Abstract
Background: Soft tissue sarcomas (STSs) are a heterogeneous group of malignant tumors that can be divided into specific reciprocal translocation associated in STSs (SRTSs) and nonspecific reciprocal translocation associated in STSs (NRTSs). Telomeres play a key role in maintaining chromosomal stability; pathological telomere elongation is found in a number of cancers. In this study, we aimed to assess telomere lengths in the two types of sarcomas. Twenty formalin-fixed paraffin-embedded (FFPE) archival tissues, namely, 10 sarcomas with characteristic translocations and 10 without characteristic translocations, were included in this study. Expression levels of special fusion gene transcripts were detected in these tumors by reverse transcription polymerase chain reaction. Telomere lengths were assessed by fluorescence in situ hybridization. Results showed that in 10 of the 10 cases of SRTSs, telomere lengths were similar to or reduced compared with the surrounding normal cells. Telomere lengths were elongated in eight of 10 cases of NRTSs, but reduced in two cases. The difference in telomere length was statistically significant in the two types of sarcomas (P = 0.001). Upon combining the P53 mutation status, we found that the telomere length was short in eight cases, and only one case demonstrated p53 mutation. However, the telomere length was long in eight cases, and p53 mutation was observed in five cases. These data suggested that p53 mutation was accompanied with long telomeres, and telomeres possibly play an important role in NRTSs. Therefore, telomere-targeting therapy may lead to novel therapeutic strategies to improve treatment of NRTS patients.
Keywords: Soft tissue sarcoma, translocation, telomere length, fusion gene, p53
Introduction
Soft tissue sarcomas (STSs) are a heterogeneous group of malignant tumors arising from mesenchymal progenitor/stem cells. They account for 5% of adult solid tumors and approximately 8%-10% of pediatric or young adult cancers. Over 50 subtypes of STSs exist based on genetic and morphological criteria. STSs can be divided into two main categories according to molecular genetics. One type is specific reciprocal translocations in STSs (SRTSs), which are characterized by simple karyotypes, including chromosomal-specific translocations and fusion gene formation, such as synovial sarcoma (SS), Ewing’s sarcoma/peripheral primitive neuroectodermal tumor (ES/pPNET), and dermatofibrosarcoma protuberans (DFSP). The other type is multiple complexes with nonspecific reciprocal translocations in STSs (NRTSs), such as pleomorphic rhabdomyosarcoma, leiomyosarcoma (LMS), undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma (UPS/MFH).
Current studies have demonstrated that gene fusions generated from these translocations are the initiating events of many SRTSs. These fusion proteins often affect transcription factors, resulting in the disruption of transcription regulation and regulation of the transcription of some genes. Several genes are involved in certain key functions of cells, such as cell proliferation [1]. These SRTSs often lack p53 alterations. By contrast, NRTSs often display p53 alterations, and p53 pathway inactivation may be a necessary common event. P53 plays a major role in regulating DNA repair, senescence, and apoptosis [2]. Inactivation of the p53 pathway is the most common genetic pathway alteration, which may be derived from the p53 mutation itself or increased expression of its downregulation [3]. In our previous study, we analyzed the p53 mutation state in STSs by polymerase chain reaction (PCR)-single strand conformation polymorphism and DNA sequencing, and found that the frequency of p53 mutations is higher in cases with NRTSs than that in cases with SRTSs [4]. Therefore, the presence of p53 mutations may play an important role in NRTSs.
Telomeres are specialized DNA-protein structures located at the ends of linear chromosomes that protect against chromosome end-to-end fusion events and prevent the ends of DNA molecules from being recognized by DNA damage mechanisms. In adult human somatic tissues, telomeres are approximately 10-15 kbp, which decreases by an average of 50-150 base pairs per cell cycle [5]. When the telomere length declines below a certain threshold, an irreversible growth arrest state is triggered, and the cell population eventually undergoes senescence and dies. Telomere shortening contributes to aging phenotypes and provides an effective tumor suppressor mechanism. Given that mutations in the p53 protein continue to divide, cells lose the ability to senesce and eventually enter extensive telomere shortening, which results in chromosomal fusion and cell death [6]. A number of studies have reported that pathological telomere elongation is found in a number of tumors. These tumor cells are believed to have escaped from this limitation by activating the telomere maintenance mechanism (TMM). Two known TMMs include catalytic activity of the enzyme telomerase or a telomerase-independent, recombination-based pathway termed alternative lengthening of telomeres (ALT). In contrast to cells with active telomerase, cells that use ALT are characterized by long, heterogeneous telomere lengths and ALT-associated PML bodies. Several studies have demonstrated that ALT is overrepresented in mesenchymal tumors [7-9], whereas epithelial tumors generally express telomerase [10]. However, telomere length is rarely reported in STSs.
In the present study, we detected fusion genes in STSs, analyzed telomere length in 20 cases of sarcomas, and combined the p53 mutation status to determine their potential role in tumor pathogenesis.
Materials and methods
Tumor samples
Twenty formalin-fixed paraffin-embedded (FFPE) samples of STSs were retrieved from the archives in the Department of Pathology, the First Affiliated Hospital of Shihezi University School of Medicine, Xinjiang, China. This study was approved by the institutional ethics committee at the First Affiliated Hospital of Shihezi University School of Medicine, and conducted in accordance with the ethical guidelines of the Declaration of Helsinki. The sample set was divided into two types as follows: 10 cases of SRTSs [one case of SS, two cases of alveolar rhabdomyosarcoma (ARMS), three cases of ES/pPNET, and four cases of DFSP] and 10 cases of NRTSs [three cases of LMS, six cases of UPS/MFH, and one case of fibrosarcoma (FS)]. H&E-stained samples were reviewed by two senior pathologists to confirm the diagnosis using standard diagnostic criteria [11]. The paraffin blocks were confirmed to contain tumor cells (at least 90%) prior to sectioning and RNA extraction.
Reverse transcription polymerase chain reaction (RT-PCR) analysis
RNA was extracted from tumor tissues according to the protocols of the manufacturer. Expression levels of the oncogenic fusion genes, PAX3/7-FKHR, EWS-Fli1, SYT-SSX1, and COLIA1-PDGFB in RMS, ES, SS, and DFSP tumors were detected using RT-PCR. The primer sequences for PAX3/7-FKHR, EWS-Fli1, SYT-SSX1, and COLIA1-PDGFB fusion transcripts; reaction mixtures; and PCR cycling conditions were as the same as previously described [12,13]. Positive, negative, and blank controls were established and run simultaneously. PCR products were purified and sequenced by Sangon Company (Shanghai, China).
Telomere-fluorescence in situ hybridization (TFISH) analysis
Pre-treatment slides were deparaffinized and then immersed in xylene for 10 min at 56°C. The latter step was repeated twice. The slides were hydrated through 100% ethanol for two times at 5 min each, and then placed in 0.2 N HCl for 20 min, followed by purified water and TBS for two soaks at 5 min each. The slides were immersed in a pre-treatment solution (concentrated proteolytic enzyme) for 20-40 min at room temperature. The slides were rinsed with TBS for 5 min twice, followed by graded ethanol. Finally, the slides were air-dried. For denaturation and hybridization, 10 µl of a Cy3-labeled telomere-specific peptide nucleic acid FISH probe (no.K5326; Dakocytomation, Denmark) was applied to the sample. The slides were coverslipped, and denaturation was performed by incubation for 8 min at 84°C. Slides were then moved to a dark closed container for hybridization at room temperature for 2-4 h. Subsequently, the slides were immersed in working solution of the rinse solution (provided, pure water diluted 1:50) to remove coverslips, followed by 5 min in pre-heated working solution of the wash solution (reagents provided, pure water diluted 1:50) and graded ethanol. After the slides were air-dried, they were counterstained with DAPI for 5 min at room temperature, coverslipped, and either imaged or stored at -20°C until used.
Microscopy
Slides were imaged with an Olympus fluorescence microscope equipped with a ×100/1.4NA oil-immersion Neofluar lens and appropriate fluorescence filter sets (Olympus BX51, Japan). TEL-FISH signals were observed at the beginning of an imaging session, and the optimum exposure time was determined within a comparison set under identical exposure times. Telomere lengths were evaluated by visual assessment of the fluorescent intensities of the telomeric signals, and directly compared with those of seemingly normal stromal cells (such as endothelial cells) within the same tissue section. Each case was scored by two authors using the following semiquantitative scoring system: (a) lesions with normal telomere intensity had signals comparable with those of normal cells; (b) lesions with short telomeres had telomere intensities obviously dimmer than the normal stroma; and (c) lesions with long telomeres demonstrated telomere signals that were brighter than that of stromal cells [14].
Statistical analysis
SPSS v. 17 software (SPSS Inc., Chicago, IL, USA) was used for statistical analyses. Parameters of interest were compared using Fisher’s exact test. Survival analyses were based on Kaplan-Meier life tables. Differences were considered statistically significant for two-sided P values less than 0.05.
Results
Expression of EWS-Fli1, PAX3-FKHR, SYT-SSX1, and COLIA1-PDGFB transcripts
The transcripts of the β-actin gene were amplifiable in all the cases, indicating the feasibility to use FFPE tissue blocks in this methodology. Three cases of ES were positive for amplification of a 277 bp fragment from EWS-Fli1 fusion transcripts. We detected PAX3-FKHR, SYT-SSX1, and COLIA1-PDGFB in ARMS, SS, and DFSP, respectively. One case of SS was positive for amplification of a 120 bp fragment from SSX1-SYT fusion transcripts. Two cases of ARMS were positive for amplification of a 147 bp fragment from PAX3-FKHR fusion transcripts. Four cases of DFSP were positive for amplification of a 103-250 bp fragment from COLIA1-PBGD fusion transcripts. By contrast, EWS-Fli1, PAX3/7-FKHR, SSX-SYT, and COLIA1-PBGD transcripts were negative in all three cases of LMS, six cases of UPS/MFH, and one case of FS.
Telomere length in STSs
We analyzed 20 cases of telomere lengths by TFISH. In all (10 of 10) SRTS cases, telomere length was similar to or reduced compared with that of surrounding nonneoplastic tissues (Table 1). The telomere length in eight cases (two cases of ES, three cases of DFSP, two cases of ARMS, and one case of SS) decreased compared with that of endothelial cells. The telomere length was normal in one case of ES and one case of DFSP. By contrast, tumor cells in eight of 10 NRTS cases displayed elongated telomeres compared with surrounding normal cells (Figure 1), of which telomere length ranged from short to elongated in three cases of MFH, one case of LMP, and one case of FS, whereas telomere length was elongated in two cases of MFH and one case of LMP. The difference in telomere length in the two types of sarcomas was statistically significant (P = 0.001, Fisher’s test).
Table 1.
Clinical characteristics, fusion gene and p53 state of 20 patients with soft tissue sarcomas
| Case | type | Sex | Age (year) | Translocation | Fusion gene | Telomere lengths | P53 mutation |
|---|---|---|---|---|---|---|---|
| 1 | ES | M | 17 | Y | EWS-FLI1 | Short-normal | N |
| 2 | ES | F | 17 | Y | EWS-FLI1 | Short-normal | N |
| 3 | ES | M | 24 | Y | EWS-FLI1 | Normal | N |
| 4 | DFSP | F | 25 | Y | COLIA1-PDGFB | Short-normal | N |
| 5 | DFSP | F | 37 | Y | COLIA1-PDGFB | Normal | N |
| 6 | DFSP | M | 35 | Y | COLIA1-PDGFB | Short-normal | N |
| 7 | DFSP | M | 28 | Y | COLIA1-PDGFB | Short-normal | N |
| 8 | SS | M | 19 | Y | SYT-SSX1 | Short-normal | N |
| 9 | ARMS | M | 16 | Y | PAX3-FKHR | Short-normal | N |
| 10 | ARMS | M | 15 | Y | PAX3-FKHR | Short-normal | Y |
| 11 | UPS | F | 33 | N | N | Short-long | Y |
| 12 | UPS | M | 69 | N | N | long | Y |
| 13 | UPS | F | 62 | N | N | long | Y |
| 14 | UPS | M | 60 | N | N | Short-long | N |
| 15 | UPS | M | 62 | N | N | Short-normal | N |
| 16 | UPS | F | 39 | N | N | Short-long | N |
| 17 | LMS | F | 39 | N | N | Short-long | N |
| 18 | LMS | F | 46 | N | N | long | Y |
| 19 | LMS | F | 48 | N | N | Short-normal | N |
| 20 | FS | M | 44 | N | N | Short-long | Y |
Note: ES: Ewing’s sarcoma, DFSP: dermatofibrosarcoma protuberans, SS: synovial sarcoma, ARMS: alveolar rhabdomyosarcoma, UPS: undifferentiated pleomorphic sarcoma, LMS: leiomyosarcoma, FS: fibrosarcoma, M: Male, F: Female, Y: Yes, N: No.
Figure 1.
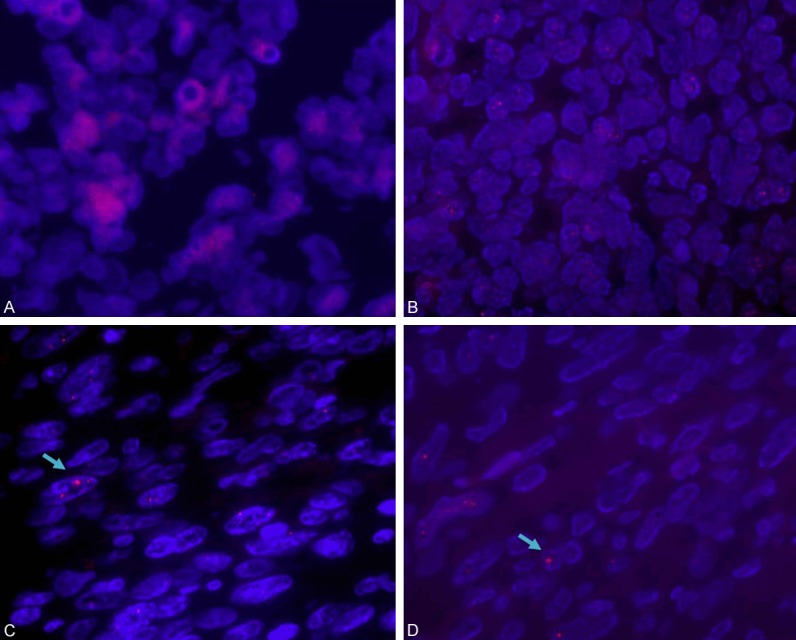
TFISH staining of sarcomas using Cy3-labeled FISH probe hybridization probe (red). A. Telomere length displaying short or normal in ES. B. Telomere length displaying short or normal in DFSP. C, D. UPS displaying from short to long telomere length and long telomeres demonstrating brighter red signals (arrow).
Interaction of telomere length, p53 mutation, and chromosomal translocation
Table 1 shows that the telomere length was short in eight cases, and p53 mutation was not found in nine cases among the SRTSs. However, the telomere length was long in eight cases, and p53 mutation was found in five cases among the NRTSs. Five cases with p53 mutation were accompanied with long telomeres.
Discussion
In recent years, advances in molecular genetics have allowed researchers to discover special fusion genes or gene mutations in some STSs, and these changes have played an important role in diagnosis, classification, prognosis, and targeted therapies. In particular, when these tumors show very little evidence of differentiation, distinguishing them from small round cell tumors is difficult. Thus, fusion gene detection has become the basis for the diagnosis of STSs. At present, sarcomas can be divided into two major genetic groups, namely, SRTSs and NRTSs. Differences clearly exist in terms of patients’ age, histological features, and molecular pathogenesis in the two types of STSs.
Generally, SRTSs break up certain genes and create gene fusions with new structures. Fusion genes resulting from these translocations encode chimeric proteins that can regulate the transcription of some genes and are usually involved in certain key functions of cells. Meanwhile, NRTSs have a high incidence of p53 mutation. Common modes of p53 pathway inactivation in sarcomas include p53 point mutations, homozygous deletion of CDKN2A, and amplification of MDM2. Therefore, p53 pathway inactivation may be a necessary common early event [15]. In our study, the frequency of p53 mutations was higher in cases with NRTSs than that in cases with SRTSs, which suggested that the presence of p53 mutations may play a leading role in NRTSs.
An increasing number of evidence showed that telomeres play an important role in tumors. Telomere studies in many human carcinomas have been conducted for diagnostic and prognostic utility [16,17]. TMM is regarded an important mechanism in evading senescence by tumor cells. At present, TMM mainly includes telomerase and ALT. In most epithelial cancers, TMM is achieved by reactivating or up-regulating telomerase activity [18]. Telomerase activity ranges from 82% to 100%, and it is correlated with telomerase activity and clinicopathological characteristics, including histological type, pathological grade, clinical stage, and prognosis. However, some tumors that are telomerase-negative maintain the length of their telomeres by one or more mechanisms referred to as ALT. About 10% of all human tumors depend on ALT for their continued growth. Some studies showed that telomerase activity in sarcomas is low, and telomere length should not be significantly different between telomerase-positive and telomerase-negative tumors, thereby indicating that the mechanism of ALT plays an important role in sarcomas [19-21]. TMM has recently been reported as a prognostic factor for patients with sarcomas, and ALT is positivity correlated with worse survival of patients with several types of sarcomas [22,23].
A previous study demonstrated telomerase activity in 70% ES tumor samples (21/30) and nine of 10 ES cell lines. ALT was observed only in the cell line without telomerase activity, and in none of the 30 ES tumor samples. The nine cases of ES patients whose tumors lacked telomerase activity did not differ significantly from the remaining patients in age, stage, EWSR1-FLI1 fusion type, prevalence of P53 point mutations, or overall survival. Among 60 osteosarcomas, ALT was observed in 38 of 60 cases (P < 0.0001). The results suggested that a predominance of telomerase activation in the absence of ALT may characterize sarcomas with specific chromosomal translocations, whereas a high prevalence of ALT is typical of sarcomas with nonspecific complex karyotypes [23].
Several scholars recently analyzed the clinicopathological features of LMSs with the ALT phenotype. TFISH revealed that 59% (51/86) of LMSs have the ALT phenotype. The ALT phenotype is associated with epithelioid/pleomorphic cell morphology, tumor necrosis, and poor differentiation. Poor differentiation, high FNCLCC grade, tumor size, and ALT phenotype were correlated with poor overall survival in univariate analysis. The ALT phenotype remained an independent prognostic factor in multivariate analysis. Therefore, the ALT phenotype in LMS is believed to be associated with aggressive histologic features and poor clinical outcome [24].
Telomere reduction or elongation has been associated with prognosis in various malignancies. Telomere length reduction, which is correlated with an adverse outcome, has been identified in breast carcinomas, prostate cancer, and multiple myelomas [25-27]. Telomere elongation is also associated with adverse outcomes, including colorectal carcinoma, head and neck cancer, and neuroblastoma [28-30].
Telomere length in STSs is rarely reported. Smadar reported short telomeres in the majority of ES (20 of 24) and long telomeres in only four tumors; they found that tumors with reduced telomere lengths result in genomic instability. In addition, telomere length reduction was determined to be the only significant predictor of outcome. Thus, they suggested that telomere length reduction in tumor cells at diagnosis can serve as a prognostic marker in ES [31]. Montgomery detected 18 cases STS (nine cases of SRTSs and nine cases of NRTSs), and found that telomere length of SRTS sarcomas is relatively normal or shorter compared with normal tissues, whereas telomere length of NRTSs dramatically increases [14]. In the present study, our results were in accordance with the findings reported by Montgomery. Telomere length in the two types of sarcomas was statistically significant, thereby supporting the idea that telomeres may possibly play an important role in nonspecific genetic alterations in STSs.
Montgomery also demonstrated that the discovery of heterogeneous telomere lengths and evidence of the ALT pathway in the majority of sarcomas with complex karyotypes supported the existence of a telomere maintenance pathway incapable of full karyotypic stabilization in pleomorphic sarcomas. Moreover, telomere dysfunction is a plausible contributor to the chromosomal aberrations found in complex sarcomas [14].
The ALT mechanism remains unclear, but several factors are believed to be involved: (a) mismatch repair defect can result in telomeric recombination, and (b) chromosomal instability leads to chromosomal end-to-end associations and breakage-fusion-bridge cycles, resulting in an increased number of complex nonreciprocal chromosomal rearrangements. Thus, the ALT mechanism was hypothesized to be one of the possible causes of karyotypic complexity [3,32].
Researchers have different views about the relationship between p53 and telomere. Some observations implied that p53 does not regulate telomerase expression or function in sarcomas [33]. Other studies suggested that p53 is associated with the telomeric complex in ALT cells. ALT is a recombination-based mechanism, and the mutation of p53 provides a permissive environment for the activation of this telomere maintenance pathway [34].
In conclusion, our results indicated that p53 and telomere may be involved in the oncogenesis of NRTSs. However, large-scale studies should be performed to evaluate the role of p53 and telomere in STSs. In the near future, a better understanding of the roles and regulation of TMMs should lead to novel therapeutic strategies to improve the treatment of sarcoma patients using telomere-targeting therapy.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81160322 and 81460404).
Disclosure of conflict of interest
None.
References
- 1.de Alava E. Molecular pathology in sarcomas. Clin Transl Oncol. 2007;9:130–44. doi: 10.1007/s12094-007-0027-2. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Vousden KH. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey SM, Murnane JP. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34:2408–17. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin L, Liu CX, Nong WX, Chen YZ, Qi Y, Li HA, Hu WH, Sun K, Li F. Mutational analysis of p53 and PTEN in soft tissue sarcoma. Mol Med Rep. 2012;5:457–61. doi: 10.3892/mmr.2011.660. [DOI] [PubMed] [Google Scholar]
- 5.Nasir L, Devlin P, Mckevitt T, Rutteman G, Argyle DJ. Telomere lengths and telomerase activity in dog tissues: a potential model system to study human telomere and telomerase biology. Neoplasia. 2001;3:351–9. doi: 10.1038/sj.neo.7900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 7.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, Shih IeM, Iacobuzio-Donahue CA, Maitra A, Li QK, Eberhart CG, Taube JM, Rakheja D, Kurman RJ, Wu TC, Roden RB, Argani P, De Marzo AM, Terracciano L, Torbenson M, Meeker AK. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–15. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulaner GA, Huang HY, Otero J, Zhao Z, Ben-Porat L, Satagopan JM, Gorlick R, Meyers P, Healey JH, Huvos AG, Hoffman AR, Ladanyi M. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 2003;63:1759–63. [PubMed] [Google Scholar]
- 9.Sanders RP, Drissi R, Billups CA, Daw NC, Valentine MB, Dome JS. Telomerase expression predicts unfavorable outcome in osteosarcoma. J. Clin. Oncol. 2004;22:3790–7. doi: 10.1200/JCO.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–11. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher , Christopher DM. International Agency for Research on Cancer. 4th edition. Lyon: IARC Press; 2013. WHO classification of tumours of soft tissue and bone World Health Organization. [Google Scholar]
- 12.Li F, Li XX, Chang B, Pang LJ, Yang JH, Hu WH, Lu TC, Li HA, Wang J, Lu HF, Sun MH, Shi DR. Diagnostic significance and clinical application of specific chimeric genes in soft tissue sarcomas by RT-PCR using paraffin-embedded tissues: a study of 103 specimens. Zhonghua Yi Xue Za Zhi. 2004;84:1518–21. [PubMed] [Google Scholar]
- 13.Liu C, Li D, Hu J, Jiang J, Zhang W, Chen Y, Cui X, Qi Y, Zou H, Zhang W, Li F. Chromosomal and genetic imbalances in Chinese patients with rhabdomyosarcoma detected by high-resolution array comparative genomic hybridization. Int J Clin Exp Pathol. 2014;7:690–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery E, Argani P, Hicks JL, DeMarzo AM, Meeker AK. Telomere lengths of translocation-associated and nontranslocation-associated sarcomas differ dramatically. Am J Pathol. 2004;164:1523–9. doi: 10.1016/S0002-9440(10)63710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borden EC, Baker LH, Bell RS, Bramwell V, Demetri GD, Eisenberg BL, Fletcher CD, Fletcher JA, Ladanyi M, Meltzer P, O'Sullivan B, Parkinson DR, Pisters PW, Saxman S, Singer S, Sundaram M, van Oosterom AT, Verweij J, Waalen J, Weiss SW, Brennan MF. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res. 2003;9:1941–56. [PubMed] [Google Scholar]
- 16.Hiyama E, Kodama T, Shinbara K, Iwao T, Itoh M, Hiyama K, Shay JW, Matsuura Y, Yokoyama T. Telomerase activity is detected in pancreatic cancer but not in benign tumors. Cancer Res. 1997;57:326–31. [PubMed] [Google Scholar]
- 17.Hiyama E, Hiyama K. Clinical utility of telomerase in cancer. Oncogene. 2002;21:643–9. doi: 10.1038/sj.onc.1205070. [DOI] [PubMed] [Google Scholar]
- 18.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 19.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, Robinson BG, Learoyd DL, Stalley PD, Bonar SF, Yu D, Pollock RE, Reddel RR. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–25. [PubMed] [Google Scholar]
- 20.Terasaki T, Kyo S, Takakura M, Maida Y, Tsuchiya H, Tomita K, Inoue M. Analysis of telomerase activity and telomere length in bone and soft tissue tumors. Oncol Rep. 2004;11:1307–11. [PubMed] [Google Scholar]
- 21.Yan P, Benhattar J, Coindre JM, Guillou L. Telomerase activity and hTERT mRNA expression can be heterogeneous and does not correlate with telomere length in soft tissue sarcomas. Int J Cancer. 2002;98:851–6. doi: 10.1002/ijc.10285. [DOI] [PubMed] [Google Scholar]
- 22.Tomoda R, Seto M, Tsumuki H, Iida K, Yamazaki T, Sonoda J, Matsumine A, Uchida A. Telomerase activity and human telomerase reverse transcriptase mRNA expression are correlated with clinical aggressiveness in soft tissue tumors. Cancer. 2002;95:1127–33. doi: 10.1002/cncr.10793. [DOI] [PubMed] [Google Scholar]
- 23.Ulaner GA, Hoffman AR, Otero J, Huang HY, Zhao Z, Mazumdar M, Gorlick R, Meyers P, Healey JH, Ladanyi M. Divergent patterns of telomere maintenance mechanisms among human sarcomas: sharply contrasting prevalence of the alternative lengthening of telomeres mechanism in Ewing's sarcomas and osteosarcomas. Genes Chromosomes Cancer. 2004;41:155–62. doi: 10.1002/gcc.20074. [DOI] [PubMed] [Google Scholar]
- 24.Liau JY, Tsai JH, Jeng YM, Lee JC, Hsu HH, Yang CY. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am J Surg Pathol. 2015;39:236–44. doi: 10.1097/PAS.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 25.Fordyce CA, Heaphy CM, Bisoffi M, Wyaco JL, Joste NE, Mangalik A, Baumgartner KB, Baumgartner RN, Hunt WC, Griffith JK. Telomere content correlates with stage and prognosis in breast cancer. Breast Cancer Res Treat. 2006;99:193–202. doi: 10.1007/s10549-006-9204-1. [DOI] [PubMed] [Google Scholar]
- 26.Fordyce CA, Heaphy CM, Joste NE, Smith AY, Hunt WC, Griffith JK. Association between cancer-free survival and telomere DNA content in prostate tumors. J Urol. 2005;173:610–4. doi: 10.1097/01.ju.0000143195.49685.ce. [DOI] [PubMed] [Google Scholar]
- 27.Wu KD, Orme LM, Shaughnessy J Jr, Jacobson J, Barlogie B, Moore MA. Telomerase and telomere length in multiple myeloma: correlations with disease heterogeneity, cytogenetic status, and overall survival. Blood. 2003;101:4982–9. doi: 10.1182/blood-2002-11-3451. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Aranda C, de Juan C, Diaz-Lopez A, Sanchez-Pernaute A, Torres AJ, Diaz-Rubio E, Balibrea JL, Benito M, Iniesta P. Correlations of telomere length, telomerase activity, and telomeric-repeat binding factor 1 expression in colorectal carcinoma. Cancer. 2006;106:541–51. doi: 10.1002/cncr.21625. [DOI] [PubMed] [Google Scholar]
- 29.Patel MM, Parekh LJ, Jha FP, Sainger RN, Patel JB, Patel DD, Shah PM, Patel PS. Clinical usefulness of telomerase activation and telomere length in head and neck cancer. Head Neck. 2002;24:1060–7. doi: 10.1002/hed.10169. [DOI] [PubMed] [Google Scholar]
- 30.Ohali A, Avigad S, Ash S, Goshen Y, Luria D, Feinmesser M, Zaizov R, Yaniv I. Telomere length is a prognostic factor in neuroblastoma. Cancer. 2006;107:1391–9. doi: 10.1002/cncr.22132. [DOI] [PubMed] [Google Scholar]
- 31.Avigad S, Naumov I, Ohali A, Jeison M, Berco GH, Mardoukh J, Stark B, Ash S, Cohen IJ, Meller I, Kollender Y, Issakov J, Yaniv I. Short telomeres: a novel potential predictor of relapse in Ewing sarcoma. Clin Cancer Res. 2007;13:5777–83. doi: 10.1158/1078-0432.CCR-07-0308. [DOI] [PubMed] [Google Scholar]
- 32.Scheel C, Schaefer KL, Jauch A, Keller M, Wai D, Brinkschmidt C, van Valen F, Boecker W, Dockhorn-Dworniczak B, Poremba C. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene. 2001;20:3835–44. doi: 10.1038/sj.onc.1204493. [DOI] [PubMed] [Google Scholar]
- 33.Razak ZR, Varkonyi RJ, Kulp-McEliece M, Caslini C, Testa JR, Murphy ME, Broccoli D. p53 differentially inhibits cell growth depending on the mechanism of telomere maintenance. Mol Cell Biol. 2004;24:5967–77. doi: 10.1128/MCB.24.13.5967-5977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milas M, Yu D, Sun D, Pollock RE. Telomerase activity of sarcoma cell lines and fibroblasts is independent of p53 status. Clin Cancer Res. 1998;4:1573–9. [PubMed] [Google Scholar]


