Abstract
Background: To investigate the inhibitory effect of midkine-binding peptides on human umbilical vein endothelial cells (HUVECs) proliferation and angiogenesis of xenograft tumor. Methods: The midkine-binding peptides were panned by Ph.D.-7™ Phage Display Peptide Library Kit, and the specific binding activities of positive clones to target protein were examined by phage ELISA. The effect of midkine-binding peptides on proliferation of HUVECs was confirmed by MTT test. The xenograft tumor model was formed in BALB/c mice with the murine hepatocarcinoma cells H22 (H22). Microvessel density (MVD) was analyzed by immunohistochemistry of factor VIII staining. Results: Midkine-binding peptides have the inhibitory effects on tumor angiogenesis, a proliferation assay using human umbilical vein endothelial cells (HUVECs) indicated that particular midkine-binding peptides significantly inhibited the proliferation of the HUVECs. Midkine-binding peptides were also observed to efficiently suppress angiogenesis induced by murine hepatocarcinoma H22 cells in BALB/c nude mice. Conclusion: The midkine-binding peptides can inhibit solid tumor growth by retarding the formation of new blood vessels. The results indicate that midkine-binding peptides may represent potent anti-angiogenesis agents in vivo.
Keywords: Midkine, angiogenesis, peptide
Introduction
The realization that angiogenesis is an essential step for tumor growth and initiation of metastasis [1] has led to significant interest in the discovery of angiogenic factors [2,3] and the development of drugs that target these molecules. Several inhibitors of angiogenesis have been identified, including angiostatin [4] and endostatin [5]. These agents are able to inhibit tumor growth by suppressing angiogenesis in xenograft models.
Midkine (MK) is a secreted heparin-binding growth factor identified as a product of a retinoic acid response gene [6,7]. The pathophysiological effects of midkine include enhanced plasminogen activator activity [8], oncogenic transformation of fibroblasts [9], antiapoptotic activity [10] and angiogenic activity [11]. MK is overexpressed in a variety of cancers such as esophageal [12], gastric [13], colon [14], pancreatic [15], lung [16] and breast cancers [17], whereas its expression is usually low or undetectable in normal adult tissues. Additionally, MK is expressed in bladder cancer and overexpression correlates with a poor outcome in patients with invasive cancers [18].
There is evidence that MK plays an important role in angiogenesis, which is a crucial event for tumor development and progression. It has been reported that enhanced tumor growth in MCF-7 breast carcinoma cells due to high levels of MK expression correlate with an increase in vascular density and endothelial proliferation. Moreover, midkine was found to induce a strong angiogenic response in a rabbit corneal assay [19]. An anti-midkine antibody was shown to inhibit the growth of Wilm’s tumor cells in vitro [20], and antisense oligo DNA to MK inhibits the growth of colon carcinoma cells in vivo [21]. Consequently, MK represents a suitable molecular target of tumor therapy.
We have confirmed that antisense oligo DNA to MK can significantly inhibit tumor growth in a human hepatocellular carcinoma (HCC) model [22] and suppress the angiogenesis both in HepG2-induced CAM and in situ human HCC tissues [23]. In contrast to large molecules such as antibodies and antisense oligo DNA, peptides are known to exhibit less toxicity and posses better pharmacokinetic properties [24]. In this study, inhibition of tumor growth and suppression of angiogenesis in a murine hepatocellular carcinoma model using peptides that bound to MK was examined.
Materials and methods
Cell lines, animals, antibodies and other reagents
Human umbilical vein endothelial cells (HU-VECs) and murine hepatocarcinoma cells H22 (H22) were purchased from American Type Culture Collector (ATCC). All cells were cultured in DMEM medium (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco). Six-week-old female BALB/c nude mice were purchased from Shanghai Bikai Experimental Animal Center and kept in the local central animal facility. The mice were housed under standard conditions and had free access to water and food. Animal procedures were performed in accordance to approved protocols and followed recommendations for proper care and use of laboratory animals. Recombinant human midkine (rh-midkine) was purchased from BioVendor Laboratory Medicine Inc. Anti-M13 phage antibody was purchased from New England Biolabs Inc. (Beverly, MA, USA). Anti-factor VIII antibody was purchased from DAKO (Denmark). All of the other reagents used were analytical grade and commercially available.
Phage peptide library and bacterial strain
The Ph.D.-7TM Phage Display Peptide Library Kit (2×1013 pfu/ml), Ph.D.-C7C™ Phage Display Peptide Library Kit (2×1013 pfu/ml) and the host bacterial strain E. coli ER2738 were purchased from New England Biolabs Inc.
Phage selection with rh-midkine and the amplification of the peptide library
The biopanning and amplification procedures were essentially performed according to procedures provided by the manufacturer. Three rounds of biopanning were performed. Briefly, 100 µl of rh-midkine (100 µg/ml in 0.1 mol/L NaHCO3, pH 8.6) was coated onto 96-well microtiter plates and incubated overnight at 4°C with gentle agitation in a humidified container. Following blocking with an appropriate buffer (TBS, pH 7.2, containing 5% BSA), each well was filled with the phage library (100 µl, approximately 2.0×1010 pfu), which was 10-fold diluted with TBST (TBS, 0.05% Tween-20) and incubated at room temperature for 1 h. To remove non-specific phage binding, the wells were washed 10 times with TBST (TBS, 0.1% Tween-20) during the first round of biopanning and the concentration of Tween-20 was raised to 0.5% in the second and third rounds of biopanning. The bound phage were eluted with 100 µl of 0.1 mol/L glycine (pH 2.2) at room temperature for 15 min and neutralized with 15 µl of 1 mol/L Tris-HCl (pH 9.1). The titer of the phage-solution was determined by infecting E. coli ER2738, followed by counting surviving colonies on LB/IPTG/X-gal agar plates after incubation at 37°C overnight. The rest of the solution was used for amplification. The amplified phage particles were purified using PEG/NaCl.
Peptide-protein binding assays
The binding activity of monoclonal phages for rh-midkine was detected by ELISA, which was essentially performed according to the manufacturer’s instructions. Briefly, 100 µl of rh-midkine (100 µg/ml in 0.1 mol/L NaHCO3, pH 8.6) was coated onto 96-well microtiter plates and incubated overnight at 4°C with gentle agitation in a humidified container. Following blocking (TBS, pH 7.2, containing 5% BSA), approximately 2×1010 pfu amplified monoclonal phages were added to each well and then incubated with rh-midkine for two hours at room temperature. The wells were washed with TBST (TBS, 0.1% Tween-20) five times and the amount of bound phages was detected with horseradish peroxidase (HRP)-conjugated anti-M13 phage antibody (1:5000, Pharmacia #27-9411-01). After the addition of the substrate, the antibody reaction was analyzed in a microtiter plate reader at 415 nm. The original phage library without selection was used as the negative control.
Peptides sequencing
Single-stranded DNA (ssDNA) was prepared from the identified phage clones as described in the kit guidelines and sequencing was carried out by Invitrogen. The primer used for sequencing was: 5’-CCCTCATGTTAGCGTAA-CG-3’. The corresponding amino acid sequences were deduced from the DNA sequences and multiple sequence alignments were performed using the BLAST software package to determine which peptides were related by sequence.
Peptide synthesis
Peptides were synthesized chemically by the standard N-(9-fluorenyl-methoxycarbonyl) (Fmoc) protocol and purified by reversed-phase high-performance liquid chromatography (RH-HPLC) (Zhongtai, Hangzhou, China). The purity of all products was > 95%.
Cell proliferation assay
HUVEC cells were seeded into 96-well plates (3000 cells/well) in 100 µl DMEM medium. The cells were treated with or without the peptides at concentrations of 15, 50 and 150 µg/ml for 72 h, 10 µl of MTT stock solution (5 mg/ml) was added into each well and incubated for another 4 h at 37°C. The solution was removed and 150 µl DMSO was added per well. After 10 min incubation at 37°C, the optical density at 570 nm was measured using an enzyme-linked immunosorbent assay reader (Bio-Rad Model 550).
Xenografts in BALB/c mice and treatment
H22 cells were suspended in DMEM at a concentration of 5×106 cells/ml. 100 µl of the suspension was injected into the subcutaneous on the right side of the back of each mouse. The mice were randomly divided into eight groups (12 mice per group). The peptides and 5-FU were injected daily into the peritoneal cavum for the seven treated groups following inoculation. The same volume of a PBS solution was injected into the mice of the negative control group. After twenty-one days of treatment, the mice were sacrificed and tumors were removed and weighted. The percentage of tumor growth inhibition was calculated as: Inhibitory rate (%) = (Wcontrol-Wtreat)/Wcontrol×100.
Immunohistochemistry analysis for microvessel formation
Tumors were fixed and frozen in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC). 5-µm-thick sections were cut and stained with hematoxylin and eosin for histopathological analysis. To analyze the microvessel formation in tumors, sections were stained with anti-factor VIII monoclonal antibody (DAKO Corp., Carpinteria, CA) and subsequently with the avidin-biotin-peroxidase (ABC) method. Positively stained vascular endothelial cells (brown) were visualized and imaged using a digital camera attached to an Olympus microscope. Microvessel density (MVD) was determined according to methods described previously [25]. Briefly, regions of the highest vessel density (“hot spot” regions) were scanned at low magnification (×40-100) and counted at higher magnification (×200). Three such “hot spot” fields were counted in each tumor section and the mean microvessel density value was recorded. Any endothelial cell or endothelial cell clusters that was clearly separated from adjacent microvessels were considered a single, countable microvessel. Positively stained vascular endothelial cells were visualized and imaged using a Magnifire camera (Olympus, Melville, NY) attached to an Olympus Provis microscope.
Statistical analysis
Data were described as the mean ± SD for general characteristics of the subjects. The SPSS13.0 software was used for all statistical analyses. Two-tailed P values with composite results P < 0.05 were considered statistically significant.
Results
Specific enrichment of positive phages
To enrich rh-midkine binding phages from the Ph.D.-C7C and Ph.D.-7 phage display libraries, three rounds of selection with the rh-midkine protein were performed. The enrichment was determined by the use of the output/input ratio of phages after each round of selection. The ratio increased approximately 10-fold after the second round of selection. After the third round of selection of the Ph.D.-C7C and Ph.D.-7 phage display libraries, the output/input ratio of phages increased 125-fold and 250-fold respectively, which indicated that both linear and cyclic peptide libraries had an obvious enrichment for the specific binding of phages to rh-midkine (Table 1).
Table 1.
Enrichment of phages for each round of selection from phage library
| rounds | Ph.D.-C7C™ phage display library | Ph.D.-7™ phage display library | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Selected phages (pfu)a (input) | Eluted phages (pfu) (output) | Ratio (%) | Selected phages (pfu) (input) | Eluted phages (pfu) (output) | Ratio (%) | |
| I | 1×1011 | 4×105 | 4×10-4 | 1×1011 | 2×105 | 2×10-4 |
| II | 3×1010 | 1×106 | 3.3×10-3 | 3×1010 | 6×106 | 2×10-3 |
| III | 2×1011 | 1×108 | 5×10-2 | 2×1011 | 1×108 | 5×10-2 |
pfu: Plaque forming units.
Estimation of binding specificities of selected phage clones
The positive clones were chosen from the samples after the third round of selection with the rh-midkine protein and the specificity was examined by phage ELISA. 29 clones bound to the target protein and were chosen for further sequencing (Figure 1).
Figure 1.
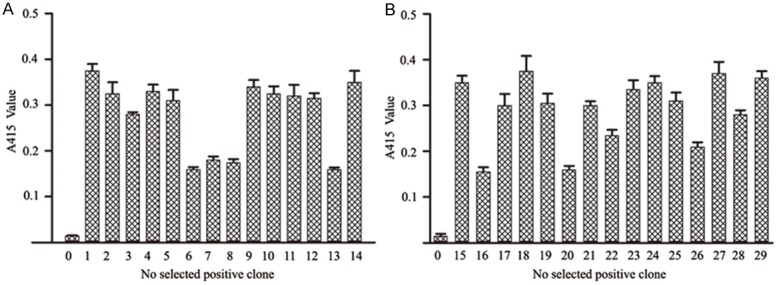
Specific binding of selected phage clones to the anti-M13 antibody. 29 clones bound to the target protein; there were 14 cyclic peptides (No. 1-14) (A) and 15 linear peptides (No. 15-29) (B). The original phage peptide library without selection, represented as No. 0, the original library without selection was used as negative control. All of the treatments were performed in triplicate.
Analyses of sequences of positive phage clones
The ssDNA was prepared for positive phage clones and sequenced and the amino acid sequences of the midkine-binding peptides were deduced from the DNA sequences. Analysis of the 29 clones from the affinity-selected phage population revealed that the majority of clones fell into six groups with different sequences (Table 2). The six peptides were designated and synthesized (Table 3).
Table 2.
Amino acid sequences deduced from the DNA sequences
| Groups | Clones | Sequences | Frequency |
|---|---|---|---|
| Cyclic peptides | Phage 1, phage 2, phage 3, phage 4, phage 10, phage 11 | ACVIHWDFIC | 6 |
| Phage 5, phage 12 | ACTWLHWWAC | 2 | |
| Phage 9, phage 14 | ACTSTAMDNC | 2 | |
| Phage 8 | ACNSHTDTNC | 1 | |
| Phage 13 | ACMPKVGLNC | 1 | |
| Phage 6 | ACKNTFANVC | 1 | |
| Phage 7 | ACTIPRMPYC | 1 | |
| Linear peptides | Phage 17, phage 19, phage 21, phage 23, phage 24 | LLWYDEI | 5 |
| Phage 15, phage 18, phage 27, phage 29 | HAIYPRH | 4 | |
| Phage 25, phage 28 | PVPRSRP | 2 | |
| Phage 16 | PISSYQR | 1 | |
| Phage 20 | RPPGYIP | 1 | |
| Phage 22 | VPFYSHS | 1 | |
| Phage 26 | RPPGYI P | 1 |
Table 3.
Name and sequence of the midkine-binding peptides
| Name | Amino acid sequence |
|---|---|
| MK-P1 | CVIHWDFIC |
| MK-P2 | CTWLHWWAC |
| MK-P3 | CTSTAMDNC |
| MK-P4 | LLWYDEI |
| MK-P5 | HAIYPRH |
| MK-P6 | PVPRSRP |
Effects of midkine-binding peptides treatment on HUVEC growth
To investigate the activity of midkine-binding peptides on the vascular endothelial cell proliferation, we analyzed the growth of HUVECs treated with midkine-binding peptides. From the results presented in Figure 2, the midkine-binding peptides were observed to significantly inhibit the proliferation of HUVECs. The inhibition level ranged from 15.6 to 76.6%. Among the six peptides, MK-P3 showed the highest inhibitory activity. The percentage of the inhibition was calculated as:
Figure 2.
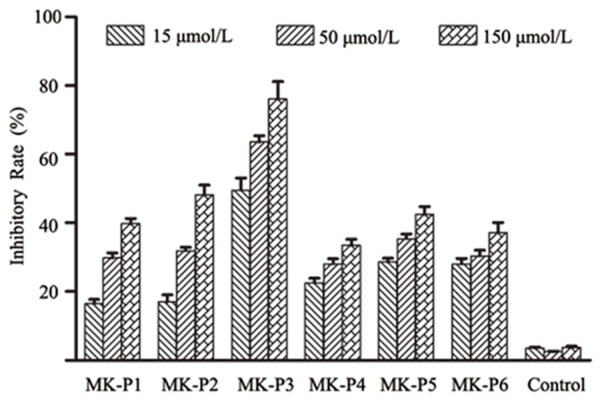
Effects of midkine-binding peptides on the proliferation of HUVECs. Midkine-binding peptides significantly inhibited the proliferation of HUVECs. Among the six peptides examined, MK-P3 showed the highest inhibitory activity, the non-specific peptide was examined as the negative control. All of the treatments were repeated six times.
Inhibitory rate (%) = (OD570control-OD570treat)/OD570control×100
Effects of midkine-binding peptides treatment on xenograft tumor growth
Treatment with the midkine-binding peptides resulted in a significant reduction in the weight of tumors when compared to PBS-treated mice (Table 4). The inhibitory efficacy of MK-P3 was 56.4%, which was the highest. Moreover, the 5-FU (5-Fluorouracil) group (positive control) had an inhibitory efficacy of 45.4% when compared to the PBS treated group.
Table 4.
Inhibitory effects of midkine-binding peptides on xenograft tumors
| Groups | n | Tumor weight (g) | Inhibitory rate (%) |
|---|---|---|---|
| PBS | 12 | 3.099 ± 0.452a | - |
| Control | 12 | 2.897 ± 0.316 | 6.5 |
| 5-FU 10 mg kg-1 | 12 | 1.692 ± 0.563* | 45.4 |
| MK-P1 0.5 mg kg-1 | 12 | 2.42 ± 0.56* | 22.0 |
| MK-P2 0.5 mg kg-1 | 12 | 2.31 ± 0.645* | 25.4 |
| MK-P3 0.5 mg kg-1 | 12 | 1.35 ± 0.88** | 56.4 |
| MK-P4 0.5 mg kg-1 | 12 | 2.31 ± 0.49* | 25.5 |
| MK-P5 0.5 mg kg-1 | 12 | 2.26 ± 0.34** | 27.0 |
| MK-P6 0.5 mg kg-1 | 12 | 2.50 ± 0.16* | 19.3 |
Data are presented as means (± SD), n = 12;
P < 0.05, compared with PBS control, the negative control group was treated with non-specific peptide;
P < 0.01, compared with PBS control, the negative control group was treated with non-specific peptide.
Effects of midkine-binding peptides treatment on tumor angiogenesis
Sections of xenograft tumors from the mice in all eight groups were stained for endothelial cell specific markers to detect the number of endothelial cells as a measure of tumor angiogenesis. A single microvessel was defined as any immunohistochemistry stained endothelial cell distinguished from adjacent tumor cells and other connective tissue elements. A representative result of immunostaining is shown in Figure 3. A high density of microvessels was observed in the untreated tumor sample, whereas treatment with the midkine-binding peptides resulted in a significant reduction in vascularization (Table 5; Figure 3).
Figure 3.
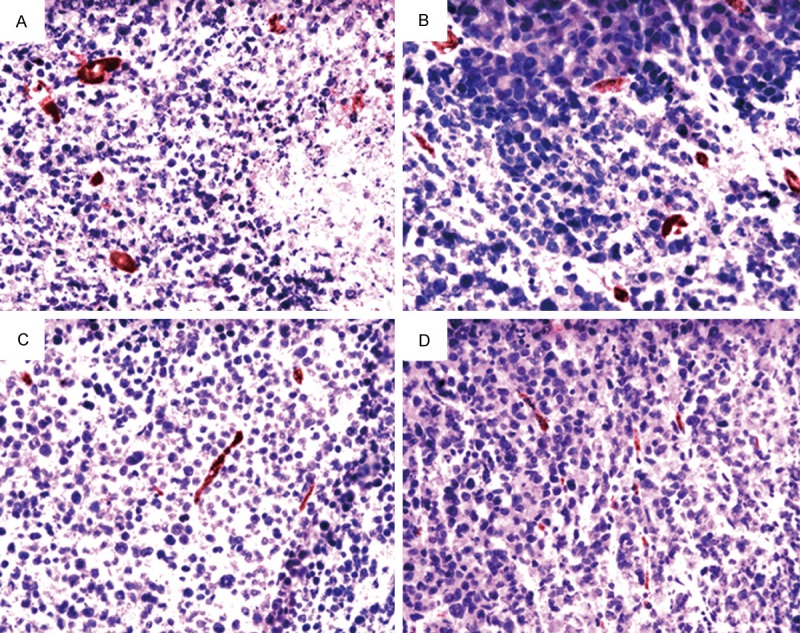
Effects of midkine-binding peptides treatment on tumor angiogenesis. The representative images of immunohistochemistry stained for microvessels (factor VIII) in a xenograft tumor (×200). A. Sections of tumors from PBS treatment. B. Sections of tumors from non-specific peptide treatment as negative control. C. Sections of tumors following treatment with MK-P3. D. Sections of tumors from treatment with MK-P.
Table 5.
Effects of midkine-binding peptides on microvessel density in tumor xenografts
| Groups | MVD |
|---|---|
| MK-P1 0.5 mg kg-1 | 18.8 ± 2.9* |
| MK-P2 0.5 mg kg-1 | 19.2 ± 2.2* |
| MK-P3 0.5 mg kg-1 | 15.3 ± 1.2** |
| MK-P4 0.5 mg kg-1 | 17.6 ± 3.0* |
| MK-P5 0.5 mg kg-1 | 15.7 ± 3.2** |
| MK-P6 0.5 mg kg-1 | 16.1 ± 2.4** |
| PBS | 25.1 ± 0.7 |
| Control | 22.7 ± 1.6 |
†Data are presented as means ± SD, n=12;
P < 0.05, compared with PBS control, the negative control group was treated with non-specific peptide;
P < 0.01, compared with PBS control, the negative control group was treated with non-specific peptide.
Discussion
Angiogenesis is a process of remodeling of a primitive vascular network into mature vasculature through sprouting, branching and differential growth of blood vessels. This complex process involves endothelial cell proliferation, chemotactic migration and functional maturation, as well as differential recruitment of supporting cells. Normal or physiological angiogenesis occurs during embryonic development, wound healing, menstruation and pregnancy. Angiogenesis is controlled and regulated by many molecules including stimulators and inhibitors. An appropriate balance between stimulators and inhibitors is pivotal for the maintenance and regulation of angiogenesis. Tumor-associated angiogenesis is critical for tumor growth and is controlled by a balance between pro- and anti-angiogenic factors. Growing evidence demonstrates the heterogeneity of tumor angiogenesis, probably arising from the vastly different microenvironments of individual tumors [26-30]. Due to the heterogeneous nature of tumor angiogenesis, effective anti-angiogenic therapies should be optimized and may require interference with multiple angiogenic pathways. A large number of natural proteins are angiogenic inhibitors and various inhibitors of angiogenesis exhibit antitumor effects [31,32]. Selection of target proteins is very important in designing and screening anti-angiogenic peptides. Novel targets could provide unique characteristics and new desirable features for further development into peptide drugs.
In this paper, we initially analyzed the effects of midkine-binding peptides on endothelial cell proliferation. The results indicated that midkine-binding peptides could retard HUVEC growth, suggesting that these peptides may also inhibit angiogenesis in vivo. Furthermore, we developed an in situ mouse hepatocellular carcinoma model to determine the influence of the midkine-binding peptides on tumor growth and angiogenesis in vivo. The results indicated that midkine-binding peptides significantly inhibited the mouse hepatocellular carcinoma model. Furthermore, midkine-binding peptides also suppressed tumor angiogenesis.
It is well established that tumor growth and metastasis depend on the induction of a new blood supply [33,34]. Angiogenic activity, as determined by MVD, has been shown to correlate with an unhealthy prognosis in a number of solid tumors [35-37]. Therefore, anti-angiogenic agents targeting either of these factors or vascular endothelial cells are promising therapeutic modalities as tumor dormancy therapies [38]. In the present study, we have addressed the potential therapeutic role of midkine-binding peptides on angiogenesis. Significant inhibition of angiogenesis was achieved by the midkine-binding peptide MK-P3, indicating that MK-P3 represents a possible effective anti-angiogenesis agent.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 30772534), the Major Science and Technology Project Fund of Huzhou City (No. 2005GS01), and the Natural Science Foundation of Zhejiang (No. Y2111181 and No. LQ13H160019).
Disclosure of conflict of interest
None.
References
- 1.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 2.Aiello L. Keeping in touch with angiogenesis. Nat Med. 2000;6:379–381. doi: 10.1038/74633. [DOI] [PubMed] [Google Scholar]
- 3.Rosen L. Antiangiogenic strategies and agents in clinical trials. Oncologist. 2000;5(Suppl 1):20–27. doi: 10.1634/theoncologist.5-suppl_1-20. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, Lane WS, Cao Y, Sage EH, Folkman J. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79:315–28. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 5.O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 6.Tomomura M, Kadomatsu K, Matsubara S, Muramatsu T. A retinoic acid-responsive gene, MK, found in the teratocarcinoma system. J Biol Chem. 1990;265:10765–10770. [PubMed] [Google Scholar]
- 7.Kadomatsu K, Huang R, Suganuma T, Murata F, Muramatsu T. A retinoic acid responsive gene, MK, found in the teratocarcinoma system is expressed in a spatially and temporally controlled manner during mouse embryogenesis. J Cell Biol. 1990;110:607–616. doi: 10.1083/jcb.110.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima S, Imui T, Muramatsu H, Kimura T, Sakakiba S, Muramatsu T. Midkine is a heat and acid stable polypeptide capable of enhancing plasminogen activator activity and neurite outgrowth extension. Biochen Biophys Res Commun. 1995;216:574–581. doi: 10.1006/bbrc.1995.2661. [DOI] [PubMed] [Google Scholar]
- 9.Kadomatsu K, Hagihara M, Akhter S, Fan QW, Muramatsu H, Muramatsu T. Midkine induces the transformation of NIH3T3 cells. Br J Cancer. 1997;75:354–359. doi: 10.1038/bjc.1997.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owada K, Sanjo N, Kobayashi T, Mizusawa H, Muramatsu T, Michikawa M. Midkine inhibits caspase-dependent apoptosis via the activation of mitogen-activated protein kinase and phosphatidylinositol 3-kinase in cultures neurons. J Neurochem. 1999;73:2084–2092. [PubMed] [Google Scholar]
- 11.Choudhuri R, Zhang HT, Donnini S, Ziche M, Bicknell R. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 1997;57:1814–1819. [PubMed] [Google Scholar]
- 12.Miyauchi M, Shimada H, Kadomatsu K, Muramatsu T, Matsubara S, Tagawa M. Frequent expression of midkine gene in esophgeal cancer suggests a potential usage of its promoter for suicide gene therapy. Jpn J Cancer Res. 1999;90:469–475. doi: 10.1111/j.1349-7006.1999.tb00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aridome K, Tsutsui J, Takao S, Kadomatsu K, Ozawa M, Aikou T, Muramatsu T. Increased midkine gene expression in human gastrointestinal cancers. Jpn J Cancer Res. 1995;86:655–661. doi: 10.1111/j.1349-7006.1995.tb02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu T, Kadomatsu K. Expression of midkine in the early stage of carcinogenesis in human colorectal cancer. Br J Cancer. 1999;79:179–184. doi: 10.1038/sj.bjc.6690030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu T, Kadomatsu K. The expression of truncates MK in human tumors. Biochem Biophys Res Commun. 1996;219:256–260. doi: 10.1006/bbrc.1996.0214. [DOI] [PubMed] [Google Scholar]
- 16.Garver RJ, Chan CS, Milner PG. Reciprocal expression of pleiotropin and midkine in normal versus malignant lung tissues. Am J Respir Cell Mol Biol. 1993;9:463–466. doi: 10.1165/ajrcmb/9.5.463. [DOI] [PubMed] [Google Scholar]
- 17.Garver RJ, Radford DM, Donis-Keller H, Wick MR, Milner PG. Midkine and pleiotrophin expression in normal and malignant breast tissue. Cancer. 1994;74:1584–1590. doi: 10.1002/1097-0142(19940901)74:5<1584::aid-cncr2820740514>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien T, Cranston D, Fuggle S, Bicknell R, Harris AL. The angiogenic factor midkine is expressed in bladder cancer, and overexpression correlates with a poor outcome in patients with invasive cancers. Cancer Res. 1996;56:2515–2518. [PubMed] [Google Scholar]
- 19.Choudhuri R, Zhang HT, Donnini S, Ziche M, Bichnell R. An angiogenic role for neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 1997;57:1841–1849. [PubMed] [Google Scholar]
- 20.Muramatsu H. Shirahama H, Yonezawa S, Maruta H, Muramatsu T. Midkine a retinoic acid-inducible growth/differentiation factor: immunochemical evidence for the function and distribution. Dev Biol. 1993;159:392–402. doi: 10.1006/dbio.1993.1250. [DOI] [PubMed] [Google Scholar]
- 21.Takei Y, Kadomatsu K, Matsuo S, Itoh H, Nakazawa K, Kubota S, Muramatsu T. Antisense oligodeoxynucleotide targeted to Midkine, a heparin-binding growth factor, suppresses tumorigenicity of mouse rectal carcinoma cells. Cancer Res. 2001;61:8486–8491. [PubMed] [Google Scholar]
- 22.Dai LC, Wang X, Yao X, Min LS, Ping JL, He JF. Antisense oligonucleotides targeting midkine inhibit tumor growth in an in situ human hepatocellular carcinoma model. Acta Pharmacol Sin. 2007;28:453–458. doi: 10.1111/j.1745-7254.2007.00532.x. [DOI] [PubMed] [Google Scholar]
- 23.Dai LC, Wang X, Yao X, Lu YL, Ping JL, He JF. Antisense oligonucleotides targeting midkine suppresses in vivo angiogenesis. World J Gastroenterol. 2007;13:1208–1213. doi: 10.3748/wjg.v13.i8.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khandare JJ, Minko T. Antibodies and peptides in cancer therapy. Crit Rev Ther Drug Carrier Syst. 2006;23:401–435. doi: 10.1615/critrevtherdrugcarriersyst.v23.i5.20. [DOI] [PubMed] [Google Scholar]
- 25.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis-correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 26.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 27.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 29.Jung YD, Ahmad SA, Akagi Y, Takahashi Y, Liu W, Reinmuth N, Shaheen RM, Fan F, Ellis LM. Role of the tumor microenvironment in mediating response to anti-angiogenic therapy. Cancer Metastasis Rev. 2000;19:147–157. doi: 10.1023/a:1026510130114. [DOI] [PubMed] [Google Scholar]
- 30.Eberhard A, Kahlert S, Goede V, Hemmerlein B, Plate KH, Augustin HG. Heterogeneity of angiogenesis and blood vessel maturation in human tumors: implications for antiangiogenic tumor therapies. Cancer Res. 2000;60:1388–1393. [PubMed] [Google Scholar]
- 31.Ruggeri B, Singh J, Gingrich D, Angeles T, Albom M, Yang S, Chang H, Robinson C, Hunter K, Dobrzanski P, Jones-Bolin S, Pritchard S, Aimone L, Klein-Szanto A, Herbert JM, Bono F, Schaeffer P, Casellas P, Bourie B, Pili R, Isaacs J, Ator M, Hudkins R, Vaught J, Mallamo J, Dionne C. CEP-7055: a novel, orally active pan inhibitor of vascular endothelial growth factor receptor tyrosine kinases with potent antiangiogenic activity and antitumor efficacy in preclinical models. Cancer Res. 2003;63:5978–5991. [PubMed] [Google Scholar]
- 32.Bergers G, Javaherian K, Lo KM, Folkman J, Hanahan D. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science (Wash, DC) 1999;284:808–812. doi: 10.1126/science.284.5415.808. [DOI] [PubMed] [Google Scholar]
- 33.Plate KH, Breier G, Risau W. Molecular mechanisms of developmental and tumor angiogenesis. Brain Pathol. 1994;4:207–218. doi: 10.1111/j.1750-3639.1994.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Folkman J. Pattern and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 35.Brawer MK, Deering RE, Brown M, Preston SD, Bigler SA. Predictors of pathologic stage in prostatic carcinoma. The role of neovascularity. Cancer. 1994;73:678–87. doi: 10.1002/1097-0142(19940201)73:3<678::aid-cncr2820730329>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Delahunt B, Bethwaite PB, Thornton A. Prognostic significance of microscopic vascularity for clear cell renal cell carcinoma. Br J Urol. 1997;80:401–404. doi: 10.1046/j.1464-410x.1997.00374.x. [DOI] [PubMed] [Google Scholar]
- 37.Bochner BH, Cote RJ, Weidner N, Groshen S, Chen SC, Skinner DG, Nichols PW. Angiogenesis in bladder cancer: relationship between microvessl density and tumor prognosis. J Natl Cancer Inst. 1995;87:1603–1612. doi: 10.1093/jnci/87.21.1603. [DOI] [PubMed] [Google Scholar]
- 38.Streeter EH, Harris AL. Angiogenesis in bladder cancer--prognostic marker and target for future therapy. Surg Oncol. 2002;11:85–100. doi: 10.1016/s0960-7404(02)00013-0. [DOI] [PubMed] [Google Scholar]


