Abstract
To investigate the clinical significance of ID1 expression in Chinese de novo AML patients. Real-time quantitative PCR was carried out to detect the status of ID1 expression in 102 de novo AML patients and 28 controls. ID1 transcript level was significantly increased in AML compared to normal controls (p=0.029). The age in the patients with high ID1 expression is significantly older than in those with low ID1 expression (p=0.044). ID1 overexpression occurred with the highest frequency in the patients with poor karyotype (7/7, 100%), lower frequency in the patients with intermediate karyotype (28/60, 47%), and the lowest frequency in the patients with favorable karyotype (12/31, 39%). Both whole AML and non-M3 patients with high ID1 expression had significantly lower rate of complete remission than those with low ID1 expression (p=0.007 and 0.038). ID1 high-expressed patients showed significantly shorter overall survival (OS) than ID1 low-expressed patients in both whole AML and non-M3 according to Kaplan-Meier analysis (p=0.007 and 0.040). However, multivariate analysis indicated that ID1 overexpression was not an independent risk factor in both whole AML and non-M3 patients. However, the adverse impact of ID1 overexpression on outcome was revealed by both Kaplan-Meier analysis and multivariate analysis in the non-M3 patients less than 60 years old. Our study reveals that ID1 overexpression may be associated with higher risk karyotype classification and act as an independent risk factor in young non-M3 patients.
Keywords: ID1, expression, prognosis, acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML), a clonal hematological malignancy, is a biologically, clinically, and etiologically heterogeneous disease [1,2]. Cytogenetic alterations and molecular biological changes play crucial roles in the pathogenesis and progression of AML. Despite the advancements in the treatment of leukemia, clinical outcome of AML remains unsatisfactory. Therefore, identifying genetic and epigenetic alterations which can recognize the patients who are at the risk of poor outcome is warranted to optimize treatment strategies. Over the past years, the prognosis of AML has been evaluated mainly based on cytogenetic analysis [3,4]. Recently, numerous genetic changes including gene mutations, deletions, amplifications and gene expression abnormalities, have been identified [5-7]. These alterations contribute to further understanding of leukemogenesis and provide more prognostic markers in AML [8,9].
ID (inhibitors of differentiation) gene encodes for a helix-loop-helix (HLH) protein, a group of dominant inhibitors of basic HLH transcriptional factors which promote cell differentiation [10,11]. ID1 (inhibitors of differentiation 1), a family member of ID genes, has been identified as a potential proto-oncogene for its role in inducing cell proliferation as well as invasion, and protecting cells against drug-induced apoptosis [11]. Overexpression of ID1 has been found in a variety of solid tumors [12-22]. However, few studies investigated the clinical relevance of ID1 expression in AML [23,24]. Therefore, the current study was intended to investigate the clinical significance of ID1 expression in Chinese de novo AML patients.
Materials and methods
Patients’ samples
Bone marrow (BM) was collected from 102 patients with de novo AML treated at the Affiliated People’s Hospital of Jiangsu University. The diagnosis and classification of AML patients were established according to the revised French-American-British (FAB) classification and the 2008 World Health Organization (WHO) criteria [25,26]. Written informed consent was obtained from all patients. The study was approved by the Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University. Karyotypes were analyzed by conventional R-banding method and karyotype risk was classified according to reported previously [27]. Treatment protocol was described previously [28]. The characteristics of AML patients were summarized in Table 1. 28 healthy donors were collected as controls. Bone marrow mononuclear cells (BMMNCs) were separated by Ficoll solution and washed twice with PBS.
Table 1.
Correlation between ID1 expression and whole AML as well as CN-AML patients parameters
| Patient’s parameters | Status of ID1 expression in whole AML | Status of ID1 expression in CN-AML | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Low (n=51) | High (n=51) | P | Low (n=27) | High (n=21) | P | |
| Sex, male/female | 29/22 | 32/19 | 0.687 | 15/12 | 16/5 | 0.224 |
| Median age, years (range) | 51 (10-93) | 60 (17-87) | 0.044 | 61 (15-86) | 61 (17-85) | 0.809 |
| Median WBC, ×109/L (range) | 5.7 (0.3-528.0) | 19.7 (1.1-185.4) | 0.062 | 10.2 (0.8-528.0) | 28.1 (1.2-136.1) | 0.330 |
| Median hemoglobin, g/L (range) | 78 (32-131) | 68 (40-138) | 0.095 | 88.5 (32-131) | 76.5 (40-138) | 0.492 |
| Median platelets, ×109/L (range) | 40 (6-140) | 42 (4-264) | 0.657 | 44.5 (6-140) | 30 (4-124) | 0.336 |
| BM blasts, % (range) | 44.0 (1.0-97.5) | 53.3 (3.0-109.0) | 0.132 | 51.8 (17.0-97.5) | 65.0 (6.0-109.0) | 0.778 |
| FAB | 0.450 | 0.542 | ||||
| M1 | 3 (6%) | 5 (10%) | 2 (7%) | 1 (5%) | ||
| M2 | 19 (37%) | 19 (37%) | 14 (52%) | 9 (43%) | ||
| M3 | 16 (31%) | 8 (16%) | - | - | ||
| M4 | 8 (16%) | 15 (29%) | 8 (30%) | 8 (38%) | ||
| M5 | 5 (10%) | 2 (4%) | 3 (11%) | 1 (5%) | ||
| M6 | 0 (0%) | 2 (4%) | 0 (0%) | 2 (9%) | ||
| WHO | 0.100 | 0.542 | ||||
| AML with t(8;21) | 3 (6%) | 4 (8%) | - | - | ||
| APL with t(15;17) | 16 (31%) | 8 (16%) | - | - | ||
| AML without maturation | 3 (6%) | 5 (10%) | 2 (7%) | 1 (5%) | ||
| AML with maturation | 16 (31%) | 15 (29%) | 14 (52%) | 9 (43%) | ||
| Acute myelomonocytic leukemia | 8 (16%) | 16 (31%) | 8 (30%) | 8 (38%) | ||
| Acute monoblastic and monocytic leukemia | 5 (10%) | 1 (2%) | 3 (11%) | 1 (5%) | ||
| Acute erythroid leukemia | 0 (0%) | 2 (4%) | 0 (0%) | 2 (9%) | ||
| Karyotype classification | 0.011 | - | ||||
| Favorable | 19 (37%) | 12 (23%) | - | - | ||
| Intermediate | 32 (63%) | 28 (55%) | - | - | ||
| Poor | 0 (0%) | 7 (14%) | - | - | ||
| No data | 0 (0%) | 4 (8%) | - | - | ||
| Karyotype | 0.033 | - | ||||
| normal | 27 (53%) | 21 (41%) | - | - | ||
| T (8; 21) | 3 (6%) | 4 (8%) | - | - | ||
| T (15; 17) | 16 (31%) | 8 (16%) | - | - | ||
| complex | 0 (0%) | 6 (12%) | - | - | ||
| others | 5 (10%) | 8 (16%) | - | - | ||
| No data | 0 (0%) | 4 (8%) | - | - | ||
| Gene mutation* | ||||||
| C/EBPA (+/-) | 6/42 (13%) | 4/44 (8%) | 0.740 | 4/23 (15%) | 3/17 (15%) | 1.000 |
| NPM1 (+/-) | 5/43 (10%) | 3/45 (6%) | 0.714 | 4/23 (15%) | 1/19 (5%) | 0.377 |
| FLT3 ITD (+/-) | 7/41 (15%) | 7/41 (15%) | 1.000 | 4/23 (15%) | 2/18 (10%) | 1.000 |
| C-KIT (+/-) | 0/48 (0%) | 0/48 (0%) | - | 0/27 (0%) | 0/20 (0%) | - |
| N/K RAS (+/-) | 4/44 (8%) | 5/43 (10%) | 1.000 | 4/23 (15%) | 2/18 (10%) | 1.000 |
| IDH1/2 (+/-) | 4/44 (8%) | 1/47 (2%) | 0.362 | 4/23 (15%) | 0/20 (0%) | 0.126 |
| DNMT3A (+/-) | 4/44 (8%) | 3/45 (6%) | 1.000 | 2/25 (7%) | 2/18 (10%) | 1.000 |
| U2AF1 (+/-) | 1/47 (2%) | 4/44 (8%) | 0.362 | 1/26 (4%) | 1/19 (5%) | 1.000 |
| CR (+/-) | 30/18 (63%) | 15/30 (33%) | 0.007 | 14/13 (52%) | 8/11 (42%) | 0.562 |
WBC, white blood cells; FAB, French-American-British classification; AML, acute myeloid leukemia; CR, complete remission;
percentage was equal to the number of mutated patients divided by total cases in each group.
RNA isolation, reverse transcription and RQ-PCR
Total RNA was isolated from the BMMNCs using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed on iCycler Thermal Cycler (Eppendorf, Hamburg, Germany). The reactions with final volume 40 μL contained 5× buffer 10 mM, dNTPs 10 mM, random hexamers 10 μM, RNAsin 80 units, and 200 units of MMLV reverse transcriptase (MBI Fermentas, Hanover, USA). The system of reverse transcription was incubated for 10 min at 25°C, 60 min at 42°C, and then stored at -20°C. Real-time quantitative PCR (RQ-PCR) was performed on a 7300 Thermo cycler (Applied Biosystems, CA, USA). The primer sequences of ID1 expression were 5’-CTCAGCACCCTCAACGG-3’ (forward) and 5’-GATCGGTCTTGTTCTCCCTC-3’ (reverse) with expected product of 199 bp. Reaction system with a volume of 20 μL was consisted of cDNA 20 ng, 0.4 μM of primers, 10 μM of AceQTM qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), and 0.4 μM of ROX Reference Dye 1 (Invitrogen, Carlsbad, CA, USA). RQ-PCR conditions were carried out at 95°C for 5 min, followed by 35 cycles at 95°C for 10 s, 62°C for 30 s, 72°C for 30 s, and 80°C for 30 s to collect fluorescence, finally followed by 95°C for 15 s, 60°C for 60 s, 99°C for 15 s, and 60°C for 15 s. Positive and negative controls were included in all assays. Housekeeping gene (ABL) was used to calculate the abundance of ID1 mRNA. Relative ID1 expression levels were calculated using the following equation: NID1= (EID1)ΔCT ID1 (control-sample)÷(EABL)ΔCT ABL (control-sample). The parameter efficiency (E) was derived from the formula E=10(-1/slope) (the slope referred to CT versus cDNA concentration plot). ΔCT reflected the disparity in CT value between control and target or reference sequences. We selected the bone marrow sample from one normal control that possessed the minimal ΔCT between ID1 and ABL transcript as control and was defined as 100% expression for ID1 transcript.
Gene mutation detection
IDH1/2, DNMT3A, N/K-RAS, C-KIT, NPM1, and U2AF1 mutations were detected by high-resolution melting analysis (HRMA) as reported previously [29-32]. All positive samples were confirmed by DNA direct sequencing. FLT3-ITD and C/EBPA mutations were detected by direct DNA sequencing [33].
Statistical analysis
Statistical analyses were performed on SPSS 17.0 software package (SPSS, Chicago, IL). Mann-Whitney’s U test was used to compare the difference of continuous variables in two groups. Pearson Chi-square analysis or Fisher exact test were employed to compare the difference of categorical variables. Receiver operating characteristic curve (ROC) and area under the ROC curve (AUC) were conducted to assess the value of ID1 expression in distinguishing AML and cytogenetically normal AML (CN-AML) patients from normal controls. Kaplan-Meier curve done by log-rank test and Cox regression backward stepwise likelihood ratio were performed to analyze the impact of ID1 expression on survival respectively. For all analyses, a two-tailed P value of 0.05 or less was determined as statistically significant.
Results
ID1 expression in normal controls and AML patients
ID1 transcript level in normal controls ranged from 0.000 to 1.000 with a median level of 0.015. The level of ID1 expression (0.000-3.536, median 0.029) was significantly increased in AML compared to normal controls (P=0.029, Figure 1). The representative electrophoresis results of RQ-PCR products were shown in Figure 2.
Figure 1.
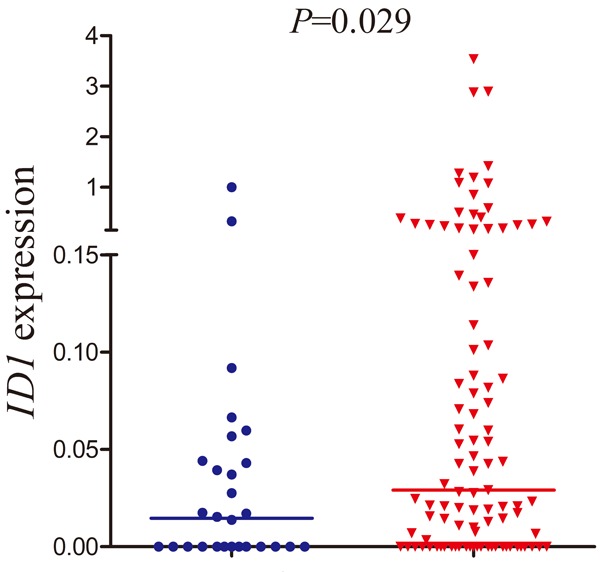
Relative expression levels of ID1 expression in AML patients and controls.
Figure 2.
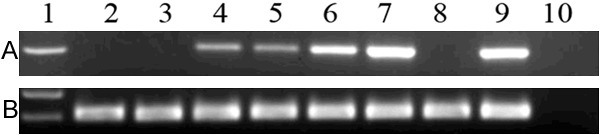
Electrophoresis results of RQ-PCR products in AML patients. 1: Gene RulerTM 100bp DNA ladder; 2-3: normal controls; 4-8: AML samples; 9: positive control; 10: negative control. A: ID1; B: ABL.
Differentiating value of ID1 expression
ROC curve was applied to evaluate the differentiating value of ID1 expression. It indicated that ID1 level might serve as a biomarker for distinguishing AML from controls (AUC=0.633, 95% CI: 0.523-0.742, P=0.032).
Clinical and laboratory characteristics of AML patients
The whole cohort of AML patients were divided into two groups at the median level of ID1 expression, and defined as low ID1 expression (ID1 low) group (<0.029) and high ID1 expression (ID1 high) group (>0.029). There were no significant differences in sex, hemoglobin (HB), platelets (PLT), and BM blasts between the ID1 high and ID1 low groups (P>0.05, Table 1). However, ID1 high cases tended to have a higher white blood cell (WBC) than ID1 low cases (P=0.062). ID1 high patients had significantly older age than ID1 low patients (P=0.044). No significant differences were found between the two groups in the distribution of both FAB and WHO subtypes. While, significant difference was observed in the distribution of karyotype classification between the ID1 high and ID1 low patients (P=0.011). ID1 overexpression occurred with the highest frequency in the patients with poor karyotype (7/7, 100%), lower frequency in the patients with intermediate karyotype (28/60, 47%), and the lowest frequency in the patients with favorable karyotype (12/31, 39%). No significant correlations were found between ID1 expression and ten gene mutations (P>0.05, Table 1). In addition, among CN-AML patients, there were no significant differences in peripheral perameters, BM blasts, FAB subtypes, and gene mutations between the ID1 high and ID1 low patients (P>0.05, Table 1).
Correlation between ID1 expression and clinical outcome
Follow-up data was obtained for 93 AML patients. After induction therapy, ID1 high patients had significantly lower rate of complete remission (CR) than ID1 low patients (33% vs. 63%, respectively, P=0.007, Table 1). Among non-M3 patients, ID1 high cases also showed significantly lower rate of CR than ID1 low cases [30% (12/40) vs. 54% (19/35), respectively, P=0.038]. However, there was no significant difference among CN-AML patients (52% vs. 42%, respectively, P=0.562, Table 1). Moreover, significantly lower CR rate was observed in ID1 high groups as compared with ID1 low groups in both whole AML and non-M3 patients less than 60 years old [48% (11/23) vs. 84% (26/31) and 40% (8/20) vs. 84% (16/19); P=0.007 and 0.008, respectively], but not in whole AML and non-M3 patients more than 60 years old (data not shown). Survival analyses were performed in 90 patients with follow-up data ranged from 1 to 92 months (median 10 months). ID1 high patients showed significantly shorter overall survival (OS) than ID1 low patients (median 5 versus 17 months, respectively, P=0.007, Figure 3A). Significant difference was also observed in non-M3 patients. The median OS in ID1 high and ID1 low cases was 5 and 12 months, respectively (P=0.040, Figure 3B). However, significant difference was not found among CN-AML patients (median 6 versus 11 months, respectively, P=0.339). Multivariate analysis including age (≤60 y vs. >60 y), WBC (≥30×109/L vs. <30×109/L), karyotype classification (favorable vs. intermediate vs. poor), four gene mutations (mutant vs. wild-type), and ID1 expression (high vs. low) variables disclosed that ID1 overexpression was not an independent risk factor in both whole AML and non-M3 patients (Table 2). However, the adverse impact of ID1 overexpression on outcome was revealed by both Kaplan-Meier analysis and multivariate analysis in the non-M3 patients less than 60 years old (Figure 3C; Table 3), but not in the whole AML patients less than 60 years old (Table 3) as well as whole AML and non-M3 more than 60 years old (data not shown).
Figure 3.
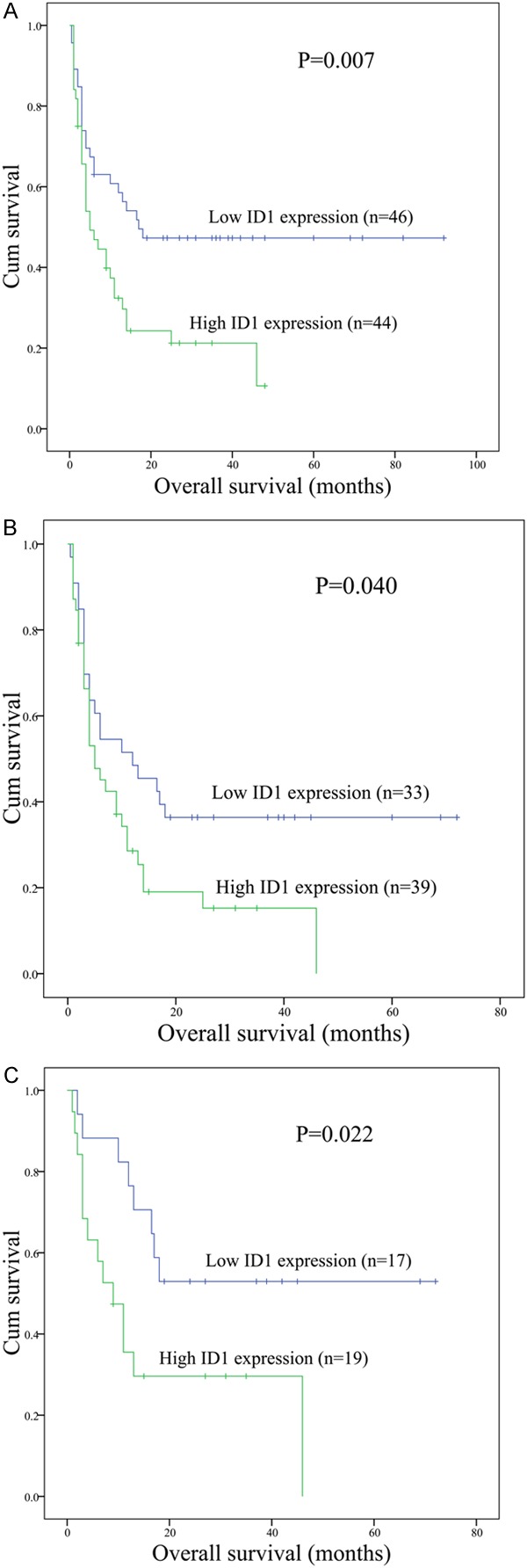
The impact of ID1 expression on overall survival of AML patients. A: All patients; B: Non-M3 patients; C: Young (age <60 years old) non-M3 patients.
Table 2.
Multivariate analyses of prognostic factors for overall survival in whole AML and non-M3 patients
| Whole AML | non-M3 | |||
|---|---|---|---|---|
|
| ||||
| hazard ratio (95% CI) | P value | hazard ratio (95% CI) | P value | |
| Age | 2.110 (1.141-3.901) | 0.017 | 2.066 (1.129-3.782) | 0.019 |
| WBC | 1.698 (0.929-3.102) | 0.085 | 1.611 (0.844-3.076) | 0.149 |
| Karyotype classifications | 2.561 (1.418-4.624) | 0.002 | 2.521 (1.254-5.071) | 0.009 |
| ID1 expression | 1.337 (0.725-2.468) | 0.352 | 1.412 (0.750-2.659) | 0.285 |
| FLT3 mutation | 0.551 (0.235-1.291) | 0.170 | 0.715 (0.290-1.761) | 0.466 |
| NPM1 mutation | 1.201 (0.338-4.266) | 0.777 | 1.010 (0.270-3.777) | 0.988 |
| C/EBPA mutation | 0.907 (0.310-2.653) | 0.859 | 0.861 (0.290-2.557) | 0.788 |
Table 3.
Multivariate analyses of prognostic factors for overall survival in young (age <60 years old) whole AML and non-M3 patients
| Whole AML | non-M3 | |||
|---|---|---|---|---|
|
| ||||
| hazard ratio (95% CI) | P value | hazard ratio (95% CI) | P value | |
| WBC | 2.738 (1.137-6.597) | 0.025 | 3.249 (1.178-8.956) | 0.023 |
| Karyotype classifications | 3.845 (1.695-8.724) | 0.001 | 3.835 (1.465-10.041) | 0.006 |
| ID1 expression | 2.114 (0.850-5.259) | 0.107 | 3.012 (1.105-8.213) | 0.031 |
| FLT3 mutation | 0.902 (0.190-4.278) | 0.897 | 1.553 (0.274-8.809) | 0.619 |
| NPM1 mutation | 0.685 (0.089-5.303) | 0.717 | 0.704 (0.084-5.877) | 0.745 |
| C/EBPA mutation | 1.149 (0.334-3.958) | 0.826 | 1.151 (0.319-4.158) | 0.830 |
Discussion
The major biological effect of ID protein is the inhibition of differentiation and maintenance of self-renewal and multipotency of stem cells, which is coordinated with continuous cell cycling [11]. ID1 proteins which could be activated by oncogenic factors are essential components of oncogenic pathways [11]. Deregulation of ID1 proteins plays a direct role in cancer initiation, maintenance, progression, and drug resistance [11]. Additionally, ID1 aberration may contribute to the initiation of myeloid malignancy [34]. Thus, ID1 may represent a potential therapeutic target for tumors including hematopoietic malignancy.
The clinical significance of ID1 aberration has been widely investigated. Although ID1 overexpression predicts poor outcome in the majority of solid tumors [11], the impact of ID1 aberration remains controversial in AML patients. Tang et al revealed that high ID1 expression independently predicted lower CR rate and shorter disease-free survival (DFS) and OS in young (age <60 y) non-M3 or cytogenetically normal patients [23]. However, Damm et al disclosed that ID1 overexpression was not an independent prognostic factor in young CN-AML patients [24]. Our results confirmed the adverse impact of high ID1 expression on prognosis in non-M3 AML patients less than 60 years. The impact of ID1 expression on outcome was not investigated in CN-AML patients less than 60 years due to the small size of case numbers.
Interestingly, our study further found the significant correlation between ID1 expression and karyotype classification and indicated that the incidence of ID1 overexpression was increased with the rising risk of karyotype. However, if M3 patients were excluded from analysis, we did not observe the significant association between ID1 expression and karyotype classification, which was in accordance with the previous investigation [23]. An early study also observed the down-regulation of ID1 expression in primary acute promyelocytic leukemia (APL) cells and NB4 cell lines, which could be rapidly induced upon all-trans retinoic acid (ATRA) treatment [35]. Moreover, ID1 overexpression inhibited proliferation and induced a G0/G1 accumulation in NB4 cells [35]. However, a later study revealed that ID1 overexpression enhanced the proliferation of primitive myeloid progenitor cells and immortalized bone marrow cells in vitro, and ID1 silencing inhibited leukemic cell line growth [34]. These results indicated that the role of ID1 in the process of leukemogenesis may be dependent on the context of different cytogenetics.
The association of ID1 expression with gene mutations has been investigated. Damm et al revealed the significantly decreased incidences of C/EBPA mut and NPM1 mut/FLT3-ITDneg in ID1 high patients [24]. Moreover, ID1 high patients showed a significantly increased frequency of FLT3-ITDmut [23,24]. Our study did not observe the correlation between these gene mutations and ID1 expression, probably due to the low frequency of these gene mutations in our cases. This difference may be attributed to the differences in ethnics and in AML subtype distribution. More cases of different races are needed to further determine the association of ID1 expression with genetic mutations.
The underlying mechanism of regulating ID1 expression was poorly studied. Although a large CpG island was identified at the 5’ region of ID1 promoter, ID1 expression silencing was not associated with its promoter methylation [36,37]. Our study further investigated the methylation status of ID1 in both normal controls and leukemic cell lines using bisulfite sequencing and manifested that ID1 promoter showed extremely low density in both normal controls and leukemic cell lines (data not shown). ID1 expression was shown to be regulated by histone acetylation of its promoter in leukemic cell lines [37]. Recently, two microRNAs (miR-29b and miR-381) have been demonstrated to play important roles in the regulation of ID1 expression in human lung adenocarcinoma [38,39]. Garzon et al disclosed the decreased expression of miR-29b in AML [40]. Furthermore, ectopic miR-29b expression could induce apoptosis and reduce cell growth in primary AML cells and cell lines, and inhibit tumorigenicity in a Xenograft leukemia model [40]. Further studies are required to explore the role of these microRNAs in regulating ID1 expression in AML patients.
In conclusion, our study suggests that ID1 overexpression may correlate with higher risk karyotype classification and serve as an independent risk factor in young non-M3 patients.
Acknowledgements
This study was supported by National Natural Science foundation of China (81270630, 81172592), Science and Technology Special Project in Clinical Medicine of Jiangsu Province (BL2012056), 333 Project of Jiangsu Province (BRA2013136), Science and Technology Infrastructure Program of Zhenjiang (SS2012003), Medical Key Talent Project of Zhenjiang, Social Development Foundation of Zhenjiang (SH2013042, SH2013082, SH2014044, SH2014086), and Jiangsu Government Scholar-ship for Overseas Studies.
Disclosure of conflict of interest
None.
References
- 1.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Smith M, Barnett M, Bassan R, Gatta G, Tondini C, Kern W. Adult acute myeloid leukaemia. Crit Rev Oncol Hematol. 2004;50:197–222. doi: 10.1016/j.critrevonc.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152. [DOI] [PubMed] [Google Scholar]
- 4.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD Cancer and Leukemia Group B (CALGB 8461) Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 5.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 6.Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002;21:3475–3495. doi: 10.1038/sj.onc.1205322. [DOI] [PubMed] [Google Scholar]
- 7.Haferlach T. Molecular genetic pathways as therapeutic targets in acute myeloid leukemia. Hematol Am Soc Hematol Educ Program. 2008:400–411. doi: 10.1182/asheducation-2008.1.400. [DOI] [PubMed] [Google Scholar]
- 8.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 9.Gulley ML, Shea TC, Fedoriw Y. Genetic tests to evaluate prognosis and predict therapeutic response in acute myeloid leukemia. J Mol Diagn. 2010;12:3–16. doi: 10.2353/jmoldx.2010.090054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein ID: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 11.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 12.Kubelac MP, Fetica B, Vlad IC, Fulop A, Popa A, Achimas-Cadariu P. The role of inhibitor of DNA-binding 1 (ID-1) protein and angiogenesis in serous ovarian cancer. Anticancer Res. 2014;34:413–416. [PubMed] [Google Scholar]
- 13.Ponz-Sarvisé M, Castañón E, Panizo-Santos A, Redrado M, López I, Rosell D, Gil-Aldea I, Calvo A, Nguewa PA, Gil-Bazo I. Differential tumor expression of inhibitor of differentiation-1 in prostate cancer patients with extreme clinical phenotypes and prognostic implications. Clin Genitourin Cancer. 2014;12:87–93. doi: 10.1016/j.clgc.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Wazir U, Jiang WG, Sharma AK, Newbold RF, Mokbel K. The mRNA expression of inhibitors of DNA binding-1 and -2 is associated with advanced tumour stage and adverse clinical outcome in human breast cancer. Anticancer Res. 2013;33:2179–2183. [PubMed] [Google Scholar]
- 15.Luo KJ, Wen J, Xie X, Fu JH, Luo RZ, Wu QL, Hu Y. Prognostic relevance of Id-1 expression in patients with resectable esophageal squamous cell carcinoma. Ann Thorac Surg. 2012;93:1682–1688. doi: 10.1016/j.athoracsur.2012.01.102. [DOI] [PubMed] [Google Scholar]
- 16.Ponz-Sarvisé M, Nguewa PA, Pajares MJ, Agorreta J, Lozano MD, Redrado M, Pio R, Behrens C, Wistuba II, García-Franco CE, García-Foncillas J, Montuenga LM, Calvo A, Gil-Bazo I. Inhibitor of differentiation-1 as a novel prognostic factor in NSCLC patients with adenocarcinoma histology and its potential contribution to therapy resistance. Clin Cancer Res. 2011;17:4155–4166. doi: 10.1158/1078-0432.CCR-10-3381. [DOI] [PubMed] [Google Scholar]
- 17.Ding R, Han S, Lu Y, Guo C, Xie H, Zhang N, Song Z, Cai L, Liu J, Dou K. Overexpressed Id-1 is associated with patient prognosis and HBx expression in hepatitis B virus-related hepatocellular carcinoma. Cancer Biol Ther. 2010;10:299–307. doi: 10.4161/cbt.10.3.12454. [DOI] [PubMed] [Google Scholar]
- 18.Yang HY, Liu HL, Liu GY, Zhu H, Meng QW, Qu LD, Liu LX, Jiang HC. Expression and prognostic values of Id-1 and Id-3 in gastric adenocarcinoma. J Surg Res. 2011;167:258–266. doi: 10.1016/j.jss.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Dong Z, Liu S, Zhou C, Sumida T, Hamakawa H, Chen Z, Liu P, Wei F. Overexpression of Id-1 is associated with tumor angiogenesis and poor clinical outcome in oral squamous cell carcinoma. Oral Oncol. 2010;46:154–157. doi: 10.1016/j.oraloncology.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Straume O, Akslen LA. Strong expression of ID1 protein is associated with decreased survival, increased expression of ephrin-A1/EPHA2, and reduced thrombospondin-1 in malignant melanoma. Br J Cancer. 2005;93:933–938. doi: 10.1038/sj.bjc.6602792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KT, Lee YW, Lee JK, Choi SH, Rhee JC, Paik SS, Kong G. Overexpression of Id-1 is significantly associated with tumour angiogenesis in human pancreas cancers. Br J Cancer. 2004;90:1198–1203. doi: 10.1038/sj.bjc.6601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindl M, Oberhuber G, Obermair A, Schoppmann SF, Karner B, Birner P. Overexpression of Id-1 protein is a marker for unfavorable prognosis in early-stage cervical cancer. Cancer Res. 2001;61:5703–5706. [PubMed] [Google Scholar]
- 23.Tang R, Hirsch P, Fava F, Lapusan S, Marzac C, Teyssandier I, Pardo J, Marie JP, Legrand O. High ID1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009;114:2993–3000. doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- 24.Damm F, Wagner K, Görlich K, Morgan M, Thol F, Yun H, Delwel R, Valk PJ, Löwenberg B, Heuser M, Ganser A, Krauter J. ID1 expression associates with other molecular markers and is not an independent prognostic factor in cytogenetically normal acute myeloid leukaemia. Br J Haematol. 2012;158:208–215. doi: 10.1111/j.1365-2141.2012.09144.x. [DOI] [PubMed] [Google Scholar]
- 25.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 26.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 27.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Lin J, Yang J, Qian J, Qian W, Yao DM, Deng ZQ, Liu Q, Chen XX, Xie D, An C, Tang CY. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37:1642–1647. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 muta-tions in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91:519–525. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Qian J, Sun A, Lin J, Xiao G, Yin J, Chen S, Wu D. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46:579–583. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Qian J, Yao DM, Lin J, Qian W, Wang CZ, Chai HY, Yang J, Li Y, Deng ZQ, Ma JC, Chen XX. U2AF1 Mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7:e45760. doi: 10.1371/journal.pone.0045760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen XM, Lin J, Yang J, Yao DM, Deng ZQ, Tang CY, Xiao GF, Yang L, Ma JC, Hu JB, Qian W, Qian J. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7:6832–40. [PMC free article] [PubMed] [Google Scholar]
- 34.Suh HC, Leeanansaksiri W, Ji M, Klarmann KD, Renn K, Gooya J, Smith D, McNiece I, Lugthart S, Valk PJ, Delwel R, Keller JR. ID1 immortalizes hematopoietic progenitors in vitro and promotes a myeloproliferative disease in vivo. Oncogene. 2008;27:5612–5623. doi: 10.1038/onc.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigten J, Breems-de Ridder MC, Erpelinck-Verschueren CA, Nikoloski G, van der Reijden BA, van Wageningen S, van Hennik PB, de Witte T, Löwenberg B, Jansen JH. ID1 and ID2 are retinoic acid responsive genes and induce a G0/G1 accumulation in acute promyelocytic leukemia cells. Leukemia. 2005;19:799–805. doi: 10.1038/sj.leu.2403699. [DOI] [PubMed] [Google Scholar]
- 36.Ryu B, Kim DS, DeLuca AM, Healey MA, Dunlap S, Fackler MJ, Herman J, Alani RM. ID1 expression is transcriptionally regulated in radial growth phase melanomas. Int J Cancer. 2007;121:1705–1709. doi: 10.1002/ijc.22875. [DOI] [PubMed] [Google Scholar]
- 37.Yu WP, Scott SA, Dong WF. Induction of ID1 expression and apoptosis by the histone deacetylase inhibitor (trichostatin A) in human acute myeloid leukaemic cells. Cell Prolif. 2008;41:86–97. doi: 10.1111/j.1365-2184.2007.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothschild SI, Tschan MP, Federzoni EA, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene. 2012;31:4221–4232. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 39.Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol. 2012;7:1069–1077. doi: 10.1097/JTO.0b013e31824fe976. [DOI] [PubMed] [Google Scholar]
- 40.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, Andreeff M, Croce CM. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]


