Abstract
Background: microRNA-26b (miR-26b) is reported to be downregulated in many human malignancies and function as a tumor suppressor. However, the roles of miR-26b expression in cervical cancer progression are unclear. The aim of this study was to investigate the clinicopathological or prognostic significance of miR-26b in human cervical cancer. Methods: A cohort of 88 paired of cervical cancer and the adjacent normal cervical epithelial tissues were collected. Quantitative RT-PCR (qRT-PCR) assay was used to detect the expression of miR-26b and its correlations with clinicopathological factors were statistically analyzed. Finally, the survival was assessed by the Kaplan-Meier method and proportional hazards model. Results: The expression level of miR-26b in cervical cancer tissues was significantly lower than that in the adjacent normal cervical tissues (P<0.001). Reduced miR-26b was observed to be significantly correlated with advanced FIGO stage, higher incidence of lymph node metastasis and recurrence of cervical cancer patients (P=0.002, 0.036 and 0.029, respectively). In addition, patients with low-miR-26b expression showed poorer recurrence-free survival (RFS) and overall survival (OS) than those with high-miR-26b expression (P=0.0043 and 0.0015, respectively). Furthermore, multivariate analyses demonstrated that low miR-26b expression was an independent prognostic factor for predicting the 5-year RFS and OS of cervical cancer patients (P=0.013 and 0.007, respectively). Conclusion: Our results showed that reduced miR-26b was correlated with tumor development and poor prognosis in human cervical cancer. The status of miR-26b expression may be a potential prognostic biomarker for cervical cancer patients.
Keywords: Cervical cancer, microRNA-26b, quantitative RT-PCR, recurrence-free survival, overall survival
Introduction
Cervical cancer remains the second most frequent malignant gynecological neoplasm worldwide. Each year, there are about 500,000 diagnoses of cervical cancer and about 200,000 deaths because of this disease [1]. Up to now, the exact causes of cervical cancer are not fully understood. In addition to infection with high-risk human papillomavirus (HPV), molecular alterations of tumor suppressor genes and/or oncogenes are also imperative for cervical carcinogenesis and progression [2]. Many studies have shown that patient age, FIGO stage, hemoglobin level, tumor size, lymph node metastasis and lymph-blood vessel invasion are independent prognostic factors for survival of cervical cancer patients [3]. However, these clinicopathological factors can not be sufficient to accurately predict prognosis of patients. Thus, identification of novel molecular biomarkers will be helpful to obtain an exact prediction of the presence and prognosis of this disease.
MicroRNAs (miRNAs), a class of small (~22 bp), non-coding RNAs, regulate gene expression at the post-transcriptional level by binding to the 3’-untranslated region (3’-UTR) of target mRNAs and causing translational inhibition and /or mRNA degradation [4,5]. Recently, miRNAs have emerged as major non-coding RNAs that may also be involved in a wide range of pivotal biological processes in both plants and animals, such as cell growth, development, proliferation, differentiation and death [6,7]. Dysregulated miRNAs has been reported to play critical roles in tumorigenesis and tumor progression [8]. Several studies have shown that unique miRNA expression profiles are present in a number of important cancers such as breast, lung, hepatcelluar, and pancreatic cancer [9-12]. The correlations of miRNAs with cervical cancer are increasingly reported. For example, by profiling miRNA expression in 10 early stage invasive squamous cell carcinomas (ISCC) and 10 normal cervical squamous epithelial specimens using TaqMan real-time qu-antitative PCR array methods, Lee’ et al identified 70 miRNAs (68 up-regulated, 2 down-regulated) with significantly different expression in the ISCCs compared with normal samples [13]. Likewise, by using a direct sequencing method to characterize the profiles of miRNAs and other small RNA segments for six human cervical carcinoma cell lines and five normal cervical samples, Lui and his colleagues observed significant expression variation of six miRNAs (let-7b, let-7c, miR-23b, miR-196b, miR-143 and miR-21) between the two groups [14]. By analysis of miRNA expression profiles in cervical cancer tissue and adjacent normal cervical tissue of 13 patients with cervical cancer, Rao’ et al also showed that 18 miRNAs were significantly upregulated (increase of ≥2×), and 19 miRNAs were significantly downregulated (decrease of ≤0.5×) in cervical cancer tissues, which was independent of lymph node involvement, vascular invasion, and pathological differentiation [15]. Also, dysregulated miRNAs are reported to regulate multiple malignant phenotypes of cervical cancer cells, including growth, invasion, chemo- or radioresistance of cervical cancer cells [16-18]. These experimental data revealed the important roles of miRNAs in cervical carcinogenesis.
In this study, we focus on miR-26b and our aim is to investigate the clinical values of this miRNA in cervical cancer. First, qRT-PCR assay was performed to detect the expression of miR-26b in 88 paired of cervical cancer and the adjacent cervical epithelial tissues. Then, the correlations of miR-26b with clinicopathological factors of patients were statistically analyzed. Finally, the Kaplan-Meier method and Cox’s proportional hazards model were used to evaluate the prognostic potential of miR-26b expression in human cervical cancer.
Materials and methods
Patients and tissues
A total of 88 paired of primary cervical cancer tissues and the adjacent normal cervical epithelial tissues from cervical carcinoma patients were collected at the Department of Pathology in Guangzhou General Hospital of Guangzhou Military Command during the period from July 2007 and March 2010. All patients had histologically proven cervical carcinoma and were clinically staged according to the International Federation of Gynaecology and Obstetrics staging criteria (FIGO, 1988). Tumor differentiation was graded following WHO criteria. Written informed consent was obtained from all patients according to the guidelines approved by the institutional research board. All specimens were immediately frozen in liquid nitrogen and stored at -70°C until use. The Guangdong Province Medical Association Society of Medicine’s Ethics Committee approved all aspects of this study in accordance with the Helsinki Declaration.
qRT-PCR detection for miRNA expression
Total RNA isolation from tissues was performed using mirVana miRNA Isolation Kit (Applied Biosystems/Ambion) according to the manufacturer’s protocol. Total miRNA from tissues was extracted by using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer’s instructions. Total RNA (2.0 μg) was used for cDNA synthesis by reverse transcription using M-MLV Reverse Transcriptase (Promega, USA) according to the manufacturer’s instructions. Expressions of mature miR-26b were quantified by miR-qRT PCR using the Hairpin-it™ miRNA qPCR Quantitation kit (GenePharma Co. Ltd, China). The expression of miRNA was defined based on the threshold cycle (Ct), and relative expression levels of miR-26b were calculated as 2-[(Ct of miR-26b)-(Ct of RNU6B)] after normalization with reference to expression of RNU6B small nuclear RNA.
Statistical analysis
All statistical analyses were performed using the SPSS 17.0 software package (SPSS, Chicago, IL, USA). The data were presented as the mean ± SD. The correlations were analyzed using the Student’s ttest, the Chi-square test and analysis of variance (ANOVA). Survival curves were calculated by the Kaplan-Meier method, and compared statistically using the log-rank test. Prognostic significance of each variable to overall survival (OS) and recurrence-free survival (RFS) were analyzed using the Cox regression model. Multivariate analysis of the prognostic factors was performed with Cox regression model. A probability value of less than 0.05 was chosen for statistical significance.
Results
Expression of miR-26b was significantly downregulated in cervical cancer tissues
To analyze the clinical significance of miR-26b expression in cervical cancer, we performed qRT-PCR to detect the expression of miR-26b in 88 paired of cervical cancer and the adjacent normal cervical epithelial tissues. As shown in Figure 1, the relative expression level of miR-26b in cervical cancer tissues was significantly higher than that in the adjacent normal cervical epithelial tissues (P<0.001). The differences among the two groups were significantly significant, suggesting that reduced miR-26b may play a critical role in cervical cancer progression.
Figure 1.
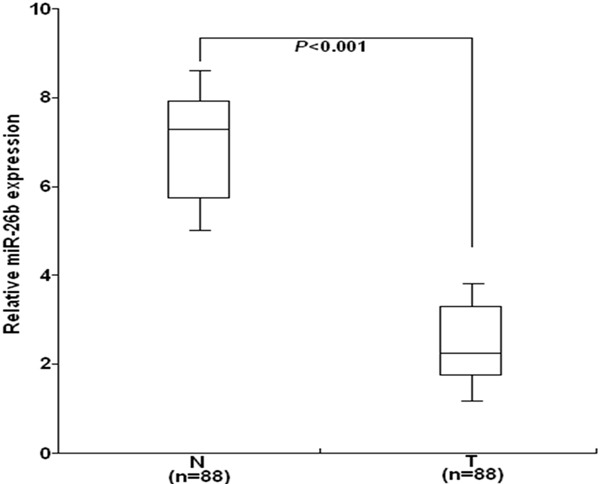
qRT-PCR detection of miR-26b expression in 88 paired of cervical cancer and the adjacent normal cervical epithelial tissues. The relative expression level of miR-26b in cervical cancer tissues was significantly higher than that in the adjacent normal cervical epithelial tissues (P<0.001). The mean and standard deviation of expression levels relative to RNU6B expression levels are shown and are normalized to the expression in the normal tissue of each matched pair. Each experiment was performed at least in triplicate. T: cervical cancer tissues; N: the adjacent normal cervical brain epithelial tissues.
Correlation between miR-26b expression and clinicopathological factors of cervical cancer patients
Then, we analyzed the correlation between miR-26b expression and clinicopathological factors of cervical cancer patients. The median value of miR-26b in all cervical cancer tissues was 2.14 and used as a cutoff value, and all cervical cancer patients were divided into two groups: high-miR-26b expression group (≥2.14; n=32) and low-miR-26b expression group (<2.14; n=56). As shown in Table 1, statistical analyses demonstrated that low miR-26b expression was observed to be significantly associated with advanced FIGO stage, higher incidence of lymph node metastasis and recurrence of cervical patients (P=0.002, 0.036 and 0.029, respectively). However, there were no significance between miR-26b expression and other clinicopathological factors of patients, such as patient age, tumor diameter, HPV infection, histological type, and tumor differentiation (P=0.742, 0.655, 0.598, 0.825 and 0.341, respectively).
Table 1.
Correlation between miR-26b expression and clinicopathological factors of cervical cancer patients
| Factors | Low miR-26b expression n=56) | High miR-26bexpression (n=32) | P-value | ||
|---|---|---|---|---|---|
|
|
|
||||
| N | % | N | % | ||
| Age (years) | 0.742 | ||||
| ≤55 | 23 | 41.1 | 12 | 37.5 | |
| >55 | 33 | 58.9 | 20 | 62.5 | |
| Tumor diameter (cm) | 0.655 | ||||
| ≤4.0 | 15 | 26.8 | 10 | 31.3 | |
| >4.0 | 41 | 73.2 | 22 | 68.7 | |
| HPV infection | 0.598 | ||||
| Negative | 30 | 53.6 | 19 | 59.4 | |
| Positive | 26 | 46.6 | 13 | 40.6 | |
| Histological type | 0.825 | ||||
| AD | 17 | 30.4 | 9 | 28.1 | |
| SCC | 39 | 69.6 | 23 | 71.9 | |
| Tumor differentiation | 0.341 | ||||
| Well | 8 | 14.3 | 7 | 21.9 | |
| Moderate | 28 | 50.0 | 11 | 34.4 | |
| Poor | 20 | 35.7 | 14 | 44.8 | |
| FIGO stage | 0.002* | ||||
| I | 18 | 32.1 | 21 | 65.6 | |
| II | 38 | 67.9 | 11 | 34.4 | |
| Lymph node metastasis | 0.036* | ||||
| Absent | 22 | 39.3 | 20 | 62.5 | |
| Present | 34 | 60.7 | 12 | 37.5 | |
| Recurrence | 0.029* | ||||
| No | 25 | 44.6 | 22 | 68.8 | |
| Yes | 31 | 55.4 | 10 | 31.2 | |
N: number;
Statistically significant difference (P<0.05).
AD: adenocarcinoma; SCC, squamous cell carcinoma.
Correlation between miR-26b expression and prognosis of cervical cancer patients
The mean duration of the follow-up period for the 88 cervical cancer patients was 74 months (range 5.12-98.5 months). To further investigate the correlation between miR-26b expression and survival of cervical cancer patients, KaplanMeier curves for RFS and OS were performed by comparing the patients with high and low miR-26 expression. As shown in Figure 2A, the 5-year RFS rate of cervical patients with low-miR-26b expression (65.8%) was significantly lower than that of patients with high-miR-26b expression (52.4%) (P=0.0043). Furthermore, the 5-year OS rate of patients with low-miR-26b expression (58.3%) was significantly lower than that of patients with high-miR-26b expression (42.8%) (P=0.0015; Figure 2B). These data demonstrated that reduced miR-200b was correlated with poor survival of cervical cancer patients.
Figure 2.
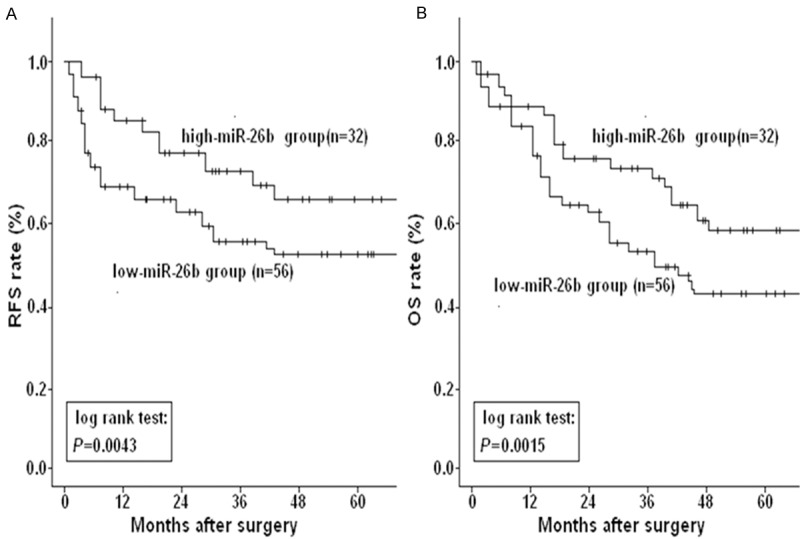
Kaplan-Meier postoperative survival curve for patterns of cervical cancer patients and miR-26b expression. A. The 5-year RFS rate of patients with low-miR-26b expression was significantly lower than that of those with high-miR-26b expression (P=0.0043). B. The 5-year OS rate of patients with low-miR-26b expression was significantly lower than that of those with high-miR-26b expression (P=0.0015). Corresponding P values analyzed by log-rank tests are indicated.
The results of multivariate analyses for RFS and OS using the Cox proportional hazards regression model were shown in Table 2. It was observed that these factors (FIGO stage, lymph node metastasis and miR-26b expression) were independently correlated with RFS of cervical cancer patients (P=0.017, 0.024 and 0.013, respectively), and miR-26b expression and lymph node metastasis were independent prognostic factors for the predicting OS of cervical cancer patients (P=0.009 and 0.007, respectively). Therefore, status of miR-26b expression may be a novel prognostic biomarker for cervical cancer patients.
Table 2.
Multivariate analysis of the correlation of RFS and OS with clinicopathological factors in cervical cancer patients
| Clinicopathological factors | 5-year RFS | 5-year OS | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age (>55 vs. ≤55 year) | 2.543 | 0.544-2.877 | 0.166 | 2.088 | 0.712-3.451 | 0.258 |
| Tumor diameter (>4.0 vs. ≤4.0 cm) | 1.308 | 0.712-1.834 | 0.203 | 1.344 | 0.825-1.877 | 0.412 |
| HPV infection (Positive vs. Negative) | 2.116 | 0.912-3.178 | 0.567 | 3.651 | 0.657-3.837 | 0.279 |
| Histology (SCC vs. AD) | 1.278 | 0.699-2.537 | 0.217 | 2.118 | 0.847-3.335 | 0.158 |
| Differentiation (Moderate + Poor vs. Well) | 1.712 | 0.884-2.088 | 0.370 | 1.444 | 0.775-2.377 | 0.188 |
| FIGO stage (II vs. I) | 2.228 | 1.699-3.125 | 0.017* | 1.807 | 0.682-2.499 | 0.226 |
| Lymph node metastasis (Yes vs. No) | 1.752 | 1.343-2.658 | 0.024* | 2.337 | 1.887-3.116 | 0.009* |
| miR-26b expression (Low vs. High) | 2.107 | 1.744-3.211 | 0.013* | 2.576 | 1.375-2.817 | 0.007* |
HR: hazard ratio; 95% CI: 95% confidence interval; RFS: recurrence-frees survival; OS: overall survival; AD: adenocarcinoma; SCC: squamous cell carcinoma.
Statistically significant difference (P<0.05).
Discussion
In the present study, we showed that the expression of miR-26b was significantly reduced in cervical cancer tissues, and reduced miR-26b expression was significantly correlated with advanced FIGO stage, higher incidence of lymph node metastasis and recurrence of patients. Also, patients with low-miR-26b expression showed poorer RFS and OS than those with high-miR-26b expression, and multivariate analysis indicated that low miR-26b expression was an independent prognostic biomarker for cervical cancer patients.
Recently, some miRNAs are reported to be correlated with prognosis of cervical cancer patients. By microarray analysis, Park’ et al identified 86 miRNAs that were dysregulated more than 2.0-fold in adenocarcinoma of the uterine cervix relative to normal tissues of the uterine cervix, and further research indicated that miR-363-3p is an independent favorable prognostic factor [19]. Wang’ et al showed that miR-31 is an independent prognostic factor and functions as an oncomir in cervical cancer via targeting ARID1A [20]. Also, Liang and his colleagues showed that the expression level of miR-215 is associated with cervical tumor progression and worse survival rate, suggesting that it may serve as a potential prognostic marker to identify patients at higher risk of recurrence [21]. Meanwhile, using an established PCR-based miRNA assay to analyze 102 cervical cancer samples, Hu’ et al identified miR-200a and miR-9 as two miRNAs that could predict patient suvival [22]. These data demonstrated that miRNAs might represent new candidate targets to be investigated for prognostic purposes in human cervical cancer. Moreover, some miRNAs are reported to function as tumor suppressors in cervical cancer. Li’ et al reported that miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer [23]. Also, miR-506 was found to act as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer [24]. At the same time, other miRNAs are reported to function as oncogenes in cervical cancer, such as miR-10a, miR-31, miR-196a, and so on [25-27]. These data demonstrated that miRNAs might represent potential molecular targets to be investigated for therapeutic purposes in cervical cancer.
miR-26 belongs to a cardiac-enriched, but noncardiac-specific, miRNA family composed of 3 members, miR-26a-1, miR-26a-2 and miR-26b. It has been reported that miR-26b is significantly downregulated in a variety of human cancers and functions as a critical regulator in carcinogenesis and tumor progression by acting as tumor suppressor gene in many human cancers. miR-26b is reported to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X [28], while this miRNA suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3 [29]. Also, miR-26b could inhibit growth and induce apoptosis in breast cancer by targeting multiple mRNAs, including CDK8, PTGS2 and SLC7A11 [30-32]. In addition, miR-26b could suppress growth of other human tumor cells, such as colon cancer, glioma and gastric cancer [33-35]. Although microRNA-26a has been reported to inhibit cell proliferation and invasion of cervical cancer cells by targeting protein tyrosine phosphatase type IVA 1 [36], the expression of miR-26b in cervical cancer and its clinicopathological or prognostic significance are unclear. To the best of our knowledge, this is the first report about whether miR-26b can be a potential prognostic biomarker for cervical cancer patients. Here, we first detected the expression of miR-26b in 88 paired of cervical cancer and the adjacent normal cervical epithelial tissues, and showed that the expression of miR-26b was significantly downregulated in cervical cancer tissues compared with the adjacent normal tissues. Then, we statistically analyzed the correlation between miR-26b expression and clinicopathological factors of cervical cancer patients. It was found that reduced miR-26b expression was significantly correlated with advanced FIGO stage and higher incidence of lymph node metastasis. Importantly, low miR-26b expression was also observed to be correlated with high recurrence rate of patients, suggesting that miR-26b could serve as a potential biomarker to identify patients at higher risk of recurrence. Furthermore, we analyzed the prognostic significance of miR-26b expression in cervical cancer. KaplanMeier curves for RFS and OS indicated that patients with low-miR-26b expression had poorer RFS and OS than those with high-miR-26b expression, and multivariate analyses demonstrated that status of miR-26b was an independent prognostic factor for both RFS and OS of cervical cancer patients. As the number of tissue samples in this study is small, further investigation of a larger case population is needed to confirm the prognostic significance of miR-26b expression in cervical cancer in future.
In conclusion, our findings indicate that the expression of miR-26b is significantly downregulated in cervical cancer, and may be a potential prognostic biomarker for cervical cancer patients. Further studies should focus on the roles of miR-26b in malignant phenotypes of cervical cancer cells, and identification of its target mRNAs will contribute to elucidating the roles of miR-26b in cervical cancer development.
Acknowledgements
Thanks to every one of the Department of Pathology in General Hospital of Guangzhou Military Command for their technique support.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 3.Moore DH. Cervical cancer. Obstet Gynecol. 2006;107:1152–1161. doi: 10.1097/01.AOG.0000215986.48590.79. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 7.Ying SY, Chang DC, Lin SL. The microRNA. Methods Mol Biol. 2013;936:1–19. doi: 10.1007/978-1-62703-083-0_1. [DOI] [PubMed] [Google Scholar]
- 8.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, Fu L, Zhang L, Wang Z, Li Z. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014;5:149. doi: 10.3389/fgene.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–594. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 11.Guo J, Friedman SL. The expression patterns and clinical significance of microRNAs in liver diseases and hepatocellular carcinoma. Curr Pharm Des. 2013;19:1262–1272. doi: 10.2174/138161213804805667. [DOI] [PubMed] [Google Scholar]
- 12.Fang Y, Yao Q, Chen Z, Xiang J, William FE, Gibbs RA, Chen C. Genetic and molecular alterations in pancreatic cancer: implications for personalized medicine. Med Sci Monit. 2013;19:916–926. doi: 10.12659/MSM.889636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 14.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 15.Rao Q, Shen Q, Zhou H, Peng Y, Li J, Lin Z. Aberrant microRNA expression in human cervical carcinomas. Med Oncol. 2012;29:1242–1248. doi: 10.1007/s12032-011-9830-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Chang L, Li Z, Gao Q, Cai D, Tian Y, Zeng L, Li M. miR-99a and -99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Med Oncol. 2014;31:934. doi: 10.1007/s12032-014-0934-3. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Ke G, Han D, Liang S, Yang G, Wu X. MicroRNA-181a enhances the chemoresistance of human cervical squamous cell carcinoma to cisplatin by targeting PRKCD. Exp Cell Res. 2014;320:12–20. doi: 10.1016/j.yexcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Ke G, Liang L, Yang JM, Huang X, Han D, Huang S, Zhao Y, Zha R, He X, Wu X. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2013;32:3019–3027. doi: 10.1038/onc.2012.323. [DOI] [PubMed] [Google Scholar]
- 19.Park H, Lee MJ, Jeong JY, Choi MC, Jung SG, Joo WD, Lee C, An HJ. Dysregulated microRNA expression in adenocarcinoma of the uterine cervix: Clinical impact of miR-363-3p. Gynecol Oncol. 2014;135:565–572. doi: 10.1016/j.ygyno.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Wang N, Zhou Y, Zheng L, Li H. MiR-31 is an independent prognostic factor and functions as an oncomir in cervical cancer via targeting ARID1A. Gynecol Oncol. 2014;134:129–137. doi: 10.1016/j.ygyno.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 21.Liang H, Li Y, Luo RY, Shen FJ. MicroRNA-215 is a potential prognostic marker for cervical cancer. J Huazhong Univ Sci Technolog Med Sci. 2014;34:207–212. doi: 10.1007/s11596-014-1260-x. [DOI] [PubMed] [Google Scholar]
- 22.Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298–3307. doi: 10.1016/j.febslet.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, Sun WW, Lou XL, Wang JH, Teng YC, Zhang ZG. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717–25. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 25.Long MJ, Wu FX, Li P, Liu M, Li X, Tang H. MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett. 2012;324:186–196. doi: 10.1016/j.canlet.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Wang N, Zhou Y, Zheng L, Li H. MiR-31 is an independent prognostic factor and functions as an oncomir in cervical cancer via targeting ARID1A. Gynecol Oncol. 2014;134:129–137. doi: 10.1016/j.ygyno.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 27.Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;110:1260–1268. doi: 10.1038/bjc.2013.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen G, Lin Y, Yang X, Zhang J, Xu Z, Jia H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer. 2014;14:393. doi: 10.1186/1471-2407-14-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. doi: 10.1186/1476-4598-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Li X, Kong X, Luo Q, Zhang J, Fang L. MiRNA-26b inhibits cellular proliferation by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7:558–565. [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13:7. doi: 10.1186/1475-2867-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX, Cao DX, He M, Chen GQ, He JR, Zhao Q. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011;585:1363–1367. doi: 10.1016/j.febslet.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Kim K, Li X, Moreno M, Sharp T, Goodheart MJ, Safe S, Dupuy AJ, Amendt BA. MicroRNA-26b represses colon cancer cell proliferation by inhibiting lymphoid enhancer factor 1 expression. Mol Cancer Ther. 2014;13:1942–1951. doi: 10.1158/1535-7163.MCT-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang Y, Cao S, Lin X. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu YP, Gui QJ, Zhang L, Li GQ. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31:1715–1722. doi: 10.3892/or.2014.3052. [DOI] [PubMed] [Google Scholar]
- 36.Dong J, Sui L, Wang Q, Chen M, Sun H. MicroRNA-26a inhibits cell proliferation and invasion of cervical cancer cells by targeting protein tyrosine phosphatase type IVA 1. Mol Med Rep. 2014;10:1426–1432. doi: 10.3892/mmr.2014.2335. [DOI] [PubMed] [Google Scholar]


