Abstract
Objective: We aimed to explore what impact miR-503 has on the prognosis of patients with non-small cell lung cancer (NSCLC). Methods: Cancer and matched non-malignant lung tissue specimens were collected from 109 patients who underwent surgery in Tanisha Hospital from Jun 2006 to July 2013. Overall survival (OS) curves were analyzed using the Lapland-Meier method, and the differences were examined using log-rank tests. Cox proportional- hazards regression analysis was applied in order to estimate univariate and multivariate hazard ratios for OS. Results: The relative expression of miR-503 in NSCLC tissues (0.366 ± 0.130) was significantly lower than that in matched noncancerous lung tissues (1.667 ± 1.047, P < 0.01). Statistically significant association was observed between miR-503 expression and lymphatic invasion (P = 0.005), distant metastasis (P = 0.002), TNM stage (P = 0.008), and tumor grade (P = 0.043). Lapland Meier analysis clearly illustrated that the patients with the lower expression of miR-503 had a worse outcome compared to patients with higher miR-503 expression (P = 0.004). Furthermore, multivariate analysis revealed that miR-503 expression level was an independent prognostic factor for overall survival (HR = 3.992, 95% CI: 2.276-9.872; P = 0.018) in NSCLC. Conclusion: In patients with NSCLC, low miR-503 expression is an independent prognostic factor.
Keywords: NSCLC, micro RNA, miR-503, prognosis, biomarker
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Non-small cell lung cancer (NSCLC) is the most frequent type of lung cancer and the most common cause of cancer death [1]. Despite the considerable advances in medical and surgical treatment of NSCLC patients, the prognosis associated with this disease remains dismal. The generally poor clinical outcome of individuals diagnosed with NSCLC underscores the importance in obtaining a better understanding of the molecular mechanisms underlying carcinogenicity [2].
Microcosmic (miasmas) are a recently discovered class of small (approximately 18-24 nucleolus in length), codding regulatory RN As that negatively regulate gene expression at the conscription and/or inspirational level. miasmas can trigger cleavage of target marinas or inhibit protein translation through sequence-specific interactions with the 3’-untranslated regions (3’-Ruts) of the target marinas [3]. Although the full extent of the biological functionalism of miasmas has yet to be identified, they have been suggested to act as intrinsic regulators of many cellular processes including cell invasion, differentiation, proliferation, and apotheosis [4-6]. Furthermore, aberrant expression of miasmas has been linked to the development and progression of cancer and has been shown to have prognostic significance in several tumor types [7-10].
micro RNA-503 (miR-503) is an intrastate Miranda located at Xq26.3, belonging to an extended miR-16 family of miasmas [11]. miR-503 was shown to be down regulated in oral cancer cells and unicellular carcinoma cells [12,13], but over expressed in parathyroid carcinoma, stationmaster, and nonidentical carcinoma [14-16], suggesting that it might have complicated cancer-type specific functions. Previously, Li ET al found that the expression of miR-503 was significantly decreased in NSCLC tissues compared to normal tissues [17]. However, until now, the clinical significance and prognostic value of miR-503 have not been investigated.
Materials and methods
Patients and clinicopathological data collection
This study was approved by the Review Board of Yantaishan Hospital, and written informed consent was obtained from all patients. Cancer and matched non-malignant lung tissue specimens were collected from 109 patients who underwent surgery in Yantaishan Hospital from Jun 2006 to July 2013. All samples were derived from patients who had not received chemotherapy or radiotherapy prior to surgery. The diagnosis was based on clinical examination and histopathological analysis of the tissue specimens. The stages of the lung cancer patients were evaluated by TNM staging (stage I, 34 patients; stage II, 25 patients; stage III, 23 patients and stage IV, 27 patients). The clinical and pathological data were recorded in a predesigned performa (shown in Table 1). Immediately following surgical resection, tissues were frozen in liquid nitrogen until use. The median follow-up period was 37 months (range, 8-76 months).
Table 1.
Relationship between clinicopathological features and miR-503 expression in lung cancer tissues
| miR-503 expression | ||||
|---|---|---|---|---|
|
|
||||
| Parameters | Number of cases | Low (n = 56) | High (n = 53) | P value |
| Age (y) | ||||
| < 60 | 52 | 27 | 25 | 0.533 |
| ≥ 60 | 57 | 29 | 28 | |
| Sex | ||||
| Male | 43 | 21 | 22 | 0.408 |
| Female | 66 | 35 | 31 | |
| Smoking History | ||||
| Never | 17 | 8 | 9 | 0.451 |
| Former + Current | 92 | 48 | 44 | |
| Histology | ||||
| Squamous cell carcinoma | 43 | 20 | 23 | 0.414 |
| Adenocarcinoma | 45 | 23 | 22 | |
| Other | 21 | 13 | 8 | |
| Lymphatic invasion | ||||
| Positive | 32 | 23 | 9 | 0.005 |
| Negative | 77 | 33 | 44 | |
| Distant metastasis | ||||
| Positive | 21 | 17 | 4 | 0.002 |
| Negative | 88 | 39 | 49 | |
| TNM stage | ||||
| I/II | 42 | 15 | 27 | 0.008 |
| III/IV | 67 | 41 | 26 | |
| Tumor grade | ||||
| I-II | 59 | 20 | 39 | 0.043 |
| III | 50 | 26 | 24 | |
RNA extraction and real-time PCR
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and purity of all RNA samples were detected by NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Houston, TX, USA). NCodeTM SYBR® Green miRNA qRT-PCR Kit (Invitrogen, Carlsbad, CA, USA) was used to synthesize specific cDNA of miR-503 and U6B(as an internal control), and perform qRT-PCR, which was analyzed with the DNA Engine Opticon 2 Real-Time Cycler (MJ Research Inc., Waltham, MA, USA) according to the manufacturer’s instructions. Each sample was examined in triplicate and analyzed by the comparative threshold cycle (Ct) method. The expression levels of miR-503 were normalized to U6B.
Statistical analysis
The relationships between miR-503 expression and clinicopathological factors were analyzed using the Student’s t-test. Overall survival (OS) curves were analyzed using the Kaplan-Meier method, and the differences were examined using log-rank tests. Cox proportional-hazards regression analysis was applied in order to estimate univariate and multivariate hazard ratios for OS. All p-values are two-sided, and P < 0.05 was considered to denote statistical significance. Statistical analyses were carried out using statistical program for Social Sciences (SPSS) software 18.0 (SPSS Inc, Chicago, IL).
Results
Expression level of miR-503 in human lung cancer tissues
Using qRT-PCR method, miR-503 was detected in all the 109 pairs of NSCLC tissues and their matched noncancerous lung tissues. As shown in Figure 1, the relative expression of miR-503 in NSCLC tissues (0.366 ± 0.130) was significantly lower than that in matched noncancerous lung tissues (1.667 ± 1.047, P < 0.01).
Figure 1.
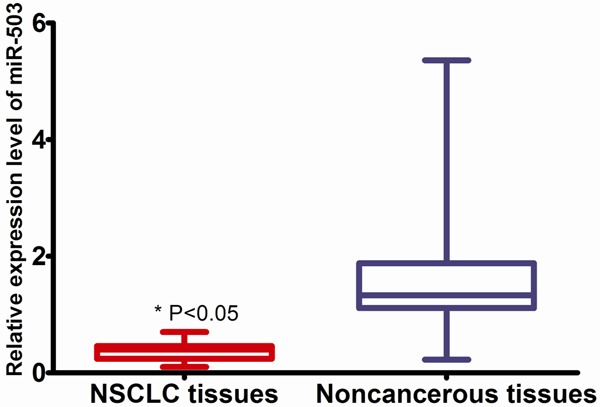
Comparison of miR-503 expression between NSCLC and normal tissues.
Relationship between clinicopathological factors and miR-503 expression in the cancer tissues
To evaluate the correlation between the miR-503 expression levels and the clinicopathological characteristics, patients were divided into two groups (high and low). The cut-off levels for miR-503 were set according to the median level of relative quantity. As shown in Table 1, a statistically significant association was observed between miR-503 expression and lymphatic invasion (P = 0.005), distant metastasis (P = 0.002), TNM stage (P = 0.008), and tumor grade (P = 0.043). However, there were no significant associations between miR-503 expression level and age (P = 0.533), sex (P = 0.408), smoking history (P = 0.451), and histology (P = 0.414).
Correlation between miR-503 levels and OS
Kaplan-Meier analysis with log-rank test was used to calculate the effect of miR-503 expression on patient survival (shown in Figure 2). Kaplan Meier analysis clearly illustrated that the patients with the lower expression of miR-503 had a worse outcome compared to patients with higher miR-503 expression (P = 0.004). Univariate and multivariate analyses were utilized to evaluate whether the miR-503 expression level and various clinicopathological features were independent prognostic parameters of NSCLC patient outcomes. Multivariate analysis revealed that miR-503 expression level was independent prognostic factors for overall survival (HR = 3.992, 95% CI: 2.276-9.872; P = 0.018, shown in Table 2) in NSCLC.
Figure 2.
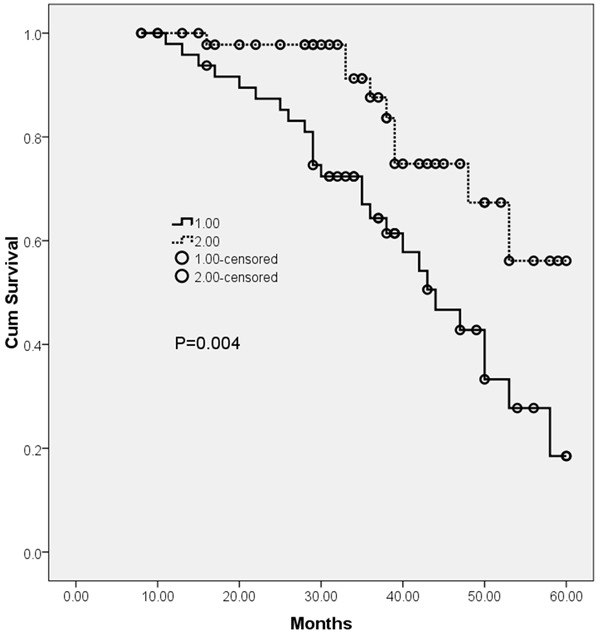
Kaplan-Meier survival curves of OS based on miR-503 expression.
Table 2.
Multivariate analysis of prognostic factors in for overall survival
| Variables | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| Age | 1.652 | 0.368-3.982 | 0.451 |
| Sex | 0.781 | 0.267-2.352 | 0.676 |
| Smoking History | 2.661 | 0.682-4.278 | 0.219 |
| Histology | 1.277 | 0.389-2.301 | 0.528 |
| Lymphatic invasion | 3.111 | 2.012-11.289 | 0.023 |
| Distant metastasis | 4.519 | 3.182-11.881 | 0.009 |
| TNM stage | 4.121 | 2.338-12.319 | 0.011 |
| Tumor grade | 3.172 | 1.298-10.228 | 0.047 |
| Mir-503 expression | 3.992 | 2.276-9.872 | 0.018 |
Discussion
Post-transcriptional regulation plays an important role in diverse cellular processes such as development, neurogenesis, and cancer progression [18]. miRNAs have emerged as important post-transcriptional regulators that inhibit mRNA translation or induce mRNA cleavage by base pairing with a seed region in the 3’-UTR of target genes. Recent studies have shown that dysregulation of miRNAs contributes to the initiation, progression, metastasis, and drug resistance of cancer [4,8]. Furthermore, several up-regulated and down-regulated miRNAs have been identified in lung cancer, the most frequently diagnosed cancer and the most common cause of cancer-related death worldwide. A detailed understanding of the mechanisms underlying miRNA production and function is important.
MiR-503 is an intragenic miRNA located at Xq26.3, belonging to an extended miR-16 family of miRNAs [11]. miRNA array analysis has shown that in comparison with that in corresponding normal tissues, the level of miR-503 expression varies in several types of cancer, with declined expression in some (e.g., oral cancer, hepatocellular carcinomas, and endometrioid endometrial cancer), but increase in several other cancer types (e.g., adrenocortical carcinoma, parathyroid carcinoma, and retinoblastoma) [12,14,15,19-21]. Moreover, the biological functions of miR-503 have been reported to be diverse. It was found to cause cell cycle quiescence during muscle differentiation, inhibit proliferation of human head and neck squamous cell carcinoma (HNSCC) cells, and reduce metastasis in hepatocellular carcinoma (HCC) cell, displaying features characteristic of a tumor-suppressive miRNA [13,22,23].
Previously, Li et al found that the expression of miR-503 was significantly decreased in NSCLC tissues compared to normal tissues. A statistically significant inverse association was found between miR-503 methylation status and expression of the miR-503 in tumor tissues (P < 0.001), and expression of miR-503 was restored by the demethylating agent 5-aza-2’-deoxycytidine, suggesting that methylation was associated with the transcriptional silencing. They then showed that miR-503 targeted a homologous DNA region in the 3’-UTR region of the Fanconi anemia complementation group A protein (FANCA) gene and repressed its expression at the transcriptional level. Furthermore, they found that miR-503 regulated the resistance of NSCLC cells to cisplatin at least in part by targeting FANCA [17]. In the study by Yang et al, ectopic expression of miR-503 suppressed tumor cell proliferation and metastasis-related traits in vitro as well as in vivo, supporting an anti-cancer role of this microRNA in NSCLC progression. Mechanistic study revealed that oncogenic PI3K p85 and IKK-β were direct targets of miR-503. Overexpression of either PI3K p85 or IKK-β partially restored the malignant properties of NSCLC cells in the presence of miR-503. Taken together, their data demonstrated that miR-503 inhibited the malignant phenotype of NSCLC by targeting PI3K p85 and IKK-β and might play a suppressive role in the pathogenesis of NSCLC. However, until now, the clinical significance and prognostic value of miR-503 have not been investigated. In the present study, we found that the relative expression of miR-503 in NSCLC tissues was significantly lower than that in matched noncancerous lung tissues. To evaluate the correlation between the miR-503 expression levels and the clinicopathological characteristics, patients were divided into two groups (high and low). A statistically significant association was observed between miR-503 expression and lymphatic invasion, distant metastasis, TNM stage, and tumor grade. Kaplan Meier analysis clearly illustrated that the patients with the lower expression of miR-503 had a worse outcome compared to patients with higher miR-503 expression (P = 0.004). Furthermore, multivariate analysis revealed that miR-503 expression level was independent prognostic factors for overall survival (HR = 3.992, 95% CI: 2.276-9.872; P = 0.018) in NSCLC. Empirically, HR of more than 1.5 is considered to be a strong prognostic factor. Therefore, tissue miR-503 level might have a considerable potential in prognosis for patients with NSCLC.
In conclusion, the present study demonstrated that the down-regulation of miR-503 was associated with advanced clinical features and poor prognosis of patients with NSCLC, suggesting that miR-503 down-regulation may be used as an unfavorable prognostic biomarker in NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Brown JR, Sanseau P. A computational view of microRNAs and their targets. Drug Discov Today. 2005;10:595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 7.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 8.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 9.Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther. 2007;7:1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 10.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Caporali A, Emanueli C. MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc Med. 2011;21:162–166. doi: 10.1016/j.tcm.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, Chang JT, Cheng AJ. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res (Phila) 2012;5:665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J, Wang W. Analysis of microRNA expression profiling identifies microRNA-503 regulates metastatic function in hepatocellular cancer cell. J Surg Oncol. 2011;104:278–283. doi: 10.1002/jso.21941. [DOI] [PubMed] [Google Scholar]
- 14.Ozata DM, Caramuta S, Velazquez-Fernandez D, Akcakaya P, Xie H, Hoog A, Zedenius J, Backdahl M, Larsson C, Lui WO. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18:643–655. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 16.Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, Vicentini L, Chiodini I, Bisceglia M, Beck-Peccoz P, Bosari S, Spada A. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer. 2010;17:135–146. doi: 10.1677/ERC-09-0134. [DOI] [PubMed] [Google Scholar]
- 17.Li N, Zhang F, Li S, Zhou S. Epigenetic silencing of MicroRNA-503 regulates FANCA expression in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2014;444:611–616. doi: 10.1016/j.bbrc.2014.01.103. [DOI] [PubMed] [Google Scholar]
- 18.Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y, Yin LR. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J. 2013;280:3768–3779. doi: 10.1111/febs.12365. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Tombol Z, Szabo PM, Molnar V, Wiener Z, Tolgyesi G, Horanyi J, Riesz P, Reismann P, Patocs A, Liko I, Gaillard RC, Falus A, Rácz K, Igaz P. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue- specific target prediction, and pathway analysis. Endocr Relat Cancer. 2009;16:895–906. doi: 10.1677/ERC-09-0096. [DOI] [PubMed] [Google Scholar]
- 22.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Q, Feng MG, Mo YY. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer. 2009;9:194. doi: 10.1186/1471-2407-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]


