Abstract
Gastric cancer was the third cause of death in China. In this study, we found that the APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3) expression was higher in gastric cancer tissues than that in normal tissues and confirmed APOBEC3B expression was correlated with the unfavorable prognosis of the patients with gastric cancer. Furthermore, APOBEC3B expression was associated with gender, tumor size, histological grade, T stage, and TNM staging of the patients with gastric cancer. Down-regulation of APOBEC3B expression in MNK28 cells could enhance the cytotoxicity of PDCD2. No editing took place in PDCD2 positive MKN28 cells with APOBEC3B shRNA. These results indicated that loss of function of PDCD2 may be partly caused by APOBEC3B-induced extensive mutagenesis.
Keywords: Gastric cancer, APOBEC3B, PDCD2, prognosis, proliferation
Introduction
The estimated cancer incident cases were 3.09 millions and 1.96 cancer deaths in 2010 with the incidence rate of 235.23/100,000 and mortality of 148.81/100,000 in China. Gastric cancer was the third cause of death in China with estimated deaths of 287,851 [1].
The seven APOBEC3 proteins are part of a larger polynucleotide cytosine deaminase family that in humans also includes activation-induced cytosine deaminase (AID) [2]. Editing activities of APOBEC3s play a critical role in restricting viral infectivity, and have also been postulated to have counteracted the actions against genome stability, mostly in terms of integration, exerted by both non-LTR (long terminal repeats) and LTR retrotransposons during evolutionary time [3,4]. A 29.5 kB deletion between exon 5 in APOBEC3A and exon 8 in APOBEC3B has recently been identified. This deletion results in complete removal of the APOBEC3B coding region and the hybrid gene contains the coding region of APOBEC3A and the 3’-untranslated region of APOBEC3B [5]. In a previous study, Burns et al. [6] provided evidence that APOBEC3B is overexpressed in breast tumors and cell lines. Prostate and renal clear cell carcinomas showed statistically significant upregulation of APOBEC3B in the tumors [7].
However, it is still unclear the roles of APOBEC3B in gastric cancer. To better understand this issue, we performed the techniques of cell biology and molecular pathology to determine the roles of the APOBEC3B in the developmental risk of gastric cancer.
Patients and methods
Tissue specimens
Tissue specimens were obtained from 236 patients without pre-surgical chemotherapy or radiotherapy in the Department of Gastroenterological Surgery, Hangzhou First People’s Hospital, School of Clinical Medicine, Nanjing Medical University between January 2009 and December 2013. The study was approved by our hospital review board, and each patient provided informed consent. Sixty-seven of 236 were histological Grade I tumors, 105 were Grade II tumors, and the remaining 64 were Grade III cases. Fresh tissue specimens and paraffin blocks were used to assess gene expression.
Cell lines and culture
Gastric cancer MKN28 cells and PDCD2 positive MKN28 cells were restored in our laboratory and grown in RPMI 1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) in a humidified incubator with 5% CO2 at 37°C.
Plasmid and transfection
PDCD2 positive MKN28 cells were transfected with APOBEC3B shRNA plasmid (sc-72515-SH, Santa Cruz biotechnology, Santa Cruz, CA, USA) using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were plated in a 24-well format in 500 µl of growth medium without addition of antibiotics and allowed to grow till 60% confluency. Both plasmid DNA and LipofectamineTM 2000 reagent were diluted in 50 µl of serum-free RPMI 1640 medium (Hyclone) medium separately and incubated for 5 min. After incubation, plasmid DNA and LipofectamineTM 2000 reagent were mixed gently and added to each well containing cells and medium.
Cell proliferation and cell cycle analysis
For the MTT assay, 1 × 104 cells/well was seeded in triplicate in 96-well plates. Optical density at 490 nm was determined directly after adding the CellTiter96 Aqueous One Solution reagent (Promega, Madison, WI, USA). For determination of cell cycle profiles, 6 × 105 cells seeded in 60-mm dishes, stained with propidium iodide (0.1 mg/ml) (Keygen, Nanjing, China), and samples were analyzed by a FACSCalibur flow cytometer (BD Biosciences, Baltimore, MD, USA).
Deep sequencing
Total DNA was extracted from cells by using a TissueGen DNA Kit (CWbiotech, Beijing, China). A portion of PDCD2 gene was amplified with the following reaction profile: 1 min at 94°C, 30 cycles of 30 s at 94°C followed by 2 min at 68°C, and followed by 3 min at 70°C. The amplicons were separated by electrophoresis on 1.5% (w/v) agarose gel, and extracted from the gel using Quiaquick Gel Extraction Kit (Qiagen, Gaithersburg, MD, USA). Purified amplicons were sequenced using GS junior bench top system (Roche, Shanghai, China) according to the manufacturer’s protocol and analyzed with equipped software, GS Amplicon Variant Analyzer.
RNA isolation and reverse transcription
Total RNA was isolated from tissue samples or cells using an RNeasy Mini Kit (Biomed, Beijing, China) and reversely transcribed into the first strand cDNA using a TaKaRa Reverse Transcription Kit (TaKaRa, Dalian, China) and oligo (dT) 15 primers (TaKaRa) according to the manufacturer’s instructions. All RNA samples were dissolved in RNAse-free water (Sigma, Shanghai, China) and their concentration and purity were evaluated by A260 nm measurement. Two micrograms of RNA extracted from each sample was subjected to reverse transcription in a total volume of 50 μl using the High-Capacity cDNA Archive Kit (PE Applied Biosystems, Foster City, CA, USA). The mixtures were incubated for 10 min at 25°C and for 2 h at 37°C. In all samples, 2 μl of the resulting complementary DNA products were used as template for PCR quantification.
qRT-PCR
The resultant cDNA was then used for qPCR amplification of PDCD2 and APOBEC3B expression, and GAPDH mRNA was used as a control. PDCD2 primers were 5’-CTGTGGAGCTGGGCTTCGCC-3’ and 5’-CAGCAGGAAGGAGAGCGGGC-3’. APOBEC3B primers were: 5’-TAGGTGCCACCCCGAT-3’ (sense) and 5’-TTGAGCATAATCTTACTCTTGTAC-3’ (antisense). GAPDH primers were 5’-AGAAGGCTGGGGCTCATTTG-3’ and 5’-AGGGGCCATCCACAGTCTTC-3’. Amplification of PDCD2, APOBEC3B, and GAPDH mRNA was performed with one cycle at 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 60 s. Relative mRNA abundance was normalized to the internal standard GAPDH by the ∆∆CT method.
Immunoblot and antibodies
Cell cultures and tissue specimens were washed twice with ice-cold PBS followed by lysis for 15 min in a buffer (20 mM sodium phosphate buffer, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, Ph = 7.4) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), sodium orthovanadate (1 mM, pH = 7.4) and a mixture of phosphatase inhibitors. Protein concentrations were measured by the BCA Protein Assay (Pierce, Rockford, IL, USA). Equal amounts (30 µg) of cell lysates were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. To detect target proteins, we incubated the membranes with antibodies against: anti-ataxia telangiectasia mutated (ATM) (Santa Cruz), anti-checkpoint kinase (Chk) 1 (Santa Cruz), anti-Chk2 (Santa Cruz), anti-phospho-S345-Chk1 (Cell Signaling Technology, Danvers, MA, USA), and anti-phospho-T68-Chk2 (Cell Signaling). HRP conjugated secondary antibodies, including anti-rabbit IgG, anti-rat IgG, or anti-mouse IgG antibodies at dilutions ranging from 1:1000 to 1:2000 (Amersham Biosciences, Needham, MA, USA), and binding results were visualized by enhanced chemiluminescence (Amersham Pharmacia, Piscataway, NJ, USA).
Statistical analysis
All the statistical analyses and graphics were performed with GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). Data were expressed as mean ± standard deviation (SD). Differences between group means were evaluated by Student’s t-test or one-way analysis of variance (ANOVA). Kaplan-Meier survival plots were generated and comparisons were made with log-rank statistics. P-values < 0.05 were considered to be statistically significant.
Results
APOBEC3B mRNA and protein were evaluated in 236 human gastric cancer specimens
The levels of APOBEC3B protein in gastric cancer tissues exhibited higher than that in matched normal tissue (P < 0.05, Figure 1A). Real-time PCR of APOBEC3B mRNA was carried out in gastric cancer specimens. The results showed that the levels of APOBEC3B mRNA were higher than normal tissue (P < 0.05, Figure 1B). Furthermore, immunohistochemical staining also confirmed that the protein levels of APOBEC3B in gastric cancer specimens were higher than that in normal tissues (Figure 1C). Both APOBEC3B mRNA and protein levels were higher in cancer tissues from Grade III stage than that from Grade I or Grade II stage (P < 0.05, Figure 1). Kaplan-Meier analysis showed that APOBEC3B expression was correlated with the unfavorable prognosis of patients with Grade I, Grade II and Grade III stage gastric cancer (P < 0.05, Figure 1D). We then analyzed the potential relationship between the expression of APOBEC3B or PDCD2 expression and the clinicopathological characteristics of these patients. APOBEC3B expression was associated with gender (P = 0.002), tumor size (P = 0.006), histological grade (P = 0.001), T stage (P = 0.005), and TNM staging (P = 0.003) of the patients with gastric cancer (Table 1). PDCD2 expression was associated with age (P = 0.014), and tumor size (P = 0.002, Table 1).
Figure 1.
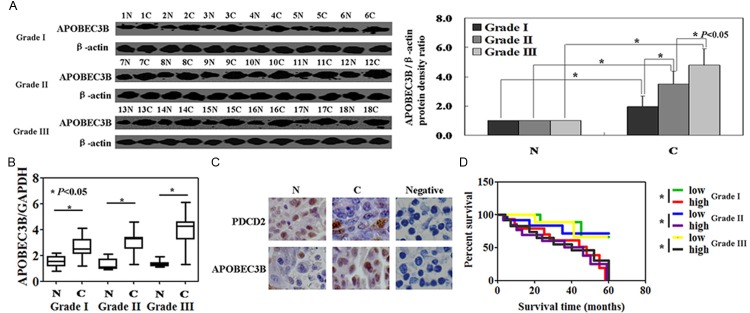
The levels of APOBEC3B protein and mRNA were measured in gastric cancer specimens using Western blot (A), real-time PCR (B), and immunohistochemical staining (C). N: normal, C: cancer, negative: negative control. (D) APOBEC3B protein and prognosis of the patients with gastric cancer by the Kaplan-Meier method.
Table 1.
Relationship between APOBEC3B or PDCD2 expression and clinicopathological parameters of patients with gastric cancer
| Clinicopathological parameters | n | APOBEC3B expression | χ2 | P value | PDCD2 expression | χ2 | P value | ||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| High | Low | High | Low | ||||||
| Gender | 9.61 | .002* | 0.29 | .590 | |||||
| Male | 163 | 99 | 64 | 55 | 108 | ||||
| Female | 73 | 60 | 13 | 28 | 45 | ||||
| Age (years) | 0.02 | .965 | 6.02 | .014* | |||||
| ≤60 | 143 | 96 | 47 | 41 | 102 | ||||
| >60 | 93 | 63 | 30 | 42 | 51 | ||||
| Tumor location | 0.15 | .929 | 0.93 | .629 | |||||
| Proximal | 137 | 91 | 46 | 49 | 88 | ||||
| Distal | 93 | 64 | 29 | 33 | 60 | ||||
| Total | 6 | 4 | 2 | 1 | 5 | ||||
| Tumor size | 7.63 | .006* | 9.88 | .002* | |||||
| ≤5.0 cm | 160 | 98 | 62 | 45 | 115 | ||||
| >5.0 cm | 76 | 61 | 15 | 38 | 38 | ||||
| Histological Grade | 10.5 | .001* | 0.16 | .686 | |||||
| G1/2 | 62 | 31 | 31 | 20 | 42 | ||||
| G3 | 174 | 128 | 46 | 63 | 111 | ||||
| T stage | 8.06 | .005* | 0.124 | .725 | |||||
| T1/2 | 47 | 23 | 24 | 15 | 32 | ||||
| T3+T4a/4b | 189 | 136 | 53 | 68 | 121 | ||||
| N stage | 2.71 | 0.10 | 1.77 | .183 | |||||
| N0 | 65 | 38 | 27 | 18 | 47 | ||||
| N+ | 171 | 121 | 50 | 65 | 106 | ||||
| M stage | 1.98 | 0.16 | 0.04 | .850 | |||||
| M0 | 213 | 140 | 73 | 75 | 138 | ||||
| M1 | 23 | 19 | 4 | 8 | 15 | ||||
| TNM staging | 14.3 | .003* | 2.49 | .478 | |||||
| I | 27 | 10 | 17 | 8 | 19 | ||||
| II | 58 | 41 | 17 | 22 | 36 | ||||
| III | 128 | 89 | 39 | 42 | 86 | ||||
| IV | 23 | 19 | 4 | 11 | 12 | ||||
Statistically significant (P < 0.05).
APOBEC3B knockdown could enhance the cytotoxicity of PDCD2 in MKN28 cells
PDCD2 positive MKN28 cells were transfected with APOBEC3B shRNA plasmid. APOBEC3B and PDCD2 expression were measured by western blot, real-time PCR, and immunofluorescence analysis. The results of Western blot analysis and immunofluorescence analysis confirmed decreased APOBEC3B protein levels and increased PDCD2 protein in MKN28 cells after transfection (Figure 2A, 2C). Real-time PCR also showed decreased APOBEC3B mRNA levels and increased PDCD2 mRNA levels in transfected cells (P < 0.05, Figure 2B).
Figure 2.
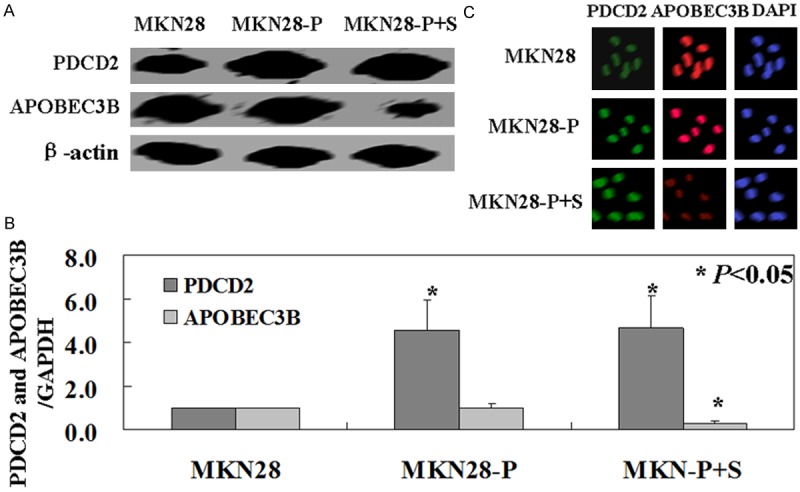
APOBEC3B knockdown in gastric cancer cell MKN28. Western blot (A), real-time PCR (B), and immunofluorescence analysis (C) were used to detect APOBEC3B and PDCD2 protein and mRNA levels in MKN28 cells. MKN28: untreated MKN28 cells, MKN28-P: PDCD2 positive MKN28 cells, MKN28-P+S: PDCD2 positive MKN28 transfected with APOBEC3B shRNA plasmid.
MTT assay showed that the proliferation rate of PDCD2 positive MKN28 cells is slightly lower than the untreated ones (Figure 3A). But the result didn’t have statistical significance (P > 0.05). The proliferation rate of PDCD2 positive MKN28 cells with APOBEC3B shRNA plasmid transfection was significant lower than PDCD2 positive MKN28 cells (P < 0.05, Figure 3A). PI staining of the cells revealed that PDCD2 positive MKN28 cells with APOBEC3B shRNA plasmid transfection were arrested in S phase of the cell cycle (P < 0.05, Figure 3B). In order to determine whether the reduced anti-tumor activities of PDCD2 correlated with mutagenesis, we sequenced PDCD2 gene in MKN28 cells. PDCD2 positive MKN28 cells with APOBEC3B shRNA did not yield any PCR products amplified at lower denaturing temperatures, suggesting that no editing took place. However, the extensive mutagenesis was observed in the MKN28 cells and PDCD2 positive MKN28 cells (Figure 3C).
Figure 3.
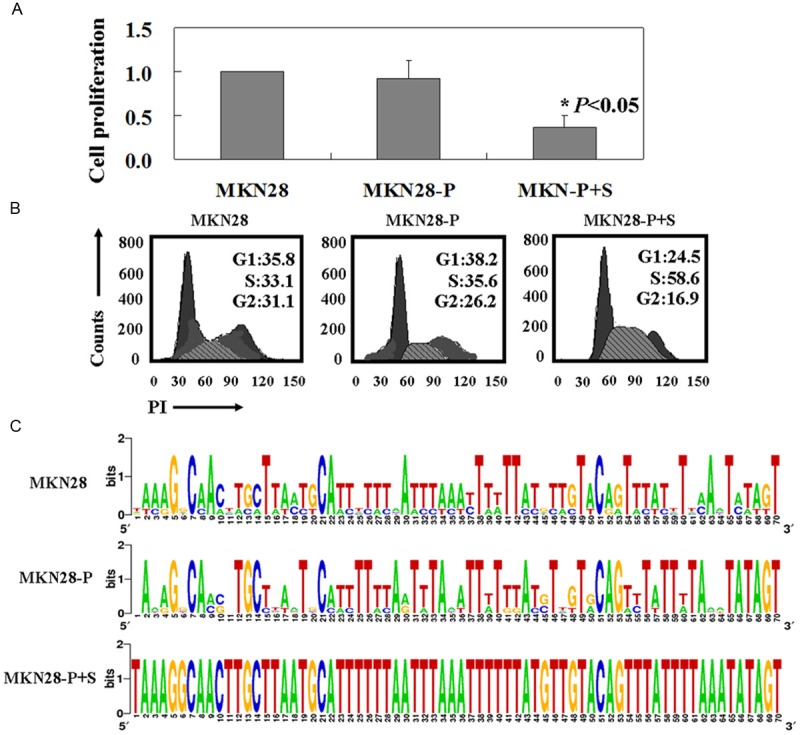
APOBEC3B knockdown could enhance the cytotoxicity of PDCD2 in MKN28 cells. A: The percentages of growth inhibition were determined by MTT assay; B: Flow cytometric cell cycle assay by using PI staining; C: Deep sequencing detected APOBEC3B-induced editing in PDCD2. MKN28: untreated MKN28 cells, MKN28-P: PDCD2 positive MKN28 cells, MKN28-P+S: PDCD2 positive MKN28 transfected with APOBEC3B shRNA plasmid.
APOBEC3B knockdown could promote PDCD2 induced activation of ATM and Chk1/2
In this study, we found that levels of Chk1 and Chk2 were not significantly changed in PDCD2 positive MKN28 cells with APOBEC3B shRNA compared with untreated MKN28 cells (Figure 4). However, the levels of p-Chk1 and p-Chk2 were significantly increased in PDCD2 positive MKN28 cells with APOBEC3B shRNA (Figure 4). Furthermore, we found that the levels of ATM, the upstream protein of Chk1 and Chk2, were also increased (Figure 4).
Figure 4.
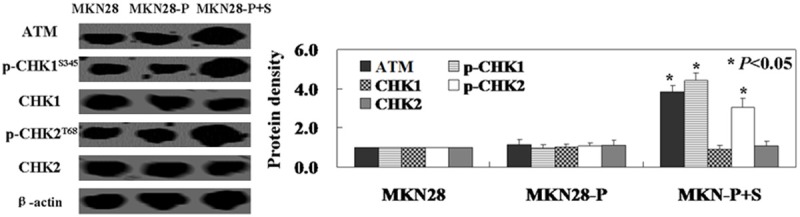
APOBEC3B knockdown could promote PDCD2 induced activation of ATM and Chk1/2. Western blot analysis was performed to detect cell cycle related proteins. No editing PDCD2 expression induced activation of ATM, P-Chk1, and P-Chk2, whereas Chk1 and Chk2 showed no changes. β-actin was used as an internal loading control. MKN28: untreated MKN28 cells, MKN28-P: PDCD2 positive MKN28 cells, MKN28-P+S: PDCD2 positive MKN28 transfected with APOBEC3B shRNA plasmid.
Discussion
APOBEC3B is overexpressed in many types of tumor tissues and cancer cell lines, such as breast cancer, renal clear cell cancer, and chondrosarcoma [6-8]. In this study, we also found that APOBEC3B is overexpressed in gastric cancer tissue and cells and confirmed APOBEC3B expression was correlated with the unfavorable prognosis of the patients with gastric cancer. Furthermore, APOBEC3B expression was associated with gender, tumor size, histological grade, T stage, and TNM staging of the patients with gastric cancer.
Jin et al. [8] found that the RUNX3 positive chondrosarcoma cells with APOBEC3B knockdown had a higher apoptotic ratio than the ones without APOBEC3B knockdown. The possible mechanism is that APOBEC3 could induce mutations in antitumor genes [8]. APOBEC3 caused C→T transitions in cervix, bladder, lung, head and neck, and breast cancers [9]. The APOBEC3 genes have been shown to deaminate 5-methylcytosine and 5-hydroxymethylcytosine, with base excision repair of the resulting mismatch providing a mechanism for active DNA demethylation [10]. In our previous study, we have found that PDCD2 showed no effects on gastric cancer cell MNK28 cells [11]. In this study, we down-regulated APOBEC3B expression in PDCD2 positive MNK28 cells by using APOBEC3B shRNA. We confirmed that APOBEC3B knockdown could enhance the cytotoxicity of PDCD2 in MKN28 cells. Furthermore, we found the mechanisms of the reduced antitumor activity of PDCD2 that caused by APOBEC3B. No editing took place in PDCD2 positive MKN28 cells with APOBEC3B shRNA. However, the extensive mutagenesis was observed in the MKN28 cells and PDCD2 positive MKN28 cells. These results indicated that APOBEC3B could inhibit the antitumor effects of PDCD2 by mutagenesis. Activation of ATM induces phosphorylation and activation of downstream targets, including Chk1 and Chk2 [12]. Our current study provided evidence that no editing PDCD2 expression induces expression of ATM, p-Chk1, and p-Chk2 proteins and S phase arrest in gastric cancer MKN28 cells.
In conclusion, this study provides evidence that APOBEC3B could interfere with PDCD2 transcription. Loss of their functions of suppressor genes may be partly caused by APOBEC3B-induced extensive mutagenesis in cancer.
Acknowledgements
This study was supported in part by grants from the Science and Technology plan for Social Development of Zhejiang Provincial Science Technology Department (2014C33236).
Disclosure of conflict of interests
None.
References
- 1.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X, He J. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26:48–58. doi: 10.3978/j.issn.1000-9604.2014.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conticello SG. The AID/APOBEC family of ucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z, Han YX, Han XR. The role of APOBEC3B in chondrosarcoma. Oncol Rep. 2014;32:1867–1872. doi: 10.3892/or.2014.3437. [DOI] [PubMed] [Google Scholar]
- 9.van Jaarsveld MT, Wouters MD, Boersma AW, Smid M, van Ijcken WF, Mathijssen RH, Hoeijmakers JH, Martens JW, van Laere S, Wiemer EA, Pothof J. DNA damage responsive microRNAs misexpressed in human cancer modulate therapy sensitivity. Mol Oncol. 2013;8:458–468. doi: 10.1016/j.molonc.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yellapa A, Bitterman P, Sharma S, Guirguis AS, Bahr JM, Basu S, Abramowicz JS, Barua A. Interleukin 16 expression changes in association with ovarian malignant transformation. Am J Obstet Gynecol. 2014;210:272, e1–10. doi: 10.1016/j.ajog.2013.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Wei W, Jin HC, Ying RC, Zhu AK, Zhang FJ. Programmed cell death 2 protein induces gastric cancer cell growth arrest at the early S phase of the cell cycle and apoptosis in a p53-dependent manner. Oncol Rep. 2015;33:103–110. doi: 10.3892/or.2014.3551. [DOI] [PubMed] [Google Scholar]
- 12.Smith GC, Cary RB, Lakin ND, Hann BC, Teo SH, Chen DJ, Jackson SP. Purification and DNA binding properties of the ataxia-telangiectasia gene product ATM. Proc Natl Acad Sci U S A. 1999;96:11134–11139. doi: 10.1073/pnas.96.20.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]


