Abstract
Previous studies have revealed several targets of miR-10b, such as syndecan-1, HOXD10, TBX5, and E-cadherin. In this study, we aimed to assess whether Krüppel-like factor 4 (KLF4) is a target gene of miR-10b in gastric cancer (GC). Targeting of KLF4 by miR-10b was confirmed by dual-luciferase reporter assays. The expression levels of miR-10b and KLF4 mRNA in 5 different gastric cancer cell lines and 65 pairs of gastric cancer tissues were detected by Real-time PCR. In addition, KLF4 protein in gastric cancer cell lines and 30 GC tissues was measured by western blotting and immunochemistry, respectively. KLF4 is a direct target gene of miR-10b in GC, and its expression is reduced by miR-10b at both mRNA and protein levels. In addition, the expression level of miR-10b was tendentiously upregulated in GC tissues while the expression levels of KLF4 mRNA and protein were decreased in gastric cancer tissues compared with normal adjacent tissue. There was a dramatically inverse correlation between the expression levels of miR-10b and KLF4 mRNA in GC (r = -0.339, P = 0.006). These findings indicate that miR-10b was upregulated in GC and may have a key role in GC pathogenesis and development through the downregulation of its target gene KLF4.
Keywords: miR-10b, gastric cancer, KLF4, target
Introduction
Gastric cancer (GC) is the second most frequent type of cancer and the third leading cause of cancer mortality following lung cancer and hepatic carcinoma in China [1]. Despite the great effort in clarifying GC occurrence and progress has been made in the several past decades, the molecular mechanism underlining GC carcinogenesis remains largely unclear. MicroRNAs (miRNAs), a class of approximately 21 to 25 nucleotide endogenous noncoding RNAs, that regulate gene expression at the post-transcriptional level by direct cleavage of the mRNA or by inhibition of protein synthesis of target genes [2,3], have been increasingly considered to play important roles in the carcinogenesis and development in various cancers [3]. Although the biological functions of most miRNAs are not yet fully elucidated, they may serve as the key regulators in the various biological processes [3-5], such as embryonic development, cellular differentiation, proliferation, apoptosis, gene regulation, as well as cancer development.
MicroRNA-10b (miR-10b), located at chromosome 2q31.1 within the HOXD gene cluster, was initially identified as a metastasis-associated miRNA in breast cancer [6,7]. Previous studies have clarified that miR-10b was upregulated in various tumors and played important biological roles in cancer occurrence and progress [8-13]. Ibrahim et al. [14] showed that miR-10b targets the syndecan-1 and promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Sun et al. [15] displayed that miR-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. In a recent study, Tian et al. [13] identified that miRNA-10b promotes migration and invasion through target gene KLF4 in human esophageal cancer cell lines. However, it remains unknown whether KLF4 is a direct and functional target of miR-10b in GC. Therefore, the purpose of this study was to determine whether KLF4 is a direct target of miR-10b in GC cells, and in which manner miR-10b regulate KLF4 expression, at the mRNA level or/and protein level? In addition, the expression pattern of miR-10b and KLF4 in GC tissues was also observed in this study.
Materials and methods
Tissue specimens
Consecutive tissue samples from 65 patients with primary gastric carcinoma were used for real-time quantitative PCR of miR-10b and KLF4 mRNA. They were obtained from Huzhou Tumor Biobank, Huzhou Centre Hospital, Huzhou, China, between 2012 and 2014. Each surgical specimen of paired GC tumor tissues and normal adjacent tissues (NATs) was instantly frozen in liquid nitrogen, and stored at -80°C until RNA extraction. For the immunohistochemical detection of KLF4, another 30 patients with primary gastric carcinoma were enrolled. Surgical specimens of each individual were fixed in 10% buffered formalin solution and embedded in paraffin wax. This study was approved by the Ethics Committee of Huzhou Centre Hospital, and all participants gave informed consent.
Cell Lines and cell cultures
Human gastric cancer cell lines MKN-45, BGC-823, SGC-7901, MGC-803, and AGS were purchased from Cell Bank of Chinese Academy of Medical Sciences (Beijing, China). All cell lines were maintained in RIPM-1640 medium supplemented with 10% fetal bovine serum (FBS, HyClone, USA) at 37°C in a humidified atmosphere of 5% CO2 (Thermo Electron Corp, USA).
Stable miRNA expression cell lines
Lentiviruses containing GFP-miR-10b mimics (miR-10b mimic) or GFP negative control (Scramble) miRNA vector were purchased from Genepharma. AGS cells were pre-seeded in a 6-well plate overnight and infected with 200 μl of virus. 24 hours after addition of viruses, infected cells were selected by adding 100 mg/ml puromycin to growth medium for 4-5 passages. Stable cell lines were verified by qRT-PCR and fluorescence microscope.
Dual-luciferase reporter assays
A wild type (3’-UTR wt) or mutant 3’-UTR (3’-UTR mut) of KLF4 were constructed using PCR, and were then inserted into the multiple cloning sites in the pRL-TK Vector (promega). In brief, the full length of KLF4-3’-UTR wt was amplified by primer F1 and R2, and the KLF4-3’-UTR mut was amplified by overlap PCR. Firstly, sequence A and sequence B were amplified by primer F1, R1 and F2, R2, respectively. Then sequence A and B was diluted by ratio 1:1 as templates and amplified by primers F1 and R2. For the luciferase assay, human 293T cells were cultured to 70-80% confluence in a six-well plate. The cells were subsequently cotransfected with pRL-TK-KLF4 3’-UTR wt or pRL-TK-KLF4-3’-UTR mut vector in combination with 100 nM miR-10b or 100 mM negative control, respectively, using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA). Following this, the cells were incubated with transfection reagent/DNA complex for 5 h, prior to being refreshed with fresh complete medium. A Dual-Luciferase® Reporter assay system (Promega Corp., Madison, WI, USA) was used to assess the luciferase activities 48 h subsequent to the cotransfection. Renilla luciferase activity was normalized to firefly luciferase activity.
RNA extraction and reverse transcription
Total miRNA was extracted from various cells or tissue specimens using mirVana PARIS Kit (Ambion), and finally eluted with 80 µl pre-heated (95°C) Elution Solution according to the manufacturer’s protocol. For reverse transcription (RT) reactions, stem-loop primers (Table 1) were applied for cDNA synthesis according to the report by Mestdagh et al [16]. The specific reverse transcription primers were listed in Table 1.
Table 1.
Reverse transcription and stem-loop primers for real-time PCR
| Gene name | Reverse transcription primers (5’-3’) | PCR primers (5’-3’) |
|---|---|---|
| miR-10b | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAAA | F: CGTCGTACCCTGTAGAACCGA |
| R: GTGCAGGGTCCGAGGT | ||
| miR-16 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCAA | F: CGCGCTAGCAGCACGTAAAT |
| R: GTGCAGGGTCCGAGGT | ||
| KLF4-wt | F: GCAGCTTCACCTATCCGATCC | |
| R: AGCACGAACTTGCCCATCA | ||
| β-actin | F: AGCGAGCATCCCCCAAAGTT | |
| R: GGGCACGAAGGCTCATCATT | ||
| KLF4-mut | F1: GCTCTAGAATCCCAGACAGTGGATATG | |
| F2: GGTGACTGGAAGTTGTGGATATGTCCCATTAAATTATATCCGTGAGTTGGGG | ||
| R1: CCCCAACTCACGGATATAATTTAATGGGACATATCCACAACTTCCAGTCACC | ||
| R2: TTGCGGCCGCGGTTTATTTAAAACTTAATTCTCACCTTG |
F: forward primer, R: reverse primer.
Quantitative real-time PCR
Quantitative Real-time PCR was conducted by using a standard SYBR Green I PCR kit (TaKaRa) on an ABI 7500 Real-Time PCR System (Applied Biosystems, USA). The PCR reactions were performed at 95°C for 10 s, and subjected to 40 cycles of 95°C for 5 s, and 60°C for 34 s, then followed by 65°C to 95°C, increment 0.5°C for 10 s to yield Melt Curve. For normalization, miR-16 [17] and β-actin were used as internal controls for the target of miR-10b and KLF4, respectively. The relative expression ratio of miR-10b or KLF4 mRNA in each paired tissue sample was calculated using the 2-ΔΔCt method [18], where ΔΔCT = ΔCTGC - ΔCTNATs. The miR-10b and KLF4 mRNA expression level was defined as being up-regulated in tumor tissue with a relative expression ratio > 1, and was defined as downregulated in tumor tissue with a relative expression ratio < 1. Quantitative real-time PCRs were performed in duplicates and the mean was calculated. Water was used as negative and quality controls, and each sample was measured in triplicate.
Western blotting analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with 1% phenylmethanesulfonyl fluoride (PMSF). A total of 50 μg protein, analyzed by a bicinchoninic acid (BCA) assay (Biyotime, China), was loaded onto an SDS-PAGE mini-gel and transferred onto polyvinylidene difluoride (PVDF) membrane. The blotting membrane was probed with diluted rabbit polyclonal KLF4 antibody (1:200, Santa Cruz) at 4°C overnight and subsequently incubated with horseradish peroxidase (HRP)-conjugated secondary goat anti-rabbit secondary antibody (1:5000, Bioworld, USA). At last, signals were visualized using ECL plus chemiluminescence substrate (Biyotime, China). β-actin (1:2000, Bioworld) was used as control.
Immunohistochemistry
Tumor specimens were fixed, embedded and sectioned at a thickness of 4 μm and stained with diluted rabbit polyclonal KLF4 antibody (1:200, Santa Cruz) using the streptavidin-peroxidase (HRP) conjugate method in accordance with the manufacturer’s manual. KLF4 staining was evaluated by two independent observers using light microscopy in a blinded fashion. KLF4 expression was evaluated semi-quantitatively [19], considering staining intensity (0 = absent; 1 = low; 2 = similar to normal epithelium; 3 = higher than normal) and percentage of positively stained cells (1 = immunostaining in ≤ 10% of cells; 2 = 11-30%; 3 = 31-60%; 4 = 61-100%). The final score was calculated by adding the intensity and percentage scores. Scores 1-4 represented absent/weak or under-expression and scores 5-7 represented no change or higher expression compared to adjacent normal.
Statistical analysis
All data were analyzed by SPSS software version 13.0 (SPCC Inc., Chicago, IL, USA). Differences between samples were analyzed using Student’s t-test. The association between expression levels of KLF4 mRNA and miR-10b was analyzed by Spearman correlation coefficient. The significance level was set to P < 0.05. All P values were two sided.
Results
MiR-10b targets KFL4 3’-UTR directly
To determined whether KFL4 is a target gene of miR-10b in GC, we generated a luciferase reporter vector that contain the KLF4 3’-UTR (3’-UTR wt) and a mutant reporter vector (3’-UTR mut) which contains the KLF4 3’-UTR with a mutation at the putative miR-10b binding site (Figure 1A). As shown in Figure 1B, we observed a marked reduction in luciferase activity in cells transfected with pre-miR-10b compared with premiR-nc transfected cells (P < 0.01). In contrast, no change of luciferase was observed in cells transfected with the mutant 3’-UTR constructs.
Figure 1.
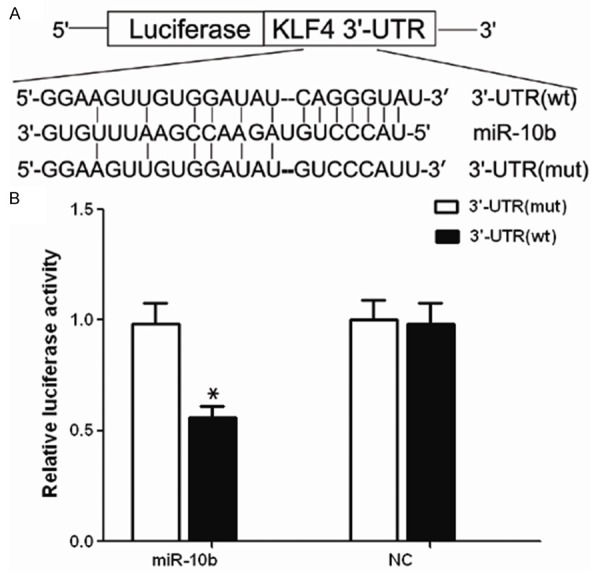
KLF4 is a target of miR-10b. A. KLF4 3’-UTR and corresponding fragments were inserted into the region immediately downstream of the luciferase gene in pRL-TK Vector and validated by DNA sequencing. The sequences of the predicted miR-10b binding sites within the KLF4 3’-UTR, including wild-type UTR or UTR segments containing mutant binding site are shown. B. Ectopic miR-10b expression inhibits wild-type but not mutant KLF4 3’-UTR reporter activity, *P < 0.001.
To further experimentally validate whether over-expression of miR-10b can lead to down-regulation of KLF4 expression, stable miR-10b overexpression cells were established using Lentivirus system. As a result, miR-10b expression was up-regulated about 125-fold in miR-10b mimic compared with that in scramble (miR-NC). KLF4 mRNA expression were observed significantly reduced in miR-10b mimic about 0.5-fold when compared with that in scramble (Figure 2A). Moreover, Western blotting showed that the enhanced miR-10b expression in miR-10b mimic significantly repressed KLF4 protein expression compared to that in scramble control (Figure 2B). All these findings indicate that KLF4 is a direct target of miR-10b and is likely to be suppressed by miR-10b through translational inhibition and mRNA degradation in AGS cells.
Figure 2.
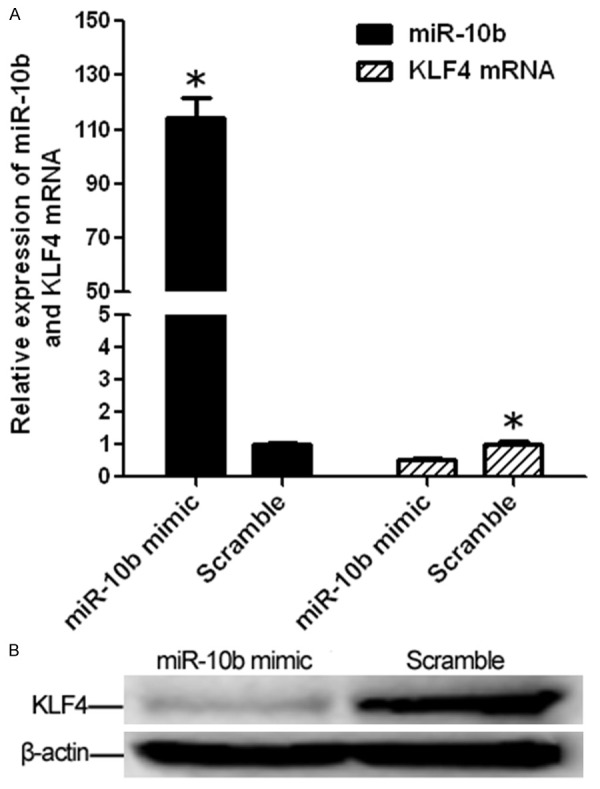
Upregulated miR-10b suppresses KLF4 mRNA and protein expression in AGS cell. A. Expression levels of miR-10b and KLF mRNA in AGS cell after transfected with miR-10b mimic using Lentivirus system were detected by real-time PCR. The miR-10b expression is significantly higher in miR-10b mimic compared with that in Scramble while the KLF4 mRNA expression is significantly decreased in miR-10b mimic compared with that in Scramble (both *P < 0.001). B. KLF4 protein was tested by western blotting assay (*P < 0.05 compared with control).
Inverse expression of miR-10b and KLF4 mRNA in GC cells
The expression levels of miR-10b and KLF4 mRNA in 5 GC cells was detected by real-time PCR method. As showed in Figure 3, the expression levels of miR-10b was substantially up-regulated in MGC-803, AGS, BGC-823, MKN-45, SGC-7901 cells while the expression levels of KLF4 mRNA was inversely down regulated in MGC-803, AGS, BGC-823, MKN-45, SGC-7901 cells one by one. These data further suggested that KLF4 mRNA was regulated by its target miR-10b in GC cell.
Figure 3.
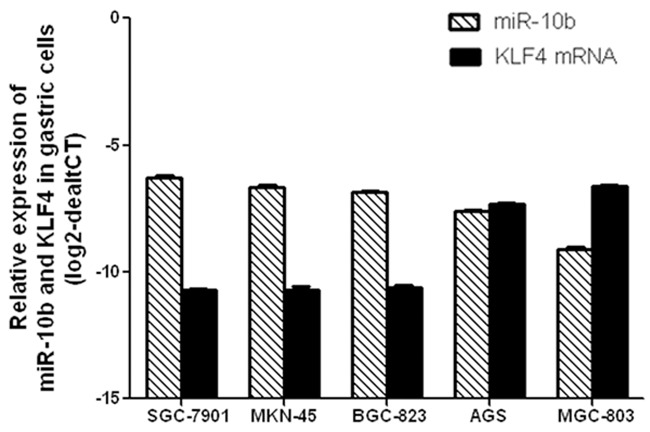
Relative expression of miR-10b and KLF4 mRNA in gastric tumor cells. The expression levels of miR-10b was substantially up-regulated in MGC-803, AGS, BGC-823, MKN-45, SGC-7901 cells while the expression levels of KLF4 mRNA was inversely down regulated in MGC-803, AGS, BGC-823, MKN-45, SGC-7901 cells one by one.
Correlation analysis of miR-10b expression and KLF4 protein expression
To ascertain whether there was any correlation between miR-10b and KLF4 levels in primary human gastric tissue specimens, SYBR Green I real-time PCR was applied to investigate the expression levels of miR-10b and KLF4 mRNA. Additionally, immunohistochemistry was used to assess KLF4 protein expression. As a result, 38 (38/65, 58.46%) cases of GC patients were up-regulated miR-10b expression and 37 cases of GC patients were down-regulated KLF4 mRNA expression (2-ΔΔct > 1) in 65 pairs of matched human gastric specimens, respectively. Using Spearman correlation analysis, miR-10b expression in GC was negative significantly related with KLF4 mRNA expression in GC (Table 2, r = -0.339, P = 0.006).
Table 2.
Correlation analysis of miR-10b expression and KLF4 mRNA expression
| miR-10b | KLF mRNA | r | P | |
|---|---|---|---|---|
|
| ||||
| High (n=28) | Low (n=37) | |||
| Up-regulated (n=38) | 11 | 27 | -0.339 | 0.006 |
| Down-regulated (n=27) | 17 | 10 | ||
In addition, Immunohistochemistry showed that KLF4 protein positive staining was mainly located in the cytoplasm and nuclei with the more prominent loss of nuclear reactivity (Figure 4). Strong staining of KLF4 expression was observed both in normal gastric tissue and adjacent tissue. Loss or weak expression of KLF4 staining was found in most gastric carcinoma or cancer nests tissue. Over all, 17 of 30 cases (56.67%) of GC patients showed KLF4 protein weak or loss expression in our study when compared with that in normal adjacent tissues, showing an approximately accordance expression with up-regulated miR-10b expression(58.46%, 38/65) in GC tissues. These data suggested an inverse correlation between miR-10b and KLF4 expression during the tumorigenesis of GC.
Figure 4.
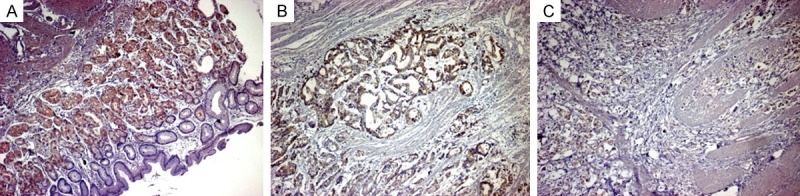
Expression of KLF4 protein in human GC tissues by immunohistochemistry. A. Strong KLF4 expression in distant non-cancerous tissues. B. Strong positive KLF4 expression in gastric cancer. C. Weakly positive KLF4 expression in gastric cancer. Original magnification, × 200.
Discussion
It is well known that an average miRNA may have more than 100 targets [20], and one mRNA might be regulated by a variety of miRNAs [21]. Previous research has identified several targets of miR-10b, such as syndecan-1 [14], HOXD10 [15], TBX5 [22], and KLF4 [13]. In our study, luciferase reporter assays, Lentivirus system, qRT-PCR, western blotting and Immunohistochemistry was used to verify whether KLF4 is a direct target of miR-10b in GC. Our data showed that KLF4 is indeed a direct target of miR-10b in GC, and its expression is regulated by miR-10b at both mRNA and protein levels. In addition, our results found an inverse correlation between the expression levels of miR-10b and KLF4 in GC tissues.
Krüppel-like factor 4 (KLF4; formerly GKLF), a zinc finger protein of the Krüppel-like factor family, has been reported to be a key regulator in cell cycle regulation, differentiation, stem cell properties and malignant transformation [23,24]. KLF4 was initially identified as a tumor suppressor in colon [25], esophageal [26], lung [27], bladder [28], colorectal [29], and gastric cancers [30,31], for its frequent loss expression pattern. Contrarily, other studies have demonstrated that KLF4 expression is increased in breast ductal cell carcinoma [32] and oral squamous cell carcinoma [33] for as yet unknown reasons. However, all these findings suggest that KLF4 has an important function in tumor development and progression. In the current study, we first performed 3’-UTR luciferase assays to confirm that KLF4 is a direct target of miR-10b in GC. Then the KLF4 mRNA and protein level was investigated in various GC cells and in high-stable expressing AGS cell established using a lenti-viral system. Our data showed that an approximately opposite expression alignment of miR-10b and KLF4 mRNA was found in GC cell lines. The levels of miR-10b expression in GC cell lines arranged in a descending order were successively SGC-7901, MKN-45, BGC-823, AGS, and MGC-803. On the contrary, KLF4 mRNA expression levels distributed in an ascending order were subsequently SGC-7901, MKN-45, BGC-823, AGS, and MGC-803. These data suggested that miR-10b may target KLF4 in mRNA regulation in GC cell lines. In addition, overexpression miR-10b in AGS cell line could depress the KLF4 mRNA expression level and KLF4 protein expression level. All these findings suggested that KLF4 is indeed a direct target of miR-10b in GC, and its expression is regulated by miR-10b both at mRNA level and protein levels.
Previous studies have demonstrated that miR-10b may act as Oncogene in many human malignancies, such breast cancer [6], glioblastoma stem cells [8] lung cancer [34], gastric cancer [35,36] and so on. However, there were also other viewpoints that miR-10b served as tumor suppressor in gastric cancer and breast caner [9,37]. In our study, 38 cases of GC patients showed upregurelated miR-10b expression in 65 GC patients, tendentiously suggesting that miR-10b is upregulated in GC. In addition, loss expression of KLF4 was observed in 17 (56.67%) of 30 GC patients, showing an accordant relationship with upregulated miR-10b in GC.
In conclusion, our study demonstrates that KLF4 is indeed a direct target of miR-10b in GC, and its expression is regulated by miR-10b both at mRNA level and protein level. In addition, our results found an inverse correlation between the expression levels of miR-10b and KLF4 in GC tissues. However, further studies enrolling larger samples of GC patients should be performed to confirm our findings, due to the small size of samples in this study. These investigations will provide important information on the potential of miR-10b and KLF4 as promising candidates for the development of effective strategies for the treatment of GC.
Acknowledgements
This work was supported by Natural Science Foundation of Zhejiang Province, China (No. Y2101444 and No. LQ14H160015). We thank the staff at Huzhou Cancer Biobank and Department of Surgery in Huzhou Centre Hospital for their assistance.
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X, He J. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26:48. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 7.Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM, Ragoussis J, Harris AL. MicroRNA-10b and breast cancer metastasis. Nature. 2008;455:E8–E9. doi: 10.1038/nature07362. [DOI] [PubMed] [Google Scholar]
- 8.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40:1553–1560. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 10.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, Tannous BA, Krichevsky AM. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B, Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, Lin C, Fujita H, Otsuka T, Aishima S, Nagai E, Oda Y, Tanaka M. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim SA, Yip GW, Stock C, Pan JW, Neubauer C, Poeter M, Pupjalis D, Koo CY, Kelsch R, Schüle R, Rescher U, Kiesel L, Götte M. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase-and E-cadherin-dependent mechanism. Int J Cancer. 2012;131:E884–E896. doi: 10.1002/ijc.27629. [DOI] [PubMed] [Google Scholar]
- 15.Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143–e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer. 2008;113:3169–3180. doi: 10.1002/cncr.23934. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 21.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Yang XY, Zhao JY, Yu LW, Zhang P, Duan WY, Chong M, Gui YH. MiR-10a and MiR-10b Target the 3’-Untranslated Region of TBX5 to Repress Its Expression. Pediatr Cardiol. 2014;35:1072–9. doi: 10.1007/s00246-014-0901-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Krüppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Deng W, Bailey SK, Nail CD, Frost AR, Brouillette WJ, Muccio DD, Grubbs CJ, Ruppert JM, Lobo-Ruppert SM. Prevention of KLF4-mediated tumor initiation and malignant transformation by UAB30 rexinoid. Cancer Biol Ther. 2009;8:289–98. doi: 10.4161/cbt.8.3.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng Z, Tao K, Xia Q, Tan J, Yue Z, Chen J, Xi H, Li J, Zheng H. Krüppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS One. 2013;8:e56082. doi: 10.1371/journal.pone.0056082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma MQ, Zhang HD, Tang P, Jiang HJ, Chen CG. Association of Kruppel-like factor 4 expression with the prognosis of esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol. 2014;7:6679. [PMC free article] [PubMed] [Google Scholar]
- 27.Hu W, Hofstetter WL, Li H, Zhou Y, He Y, Pataer A, Wang L, Xie K, Swisher SG, Fang B. Putative tumor-suppressive function of Krüppel-like factor 4 in primary lung carcinoma. Clin Cancer Res. 2009;15:5688–5695. doi: 10.1158/1078-0432.CCR-09-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 29.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Krüppel- like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 31.Hsu LS, Chan CP, Chen CJ, Lin SH, Lai MT, Hsu JD, Yeh KT, Soon MS. Decreased Kruppel-like factor 4 (KLF4) expression may correlate with poor survival in gastric adenocarcinoma. Med Oncol. 2013;30:632. doi: 10.1007/s12032-013-0632-6. [DOI] [PubMed] [Google Scholar]
- 32.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 33.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT, Ruppert JM. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 34.Liu Y, Li M, Zhang G, Pang Z. MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur J Med Res. 2013;18:41. doi: 10.1186/2047-783X-18-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40:1553–1560. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 36.Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX, Tao HQ, Wang HJ, He XJ. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum Pathol. 2013;44:1278–1285. doi: 10.1016/j.humpath.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Kim K, Lee HC, Park JL, Kim M, Kim SY, Noh SM, Song KS, Kim JC, Kim YS. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6:740–751. doi: 10.4161/epi.6.6.15874. [DOI] [PubMed] [Google Scholar]


