the large RNA extracted from the cells. The cDNA was used for the amplification of SDC1 gene and the β-actin gene was used as an endogenous control for the PCR reaction. PCR was performed under the following conditions: 94°C for 4 min followed by 40 cycles of 94°C for 1 min, 56°C for 1 min, 72°C for 1 min. SYBR Premix Ex Taq™ Kit (TaKaRa) was used following the manufacturer’s instructions, and the real-time PCR was performed and analyzed by 7300 Real-Time PCR system (ABI). All primers were purchased from AuGCT Inc.
Western blotting
Cultured cells were lysed by RIPA (0.1%SDS, 1% Triton X-100, 1 mM MgCl2, 10 mM Tris-HCl (pH 7.4) in 4°C for 25 min. Collected the lysates and cleared by centrifugation, and protein concentration was determined. Total cell lysates (50 μg) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were electroblotted onto nitrocellulose membranes. Nonspecific binding sites of membranes were saturated with 5% skim milk in TBST solution (100 mmol/L Tris-Cl, pH7.5, 150 mmol/L NaCl, 0.1% Tween 20) and incubated for 2 hours with antibodies at room temperature. The following antibody was used: anti-SDC1 and anti-GAPDH. After 4 washes with TBST, the filters were incubated with goat anti-mouse peroxidase-conjugated secondary antibody (Sigma) in 5% skim milk in TBST solution for 1 hour at room temperature; reactions were developed using enhanced chemoluminiscence (Perkin Elmer, USA).
Cell proliferation assay and colony formation assay
SKOV3 and OVCAR3 cells were seeded in 96-well plate at 6,000, 7,000 and 8,000 cells per well the day before transfection. The cells were transfected with miR-302a or control vector 0.2 µg per well. MTT assay was used to measure the viable, proliferating cells at 12 h, 24 h, 48 h after transfection. The absorbance at 570 nm was measured using a μQuant Universal Microplate Spectrophotometer (Bio-tek Instruments). After transfection, SKOV3 and OVCAR3 cells were counted and seeded in 6-well plates (in triplicate) at 50, 60 and 75 cells per well. Fresh culture medium was replaced every 3 days. Colonies were counted only if they contained more than 50 cells, and the number of colonies was counted from the 6th day after seeding and then the cells were stained using crystal violet. The rate of colony formation was calculated with the equation: colony formation rate = (number of colonies/number of seeded cells) ×100%.
Flow cytometry analysis
After 48 hours transfection as earlier described, the cells were harvested and washed twice with PBS. Washed cells were resuspended in 0.6 mL PBS, and fixed by the addition of 1.4 mL 100% ethanol at 4°C overnight. The fixed cells were rinsed twice with PBS, and resuspended in propidium iodine (PI) solution, including 50 mg/mL PI and 50 mg/mL RNaseA (Sigma) in PBS without calcium and magnesium, and incubated at 37°C for 30 minutes in the dark. Stained cells were passed through a nylonmesh sieve to remove cell clumps and analyzed by a FACScan flow cytometer and Cell Quest analysis software (Becton Dickinson, San Jose, CA, USA). Flow cytometry analysis was repeated 3 times.
Statistical analysis
Data are expressed as means ± standard deviation (SD), and P < 0.05 is considered as statistically significant by Students-Newman-Keuls test.
Results
MiR-302a expression level in human ovarian cancer cells and their correlation analysis with clinicopathological characteristics
We use quantitative Real-time PCR to detect miR-302a differential expression level in 4 kinds of human ovarian cancer cells, C13K, 3AO, SKOV3 and OVCAR3. The results showed that, the expression level of miR-302a in human ovarian cancer cells was significantly lower than the normal cells (Figure 1A). Staging of ovarian cancer is based on clinical and radiologic examination. Ovarian cancer at its early stages (I/II) is difficult to diagnose until it spreads and advances to later stages (III/IV) s because most symptoms are nonspecific and thus of little use in diagnosis. Most patients present with Stage III or IV disease. Stage I is a small tumor completely confined to ovary. Stage II is a tumor has pelvic extension (must be confined to the pelvis) or primary peritoneal tumor, involves one or both ovaries. Stage III is cancer found outside the pelvis or in the retroperitoneal lymph nodes, involves one or both ovaries. Stage IV is any distant metastasis (i.e. outside of the peritoneum). The expression level of miR-302a was associated with TNM stage. MiR-302a has lower expression level in the stage III/IV than stage I/II (Figure 1C). These data suggested that alterations of miR-302a could be involved in ovarian cancer progression.
Figure 1.
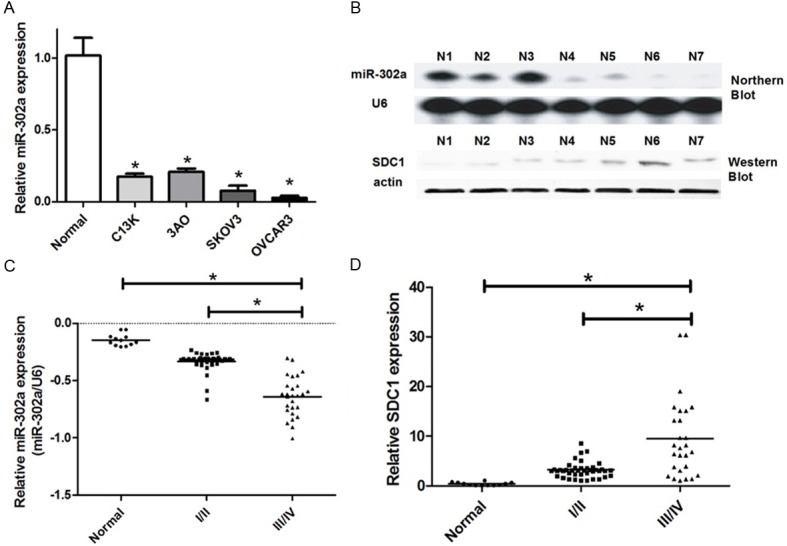
Identification of differential expression of miR-302a in human ovarian cancer. A. We use quantitative Real-time PCR to detect miR-302a differential expression level in human ovarian cancer cells (C13K, 3AO, SKOV3 and OVCAR3) and normal cells. U6 snRNA was regarded as an endogenous normalizer and the relative miR-302a expression level of the 4 kinds of human ovarian cancer cells (means ± SD) is shown (*P < 0.05). B. We use Northern blot and Western blot to detect miR-302a and SDC1 differential expression level in 7 human ovarian cancer tissue samples (*P < 0.05). C. We use quantitative Real-time PCR to detect miR-302a differential expression level in human ovarian cancer stage III/IV tissue samples and the stage I/II ones (*P < 0.05). D. We use Western blot to detect SDC1 differential expression level in human ovarian cancer stage III/IV tissue samples and the stage I/II ones (*P < 0.05).
Overexpression of miR-302a suppresses ovarian cancer cells proliferation in vitro
In order to study the effects of miR-302a on ovarian cancer cells proliferation, we constructed a overexperssion vector: miR-302a. After transfection of ovarian cancer cells, we test the validity of miR-302a ectopic expression by quantitative Real-time PCR in SKOV3 and OVCAR3, respectively. The results revealed that miR-302a expression level was significantly higher than the control group (Figure 2A). To test the effects of miR-302a on ovarian cancer cells proliferation, we investigated cell growth by colony formation assay and MTT assay. The colony formation rate of SKOV3 and OVCAR3 cells transfected with miR-302a were significantly lower than the control group (Figure 2B, 2C). We performed MTT assay to further confirm the effects of miR-302a on cell proliferation. We found that miR-302a could obviously suppressed SKOV3 and OVCAR3 cells growth (Figure 2D). These two experiments showed that miR-302a played a role in suppressing cell growth and proliferation in ovarian cancer cells. Up-regulating the miR-302a, cell viability and proliferation were significantly inhibited.
Figure 2.
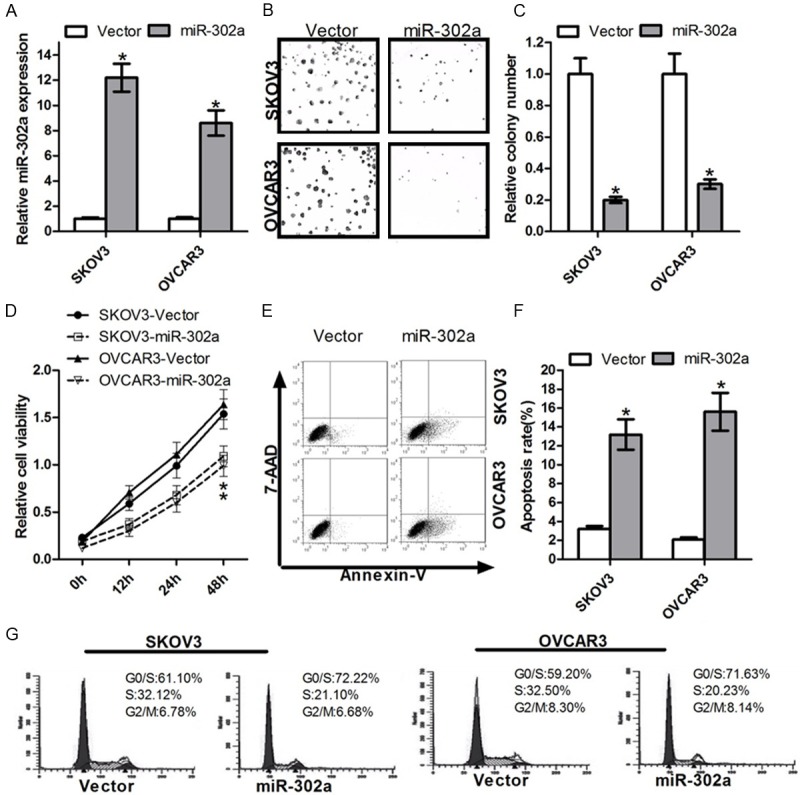
Overexpression of miR-302a enhances ovarian cancer cells proliferation and promotes the cell cycle. A. The relative level of miR-302a expressed in OC cells after the transfection with miR-302a or control vector. B. The cell independent growth activity in vitro was assessed by colony formation assay. OC cells were transfected with miR-302a or control vector. The colony formation assay was shown. C. OC cells were transfected with miR-302a or control vector, and then seeded in 6-well plates. Colonies were counted only if they contained more than 50 cells, and the number of colonies was counted from the 6th day after seeding. The number of colonies was counted from the 6th day after seeding. The colony formation rate was calculated and was shown (*P < 0.05). D. OC cells were transfected with miR-302a or control vector. Cell growth activity was determined at 12 h, 24 h and 48 h post-transfection by MTT assay. Values are means ± SD of three duplications and the relative cell growth activity is shown (*P < 0.05). E. The effect of miR-302a on apoptosis was examined by FCM analysis. OC cells were transfected with miR-302a mimics or control, and then the medium was replaced with serum-free DMEM for 48h. F. OC cells after transfected were analyzed for apoptotic rate after staining with Annexin V-FITC and PI. Data represent means ± S.D. from four independent experiments (*P < 0.05). G. The histogram showed the percentages of OC cells after miR-302a transfection in G1/S, S and G2/M phases (n=3, mean ± SD).
MiR-302a mediates cell cycle arrest and potentiates apoptosis in human ovarian cancer cells
To validate whether miR-302a is able to influence apoptosis, Flow cytometry assay was performed (Figure 2E). The results indicated that the significant increase in the apoptosis was observed in the SKOV3 and OVCAR3 cells transfected with miR-302a (Figure 2F). These results strongly suggested that introduction of miR-302a could inhibit human ovarian cancer cells growth by promoting apoptosis of cancer cells. To confirm that the expression of miR-302a can cause G1/S arrest, SKOV3 and OVCAR3 cells transfected with miR-302a mimics were synchronized at the G1/S transition by serum starvation and hydroxyurea (HU). DNA content was examined from the time of HU release. The results showed that all cells transfected with miR-302a mimics began to arrest at G1 phase and inhibited the transfection from G1 phase to S phase (Figure 2G).
MiR-302a directly inhibits expression of SDC1 via its 3’UTR
We used bioinformatics methods to predict miR-302a potential target genes. The 3’UTR region of SDC1 mRNA contains miR-302a complementary binding sites (Figure 3A). Luciferase reporter assay has been widely used in verification of miRNA target genes [47,48]. To investigate whether SDC1 can be directly targeted by miR-302a, we performed luciferase reportor assay, engineering luciferase reporters, that have either the wild-type 3’UTR of SDC1, or the mutant UTR with a 4 base pair for site-directed mutagenesis in the complementary seed sequence (Figure 3A). First, OVCAR3 cells were transfected with SDC1-wt, miR-302a and control mimics. The results showed that, compared with the control group, co-transfected with miR-302a, the fluorescent EGFP expression were significantly lower (Figure 3B), indicating that overexpression of miR-302a enhanced miR-302a binding to its target gene SDC1 mRNA 3’UTR, so that luciferase activities were decreased. In contrast, mutant reporters were not repressed by miR-302a (Figure 3B). After a 4-base pair mutant of miR-302a, the luciferase activities were not decreased as well (Figure 3C). These all results suggested that, miR-302a could combine with the specific SDC1 mRNA 3’UTR binding sites and play a role in inhibiting the expression of SDC1 gene.
Figure 3.
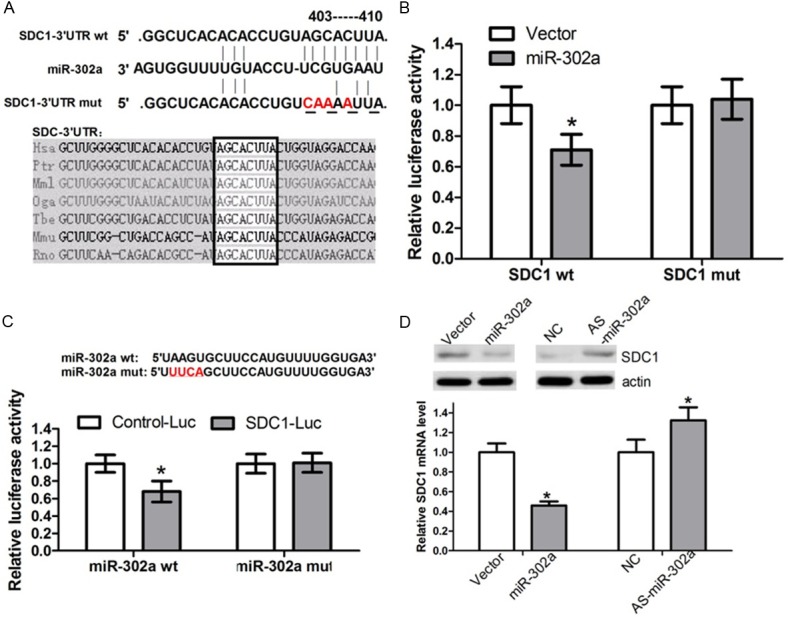
SDC1 is a directly target gene of miR-302a. A. The predicted binding sites of miR-302a on SDC1 mRNA is shown. The mutant UTR with a 4 base pair for site-directed mutagenesis in the complementary seed sequences. B. OC cells were transfected with the wild type of SDC1 reporter vector as well as miR-302a or control vector. MiR-302a suppressed the EGFP fluorescence intensity of SDC1-wt (*P < 0.05), the group transfected with SDC1-mut was not significant different to the group. C. OC cells were transfected with the mutant miR-302a or miR-302a. MiR-302a-mut could not able to significantly suppress the EGFP fluorescence intensity of SDC1-Luc (*P < 0.05). D. OC cells were transfected with miR-302a and control vector, the expression of SDC1 mRNA and protein expression level were measured by quantitative RT-PCR and Western blot. β-actin mRNA was regarded as an endogenous normalizer and the relative SDC1 mRNA expression level is shown (*P < 0.05). GAPDH protein was regarded as endogenous normalizer and the relative SDC1 protein quantity is shown (*P < 0.05).
MiR-302a plays a negative regulatory role at SDC1 post-transcriptional level
MiRNAs regulate the target genes at the post-transcriptional level by binding their target genes 3’UTR to silence the gene function. We transfected OVCAR3 cells with miR-302a, in order to examine whether miR-302a depress endogenous SDC1 through translational repression, the expression of SDC1 protein was examined by Western blot. The results showed that overexpression of miR-302a made the expression level of SDC1 protein decreased (Figure 3D), suggesting that miR-302a negatively regulates endogenous SDC1 protein expression through translational repression mechanism. Meanwhile, high expression level of miR-302a in OVCAR3 cells could also decrease the endogenous SDC1 mRNA level (Figure 4C). Furthermore, in the 7 pairs of human ovarian cancer tissues, we found the expression level of SDC1 had a negative correlation with miR-302a expression (Figure 1B). All these data suggest that miR-302a negatively regulates the expression of SDC1 through mRNA cleavage mechanism at the post-transcriptional level.
Figure 4.
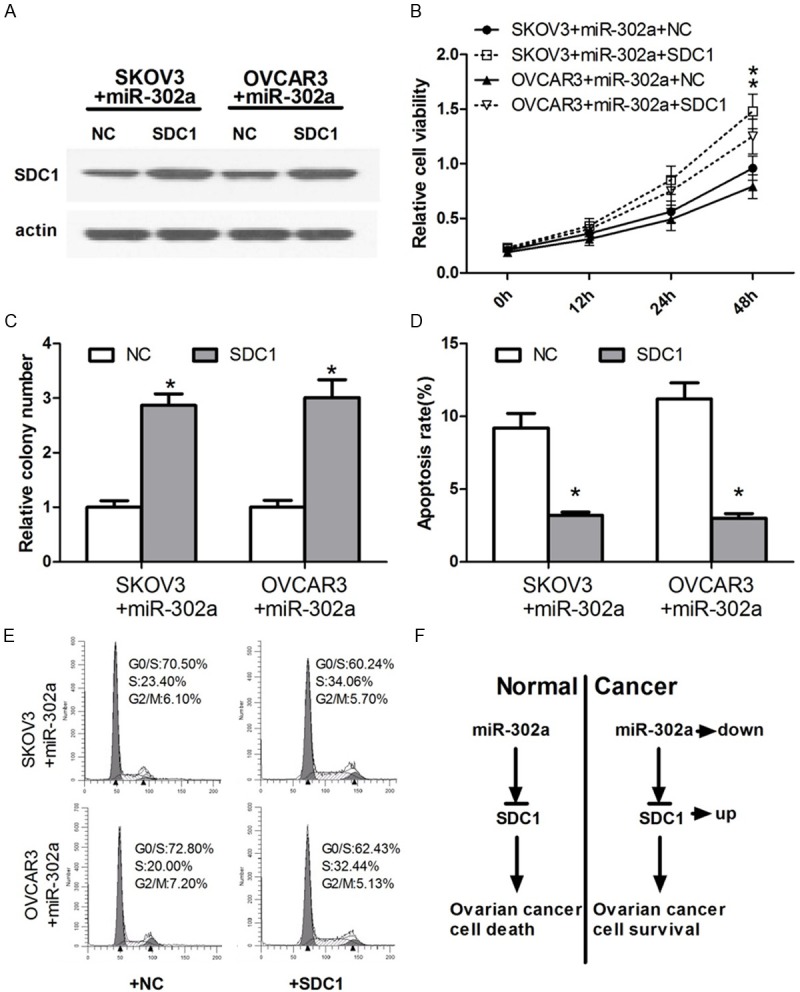
Overexpression of SDC1 could rescue the effects which miR-302a upregulated and led to cell survival in human ovarian cancer. A. We use Western blot to detect the SDC1 differential expression level after co-transfection with miR-302a and SDC1 in human ovarian cancer cells. B. OC cells were co-transfected with miR-302a and SDC1. Cell growth activity was determined at 12 h, 24 h and 48 h post-transfection by MTT assay. Values are means ± SD of three duplications and the relative cell growth activity is shown (*P < 0.05). C. OC cells were co-transfected with miR-302a and SDC1, and then seeded in 6-well plates. Colonies were counted only if they contained more than 50 cells, and the number of colonies was counted from the 6th day after seeding. The number of colonies was counted from the 6th day after seeding. The colony formation rate was calculated and was shown (*P < 0.05). D. The effect of co-transfected with miR-302a and SDC1 on apoptosis was examined by FCM analysis. OC cells after transfected were analyzed for apoptotic rate after staining with Annexin V-FITC and PI. Data represent means ± S.D. from four independent experiments (*P < 0.05). E. The histogram showed the percentages of OC cells after co-transfected with miR-302a and SDC1 in G1/S, S and G2/M phases (n=3, mean ± SD). F. Upregulation of SDC1 could block the effects which miR-302a upregulated and led to cell survival in human ovarian cancer.
MiR-302a repressed OC cells proliferation and promoted apoptosis by negatively regulating SDC1 expression
Previous studies show that miR-302a plays an important role in suppressing tumor cell proliferation and promoting the apoptosis. Accordingly, we detected whether SDC1 affects ovarian cancer cell growth. We constructed the SDC1 plasmid to up regulate the expression level of SDC1 and then identified the expression of SDC1 protein by Western blot. While we enhanced the expression level of miR-302a in OC cells, and then transfected with SDC1, Western blot displayed an increase protein level of SDC1 (Figure 4A). The SDC1 plasmid was confirmed effectively increase the SDC1 protein expression level. We further performed a “rescue” study in OC cells, namely, co-transfection of miR-302a mimics and SDC1 partially reverse the growth inhibition effects of OC cells from miR-302a mimics (Figure 4B, 4C). Additionally, the co-transfection with miR-302a and SDC1 showed a similar result in the “rescue” apoptosis assay (Figure 4D, 4E). These data suggests that miR-302a repressed OC cells proliferation and promoted apoptosis in OC cells through targeting SDC1.
Discussion
Transformation process of malignant tumors is regulated by the synergy of multiple genes, including overexpression of oncogenes and low expression or even loss of function of tumor suppressor genes. Recent research uncovered that the regulation of oncogenes and tumor suppressor genes was not only in the transcriptional level, but also in the post-transcriptional level, which was more important and accurate. MicroRNAs (miRNA), as important regulation factors, participate gene expression in human carcinogenesis. In recent years, the research about miRNAs has been more in-depth. MiRNA-mediated post-transcriptional gene silencing (PTGS) and the relevance with tumor formation have become the focus of attention. Tumor cells and normal cells have a significant difference in miRNA expression profiling. Most miRNA genes locate in chromosomal regions frequently display amplification, deletion or translocation in human cancers. Detection of differentially expression of miRNAs in human ovarian cancer, to determine the role of miRNAs in cancer mechanism and function of their target genes, provide a new direction for the diagnosis and treatment of in human ovarian cancer.
In the present study, we tried to identify a novel miRNA which regulates the expression of SDC1, and evaluate its effects on cell phenotype using OC cells. Initially, we used real-time PCR to find that miR-302a was significantly down-regulated in human OC cells, compared with the normal cells. The results suggested that alterations of miR-302a could be involved in ovarian cancer progression. Therefore, we hypothesized that miR-302a was a negative factor of carcinogenesis in human ovarian cancer cells due to the low expression levels in human ovarian cancer. We calculated the cell growth viability through the MTT and colony formation assay to detect the relationship between miR-302a and the growth capacity of ovarian cancer cell line SKOV3 and OVCAR3. The cell growth viability of OC cells transfected with the miR-302a was significantly decreased when compared to control group (Figure 2C, 2D). We further showed that overexpressed miR-302a in OC cells induced G1 arrest, suppressed cell proliferation, and induced apoptosis. These data indicate that miR-302a may act as a tumor suppressor to inhibit cell proliferation by blocking the G1/S transition of OC cells. In other words, reduced miR-302a expression in OC cells and tissues may promote cell proliferation by activating the cell cycle (Figure 2F, 2G).
Secondly, bioinformatics analyses predicted a miR-302a binding site on the SDC1 transcript. Experimental evidence indicated that SDC1 was indeed a target of miR-302a. On one hand, the ability of miR-302a to regulate SDC1 expression was likely direct because it bound the 3’UTR of SDC1 mRNA complementarily to the miR-302a seed region. The EGFP fluorescence intensity of SDC1-wt was specifically responsive to miR-302a overexpression (Figure 3B). Furthermore, mutation of the miR-302a binding site abolished the effect of miR-302a on the regulation of EGFP fluorescence intensity. On the other hand, the endogenous SDC1 protein expression was decreased in OC cells transfected with miR-302a (Figure 3D). These results suggested that miR-302a regulated SDC1 protein expression at the post transcription level.
Syndecan 1 (SDC1) is a member of a transmembrane heparan sulfate proteoglycan family, which expresses in epithelia, and plays a critical role in cellular processes including differentiation, cell adhesion, migration and invasion, and angiogenesis [49-51]. Functions have been ascribed to the extracellular domain that carries glycosaminoglycan (GAG) side chains, to the transmembrane domain and to the cytoplasmic domain that transduces signals from extracellular ligand binding. Initial reports demonstrated that expression of syndecan-1 in vitro was associated with maintenance of epithelial morphology, anchorage-dependent growth, and inhibition of invasiveness [52]. This “tumor suppressor” function was subsequently supported by immunohistochemical studies that correlated loss of syndecan-1 with reduced survival in patients with malignant mesothelioma [53], squamous cell carcinoma of the head and neck [54], and laryngeal cancer [55]. Altered SDC1 expression has been reported gradually in a number of malignant tumor types and has been associated with differentiation stage and grade [56-58]. Marzioni D et al [59], have reported a positive correlation of SDC-1 with fibroblast growth factors (FGFRs) in bladder tumors, these factors are thought to be key molecules in low-grade BCa. Shimada et al [60] have investigated the biologic role of SDC-1 in human BCa cells. In their study, the BCa cell lines, UMUC2 and UMUC3 had SDC-1 expression silenced by siRNA transfection, which led to an induction of apoptosis in vitro and a reduction in mouse orthotopic bladder tumor growth. In addition, studies in breast and gastric cancer have demonstrated an association between increased stromal SDC1 expression, loss of cancer cell SDC1 expression, and an adverse clinical outcome [61-63]. But the association between SDC1 status and human ovarian cancer has not been extensively studied. To our knowledge, our study is the latest study to date to evaluate the effect between miRNA and SDC1 in human ovarian cancer cells. A high expression level of SDC1 could repress OC cells proliferation and promoted apoptosis in OC cells.
In summary, we demonstrated that miR-302a played an important role in the regulation of SDC1 gene expression. The effect of miRNAs on OC cells expression occurred at both mRNA and transcription levels, and at least in part through targeting SDC1. However, we emphasize that miR-302a may be capable of controlling tumor-specific gene(s), consequently favoring cell apoptosis. Therefore, our study suggests that unregulation of miR-302a may provide a better strategy to block tumor proliferation and cell cycle.
Disclosure of conflict of interest
None.
References
- 1.Bell D, Berchuck A, Birrer M. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 4.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Johnson SM, Grosshans H, Shingara J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 9.Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, Yang KD, Cheng JT, Huang CC, Liu RT. miR-146b is Highly Expressed in Adult Papillary Thyroid Carcinomas with High Risk Features Including Extrathyroidal Invasion and the BRAF(V600E) Mutation. Thyroid. 2010;20:489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 10.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 11.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 12.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 13.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 14.Li llL, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 15.Cimmino A, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 17.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, Osada H, Takahashi T. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 19.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 22.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 25.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 26.Na Xu, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 27.Di Leva G, Calin GA, Croce CM. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res C Embryo Today. 2006;78:180–189. doi: 10.1002/bdrc.20073. [DOI] [PubMed] [Google Scholar]
- 28.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 29.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 31.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 33.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 36.Barroso-del Jesus A, Lucena-Aguilar G, Sanchez L, Ligero G, Gutierrez-Aranda I, Menendez P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011;25:1497–1508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Deng S, Zhao Z, Zhang H, Xiao J, Song W, Gao F, Guan Y. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol Biol Rep. 2011;38:2155–2160. doi: 10.1007/s11033-010-0343-4. [DOI] [PubMed] [Google Scholar]
- 38.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluri- potent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo CH, Deng JH, Deng Q, Ying SY. A novel role of miR-302/367 in reprogramming. Biochem Biophys Res Commun. 2012;417:11–16. doi: 10.1016/j.bbrc.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 42.Lipchina I, Studer L, Betel D. The expanding role of miR-302-367 in pluripotency and reprogramming. Cell Cycle. 2012;11:1517–1523. doi: 10.4161/cc.19846. [DOI] [PubMed] [Google Scholar]
- 43.Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70:9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- 44.Akhavantabasi S, Sapmaz A, Tuna S, Erson-Bensan AE. miR-125b targets ARID3B in breast cancer cells. Cell Struct Funct. 2012;37:27–38. doi: 10.1247/csf.11025. [DOI] [PubMed] [Google Scholar]
- 45.Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, Virolle T. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 49.Couchman JR, Pataki CA. An introduction to proteoglycans and their localization. J Histochem Cytochem. 2012;60:885–897. doi: 10.1369/0022155412464638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McQuade KJ, Rapraeger AC. Syndecan-1 transmembrane and extracellular domains have unique and distinct roles in cell spreading. J Biol Chem. 2003;278:46607–46615. doi: 10.1074/jbc.M304775200. [DOI] [PubMed] [Google Scholar]
- 51.Teng YH, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012;31:3–16. doi: 10.1016/j.matbio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inki P, Jalkanen M. The role of syndecan-1 in malignancies. Ann Med. 1996;28:63–67. doi: 10.3109/07853899608999076. [DOI] [PubMed] [Google Scholar]
- 53.Kumar-Singh S, Jacobs W, Dhaene K, Weyn B, Bogers J, Weyler J, Van Marck E. Syndecan-1 expression in malignant mesothelioma: correlation with cell differentiation, WT1 expression, and clinical outcome. J Pathol. 1998;186:300–305. doi: 10.1002/(SICI)1096-9896(1998110)186:3<300::AID-PATH180>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 54.Anttonen A, Kajanti M, Heikkila P, Jalkanen M, Joensuu H. Syndecan-1 expression has prognostic significance in head and neck carcinoma. Br J Cancer. 1999;79:558–564. doi: 10.1038/sj.bjc.6690088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pulkkinen JO, Penttinen M, Jalkanen M, Klemi P, Grenman R. Syndecan-1: a new prognostic marker in laryngeal cancer. Acta Otolaryngol. 1997;117:312–315. doi: 10.3109/00016489709117794. [DOI] [PubMed] [Google Scholar]
- 56.Conejo JR, Kleeff J, Koliopanos A, Matsuda K, Zhu ZW, Goecke H, Bicheng N, Zimmermann A, Korc M, Friess H, Büchler MW. Syndecan-1 expression is up-regulated in pancreatic but not in other gastrointestinal cancers. Int J Cancer. 2000;88:12–20. doi: 10.1002/1097-0215(20001001)88:1<12::aid-ijc3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 57.Contreras HR, Ledezma RA, Vergara J, Cifuentes F, Barra C, Cabello P, Gallegos I, Morales B, Huidobro C, Castellón EA. The expression of syndecan-1 and -2 is associated with Gleason score and epithelial-mesenchymal transition markers, E-cadherin and beta-catenin, in prostate cancer. Urol Oncol. 2010;28:534–540. doi: 10.1016/j.urolonc.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 58.Shah L, Walter KL, Borczuk AC, Kawut SM, Sonett JR, Gorenstein LA, Ginsburg ME, Steinglass KM, Powell CA. Expression of syndecan-1 and expression of epidermal growth factor receptor are associated with survival in patients with nonsmall cell lung carcinoma. Cancer. 2004;101:1632–1638. doi: 10.1002/cncr.20542. [DOI] [PubMed] [Google Scholar]
- 59.Marzioni D, Lorenzi T, Mazzucchelli R, Capparuccia L, Morroni M, Fiorini R, Bracalenti C, Catalano A, David G, Castellucci M, Muzzonigro G, Montironi R. Expression of basic fibroblast growth factor, its receptors and syndecans in bladder cancer. Int J Immunopathol Pharmacol. 2009;22:627–638. doi: 10.1177/039463200902200308. [DOI] [PubMed] [Google Scholar]
- 60.Shimada K, Nakamura M, De Velasco MA, Tanaka M, Ouji Y, Miyake M, Fujimoto K, Hirao K, Konishi N. Role of syndecan-1 (CD138) in cell survival of human urothelial carcinoma. Cancer Sci. 2010;101:155–160. doi: 10.1111/j.1349-7006.2009.01379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanley MJ, Stanley MW, Sanderson RD, Zera R. Syndecan-1 expression is induced in the stroma of infiltrating breast carcinoma. Am J Clin Pathol. 1999;112:377–383. doi: 10.1093/ajcp/112.3.377. [DOI] [PubMed] [Google Scholar]
- 62.Barbareschi M, Maisonneuve P, Aldovini D, Cangi MG, Pecciarini L, Angelo Mauri F, Veronese S, Caffo O, Lucenti A, Palma PD, Galligioni E, Doglioni C. High syndecan-1 expression in breast carcinoma is related to an aggressive phenotype and to poorer prognosis. Cancer (Phila) 2003;98:474–483. doi: 10.1002/cncr.11515. [DOI] [PubMed] [Google Scholar]
- 63.Wiksten JP, Lundin J, Nordling S, Lundin M, Kokkola A, von Boguslawski K, Haglund C. Epithelial and stromal syndecan-1 expression as predictor of outcome in patients with gastric cancer. Int J Cancer. 2001;95:1–6. doi: 10.1002/1097-0215(20010120)95:1<1::aid-ijc1000>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]


