Abstract
Approximately 15% of gastrointestinal stromal tumors (GIST) do not express KIT mutations and of these about 5 to 7% harbor mutations in PDGFRA. DOG1 was specifically expressed in GISTs. These cases require special attention for PDGFRA and DOG1 mutational status. Hundred cases of GIST were diagnosed between August 2007 and October 2012 at the First Affiliated Hospital of Guangxi Medical University. DNA from tumor tissues and normal adjacent tissues was isolated and amplified for the 22 exons of PDGFRA and 26 exons of DOG1. Each PCR product was sequenced. Amino acid sequences were inferred from DNA and aligned to GenBank reference sequences to determine the position and type of mutations. Overall, 16.0% of the samples had a mutation in PDGFRA, and GISTs with mutations in the DOG1 gene were not found. Of the mutations detected, they were in PDGFRA exon 18 (8 cases, 8%), PDGFRA exon 12 (5 cases, 5%), PDGFRA exon 14 (1 cases, 1.0%), PDGFRA exon 11 (1 cases, 1.0%), and PDGFRA exon 8 (1 cases, 1.0%). Of these, Y392S, L521P and T632K mutant occurred in PDGFRA exon 8, exon 11 and exon 14, respectively. The mutation of PDGFRA has been considered as another causative genetic event as PDGFRA mutations were found in most GISTs lacking a KIT mutation. PDGFRA mutations occurred preferentially in exon 18 and exon 12. Mutations occurring in PDGFRA exon 8 (Y392S), exon 11 (L521P) and exon 14 (T632K) also were first identified. The over-expression of DOG1 was not related to DOG1 gene mutation.
Keywords: Gastrointestinal stromal tumor, mutation, platelet-derived growth factor alpha, DOG1
Introduction
GISTs are the most frequent sarcoma [1] that arises from the interstitial cells of Cajal (ICC) [2,3]. GIST has an estimated annual incidence worldwide of approximately 10-20 per million individuals [4]. ICC can be immunohistochemically identified by antihuman KIT (CD117) antibody since KIT is strongly expressed in most GISTs [5,6]. KIT is a transmembrane tyrosine kinase which serves as a receptor for stem cell factor [7]. The pathogenesis of GISTs is by activation of the KIT receptor tyrosine kinase resulting from the mutations occurring in two oncogenes, KIT and PDGFRA [3,8]. These mutations can cause the receptors to get constitutively activated, leading to the dysfunction of cellular signalling pathways and uncontrolled cell growth and proliferation [9]. Approximately, 85% of GIST tumors were found to have an active mutation in the KIT proto-oncogene [10], this affects the diagnosis of GIST in patients who may benefit from treatment with receptor tyrosine kinase inhibitors. Another close homologous tyrosine kinase PDGFRA was seen in 5% to 7% of GISTs [11]. KIT and PDGFRA are mutually exclusive, and they are involved in similar cellular signaling pathways which results in GIST oncogenesis, but act at different receptor site [12]. Also, 5% of GISTs have no detectable kinase mutations [13]. Discovered on GIST-1 (DOG-1) was reported to have a high sensitivity and specificity in GISTs [14]. DOG1 is identified as a gene in the CCND1-EMS1 locus on human chromosome 11q13, which is a sensitive immunohistochemical marker for GIST. The pathogenesis of DOG-1 in GISTs is still unknown.
In this study, paraffin-embedded tumor tissues from 100 cases of GIST were subjected to DNA extraction, PCR amplification, DNA sequencing and prediction of mutation. The results of this study contributes to gain a better understand the mutations of PDGFRA and screen the mutation in DOG1 gene to explain the over-expression mechanism in GIST. This may provide a better target for more effective therapy in patients with GIST based on molecular analysis. It is often considered that molecular analyses are necessary to confirm the diagnosis of GIST and to determine further therapeutic strategy for the patients [15].
Materials and methods
This study was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University and written informed consent was obtained from every participant.
Patient information
100 GIST patients were diagnosed between August 2007 and October 2012 at the First Affiliated Hospital of Guangxi Medical University. The samples were anonymous and without identifying any personal information. All patients underwent surgical resection through laparotomy and the final diagnosis was obtained from the analysis of clinicopathological findings. The morphological diagnosis was confirmed by standard H&E staining and immunoreactivity to KIT (CD117) on routinely formalin-fixed paraffin-embedded specimens. Our study was approved by the Ethics Committee of the First Affi-liated Hospital of Guangxi Medical University (NO. 2011 KY022).
DNA extraction from tumor tissues and normal adjacent tissues
After manual microdissection of tumor tissues and normal adjacent tissues, DNA was extracted from formalin-fixed paraffin-embedded tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen, Mainz, Germany). Paraffin was removed by dissolving in xylene. Samples were lysed under denaturing conditions with proteinase K. Incubation at 90°C reversed formalin cross-linking. Residual contaminants were washed away. Pure DNA was eluted from the membrane. The purity of DNA is determined by UV spectrophotometry.
PCR amplification of the PDGFRA and DOG1 gene
The primers for the exons of PDGFRA and DOG1 gene were designed from the earlier submitted sequences from the Genbank. All exons of PDGFRA and DOG1 gene were amplified by polymerase chain reaction (PCR) using the primers detailed in Table 1. PCR was performed in a total volume of 50 μL containing 1000 ng of template DNA, 25 μL 2×Taq Plantinum PCR MasterMix (Tiangen, Beijing, China), 2 μL of 10 μM primer1, and 2 μL of 10 μM primer2 (Table 1).
Table 1.
Primers used for PCR
| PDGFRA EXON | PRIMER | DOG1 EXON | PRIMER |
|---|---|---|---|
| EXON 1 FORWARD | gccccattgattctttcatc | EXON 1 FORWARD | cggaaaatctgaccggcg |
| EXON 1 REVERSE | aactgccactggagagcatt | EXON 1 REVERSE | tgagctcttggttgggctc |
| EXON 2 FORWARD | gggtgaatctagtggggctt | EXON 2 FORWARD | agtgagtggatgaagggagc |
| EXON 2 REVERSE | acaggagagacaggaagagaag | EXON 2 REVERSE | ggatggcctcgatctcttga |
| EXON 3 FORWARD | agtggggcaattcttctgga | EXON 3 FORWARD | ttgtggtggcctctgaagat |
| EXON 3 REVERSE | acgcaccttatgattttgcct | EXON 3 REVERSE | acatggtctttgcaggggta |
| EXON 4 FORWARD | ctgaggaatgcggtgttctg | EXON 4 FORWARD | gtgggatggttctctgacca |
| EXON 4 REVERSE | atgtgcccatcagtgacaga | EXON 4 REVERSE | gaacacgctcttctttgggg |
| EXON 5 FORWARD | tagcctcccaccttgtcaac | EXON 5 FORWARD | gtctgttccaaatgcccagg |
| EXON 5 REVERSE | ggttgacagcttccaactgg | EXON 5 REVERSE | cacagacaattagagccggc |
| EXON 6 FORWARD | atgtcagttgtccatgctgc | EXON 6 FORWARD | acacgtcactagtcaagggg |
| EXON 6 REVERSE | agtgacctgtctttccccag | EXON 6 REVERSE | caatgctgatctcaagcccc |
| EXON 7 FORWARD | tcattcagaagtcaggccgt | EXON 7 FORWARD | ttctgccaatcgagcatgtg |
| EXON 7 REVERSE | ttttcttccatgaccgggga | EXON 7 REVERSE | ttcagaggaagtcgaggcag |
| EXON 8 FORWARD | ccggagtgttttgaatgcca | EXON 8 FORWARD | tgttgccccaggatgatctt |
| EXON 8 REVERSE | acatgcagtccgactaccaa | EXON 8 REVERSE | tgctccgctccatcaactta |
| EXON 9 FORWARD | tcccaactccttgccatctt | EXON 9 FORWARD | tctttgcatcccgtgagagt |
| EXON 9 REVERSE | tctgccttgggaccttcatt | EXON 9 REVERSE | gcttcatctaatgctggccc |
| EXON 10 FORWARD | cagacacagccacactacct | EXON 10 FORWARD | ctgcttctgggatgagggaa |
| EXON 10 REVERSE | gtgtgcaagggaaaagggag | EXON 10 REVERSE | atgtcctttcctcctgacgg |
| EXON 11 FORWARD | tggtgctgttggtgattgtg | EXON 11 FORWARD | cccgtcccattgtgtgtttt |
| EXON 11 REVERSE | gtgtgcaagggaaaagggag | EXON 11 REVERSE | aagacagacagaagcagcct |
| EXON 12 FORWARD | ctcctttctcccgtctgtgt | EXON 12 FORWARD | agaatcgcttgaacccagga |
| EXON 12 REVERSE | ccaacatacaggcagcaaga | EXON 12 REVERSE | tgtccatttcctctccaccc |
| EXON 13 FORWARD | gcactgaggccaagtagcta | EXON 13 FORWARD | tcaccctcaagcagcagtaa |
| EXON 13 REVERSE | atgtgtggggatggagagtg | EXON 13 REVERSE | agggtgctggaaggaaagaa |
| EXON 14 FORWARD | actctccatccccacacatg | EXON 14 FORWARD | tgttctcagggccagttcat |
| EXON 14 REVERSE | aagactggacagggtggttt | EXON 14 REVERSE | taccatggcagagttgagca |
| EXON 15 FORWARD | ccagttagctcccatgccta | EXON 15 FORWARD | tttaaaacgcccagaacccg |
| EXON 15 REVERSE | gaactggttgtgcagacctg | EXON 15 REVERSE | tggcatgcatgttaacgagg |
| EXON 16 FORWARD | cctcctccctgattcaagca | EXON 16 FORWARD | gcttcttggaaccctatgcg |
| EXON 16 REVERSE | gtccacactccactcactga | EXON 16 REVERSE | aagccaatcacttccttgcg |
| EXON 17 FORWARD | ctctccttcatcccctacgc | EXON 17 FORWARD | ccatctgtctctggtgggtt |
| EXON 17 REVERSE | caatcagggagacagagggg | EXON 17 REVERSE | acccattgagaaccagggag |
| EXON 18 FORWARD | tgtggccgtgatagctgtaa | EXON 18 FORWARD | ctccctggttctcaatgggt |
| EXON 18 REVERSE | cctggggcttgaaagaacac | EXON 18 REVERSE | tggggaaaggctgaagatgt |
| EXON 19,20 FORWARD | aaaaccagtgcttcaaggct | EXON 19 FORWARD | ttgccttaatcccctcctgg |
| EXON19,20 REVERSE | gctcctctagaactgcggaa | EXON 19 REVERSE | tgcacccgtccctcattaat |
| EXON 21 FORWARD | tggccccttcttcatgttct | EXON 20 FORWARD | acaccccagcatgaaaacac |
| EXON 21 REVERSE | tctgcaacttgtccctgagt | EXON 20 REVERSE | cctccttctctccctgcttc |
| EXON 22 FORWARD | gctttcgtttgtctctgggg | EXON 21 FORWARD | gcaacactgtgaaaccccat |
| EXON 22 REVERSE | gacatcaggtttcaagaggga | EXON 21 REVERSE | tgcatctctcttctccctgc |
| EXON 22 FORWARD | ggtccatcccaagagctcat | ||
| EXON 22 REVERSE | caaacccagcacgcactc | ||
| EXON 23 FORWARD | cctcaacagcaaagccatgt | ||
| EXON 23 REVERSE | ccaaggtccctgatgtcact | ||
| EXON 24 FORWARD | accctgcatccattcactca | ||
| EXON 24 REVERSE | cacaacgcccggctatttaa | ||
| EXON 25 FORWARD | agtaggctctgcactcttgg | ||
| EXON 25 REVERSE | agcaagttctccaagcctct | ||
| EXON 26 FORWARD | aaagtcggccaacatttccc | ||
| EXON 26 REVERSE | tgccgtattaccagccatca |
DNA sequencing
Amplified products were separated in 2% agarose gel electrophoresis to confirm correct amplification. Each PCR product was sequenced by using an ABI DNA Analyzer (Applied Biosystems, Foster City, CA). Mutated amino acids within the sequences were determined based on a comparison to Genbank reference sequences.
Results
100 GIST cases had been examined, 16 cases (16%) had PDGFRA mutations tumor tissues (Table 2; Figure 1). No mutations were detectable in normal adjacent tissues of the 100 cases .The majority of PDGFRA mutations were observed in exon 18 (8 cases, 8%) and in exon 12 (5 cases, 5%). Exon 8, 11 and 14 mutations were seen in 3 cases, respectively. The missense mutation occurring in PDGFRA exon 8 led to the substitution of Tyr to Ser in codon 392. The missense mutation occurring in PDGFRA exon 11 led to the substitution of Leu to Pro in codon 521. The missense mutation in PDGFRA exon 14 led to the substitution of Thr to Lys in codon 632. Of the 8 PDGFRA exon 18 mutations, 5 (50%) were in codon 842, and 4 were in D842V substitutions. Across exon 18, 5 substitutions, 2 deletions, and 1insertion were observed. The patients with single amino acid substitutions encoded by the PDGFRA exon 18 were in codons 814 and 842 and the substitutions were D842V (n=4), and C814R (n=1), respectively. The amino acid deletions in exon 18 was R817del (n=1), D842-845Hdel (n=1). Of the 5 PDGFRA exon 12 mutations, 4 substitutions and 1 insertion was seen. These substitutions were P553Q (n=1), I562L (n=1), L595V (n=1), and S566I (n=1), respectively. Ex-on 8, 11 and 14 mutations identified in 3 cases respectively, all of which were substitutions. Of the 16 PDGFRA mutations, 11 tumors originated from the stomach (7 with PDGFRA exon 18, 3 PDGFRA exon 12 and 1 PDGFRA exon 14), 3 from the small bowel (2 with PDGFRA exon 12 and 1 PDGFRA exon 8) and 2 from the peritoneum (1 with PDGFRA exon 18 and 1 PDGFRA exon 11). DOG1 gene has 26 exons. No DOG1 mutations were found in 100 patients.
Table 2.
Characteristics of patients with PDGFRA gene mutations
| Case no. | Tumor site | Exon | Nucleotide change | Amino acid change |
|---|---|---|---|---|
| 9 | Stomach | 18 | GAC→GTC | Asp→Val (D842V) |
| 11 | Stomach | 18 | TGT→CGT | Cys→Arg (C814R) |
| 15 | Stomach | 18 | GAC→GTC | Asp→Val (D842V) |
| 19 | Stomach | 18 | CCGTG→CG | Arg817del |
| 25 | Stomach | 18 | GAC→GTC | Asp→Val (D842V) |
| 29 | Stomach | 18 | AGACATCATGCATG→AG | AspIleMetHis842_845del |
| 49 | Stomach | 18 | GAC→GTC | Asp→Val (D842V) |
| 77 | Peritoneum | 18 | CGTGAT→CGTCGAT | ArgAsp→ArgArg817ins |
| 23 | Stomach | 12 | CCG→CAG | Pro→Gln (P553Q) |
| 44 | Stomach | 12 | ATT→CTT | Ile→Leu (I562L) |
| 52 | Stomach | 12 | CTT→GTT | Leu→Val (L595V) |
| 81 | Small bowel | 12 | AGC→ATC | Ser→Ile (S566I) |
| 91 | Small bowel | 12 | GAAATT→GACAAT | GluIle→AspAsn556ins |
| 43 | Stomach | 14 | ACG→AAG | Thr→Lys (T632K) |
| 61 | Peritoneum | 11 | CTG→CCG | Leu→Pro (L521P) |
| 46 | Small bowel | 8 | TAT→TCT | Tyr→Ser (Y392S) |
Figure 1.
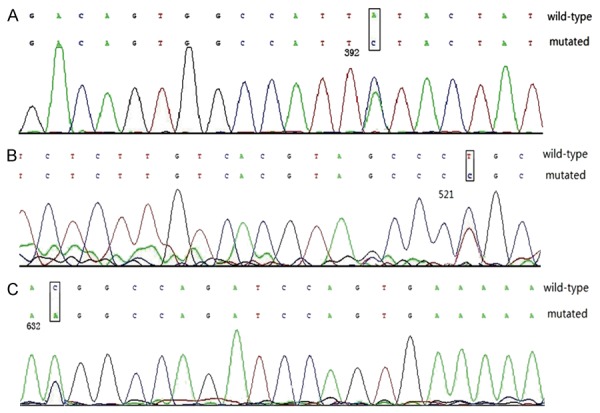
Demonstration of PDGFRA mutations A: Showing PDGFRA exon 8 Y392S; B: Showing PDGFRA exon 11 L521P; C: Showing PDGFRA exon 14 T632K.
Discussion
The present study is the first in China to analyze all exons of PDGFRA and DOG1 mutations associated with GISTs. Recently, the PDGFRA mutations have been considered to be causative genetic events. PDGFRA mutations were found in most GISTs lacks a KIT mutation. KIT mutations were observed in 75% to 80% of GISTs [16,17]. PDGFRA mutations are identified in 22.5% of GISTs [17,18]. In this study, PDGFRA mutations were observed in 16 (16%) out of 100 cases. All the 16 GISTs showed a missense mutation. Mutations involved exons 18, 12, 14, 11 and 8. The mutations in PDGFRA exons 18 and 12 that have been frequently observed in other studies were also identified in this study. The majority of PDGFRA mutations occur in exon 18 [17]. In this study, we found that PDGFRA mutations in exons 18 were identified in 8 (50%) of 16 cases. The most common PDGFRA mutation is D842V in exon 18. Heinrich et al [8] found that when PDGFRA mutation is D842V, it activates phosphorylation cascades independently of the presence of the ligands. PDGFRA mutations occurring in exon 12 are very rare [17]. However, we found that PDGFRA mutations in exons 12 were identified in 5 (31.2%) of 16 cases in our study. Lasota et al reported that all exon 12 PDGFRA mutations in GISTs were clustered between 560 and 577 PDGFRA amino-acid residues, and this region is considered as a mutational “hot spot” for GIST [19]. In this study, the PDGFRA mutations in exons 12 were clustered between 553 and 595 PDGFRA amino-acid residues. No PDGFRA exon 8 and 11 mutant, which was identified in this study, was found in the earlier studies. The mutations in PDGFRA exon 8 (Y392S; TAT→TCT) and PDGFRA exon 11 (L521P; CTG→CCG) are accidental. Mutations have been detected at much lower frequencies in PDGFRA exons 14 [16]. In our study, the PDGFRA exons 14 mutation was identified in only one sample. The mutation in PDGFRA exons 14 (T632K; ACG→AAG) has not been previously identified. The type and location of PDGFRA mutations in GIST can be used to predict the response to imatinib treatment [20]. GISTs with KIT exon 11 mutations exhibit better therapeutic outcomes with imatinib therapy in comparision with wild-type tumors and any other mutations [21]. D842V in exon 18, the most common PDGFRA mutation, is resistant to imatinib [8,22].
DOG1 (FLJ10261, locus on human chromosome 11q13), encoding a hypothetical protein, has been found to be specifically expressed in GISTs. Although the over-expression of DOG1 in GIST has been recently identified, the biological function and the mechanism in GIST are still unknown. Two possible over-expression mechanisms were reported by West et al [23]. ICCs were immunoreactive for DOG1, as in KIT. The protein may be a transmembrane tyrosine kinase which serves as a receptor for stem cell factor. The binding of stem cell receptor to the protein results in activation of tyrosine kinase and downstream intracellular signal transduction pathways. On the other hand, DOG1 is a possible marker of the GIST, which is irrelevant to the kinase type III signal transduction pathways [24]. Miwa et al [25] tried to find the mutation of DOG1 gene to explain the over-expression mechanism in GIST. In their studies, 26 exons of DOG1 from 4 cases of GIST were selected for the molecular analysis and no DOG1 mutation was found. In our studies, 26 exons of DOG1 from 100 cases of GIST were observed and no DOG1 mutation was found. Sequence analysis did not show any evidence of DOG1 gene mutation, so we believed that the over-expression mechanism of DOG1 was not related to DOG1 gene mutation. DOG1 was highly expressed in KIT-and PDGFRA-mutant GISTs [13]. DOG1 identifies the vast majority of both KIT- and PDGFRA-mutated GISTs. This may be of clinical value in identifying candidates for imatinib therapy. DOG1 may also be a potential therapeutic target.
Acknowledgements
This study was supported by national natural science foundation of china (Grant No. 81160274).
Disclosure of conflict of interest
None.
References
- 1.Cassier PA, Ducimetiere F, Lurkin A, Ranchere-Vince D, Scoazec JY, Bringuier PP, Decouvelaere AV, Meeus P, Cellier D, Blay JY, Ray-Coquard I. A prospective epidemiological study of new incident GISTs during two consecutive years in Rhone Alpes region: incidence and molecular distribution of GIST in a European region. Br J Cancer. 2010;103:165–170. doi: 10.1038/sj.bjc.6605743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mettinen M, Lasota J. Gastrointestinal stromal tumors-definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Archiv. 2001;438:1–12. doi: 10.1007/s004280000338. [DOI] [PubMed] [Google Scholar]
- 3.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 4.Goettsch WG, Bos SD, Breekveldt-Postma N, Casparie M, Herings RM, Hogendoorn PC. Incidence of gastrointestinal stromal tumours is underestimated: results of a nation-wide study. Eur J Cancer. 2005;41:2868–2872. doi: 10.1016/j.ejca.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Miettinen M, Sobin LH, Sarlomo-Rikala M. Immunohistochemical spectrum of GISTs at different sites and their differential diagnosis with a reference to CD117 (KIT) Modern Pathology. 2000;13:1134–1142. doi: 10.1038/modpathol.3880210. [DOI] [PubMed] [Google Scholar]
- 6.Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol. 1998;152:1259. [PMC free article] [PubMed] [Google Scholar]
- 7.Gramza AW, Corless CL, Heinrich MC. Resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:7510–7518. doi: 10.1158/1078-0432.CCR-09-0190. [DOI] [PubMed] [Google Scholar]
- 8.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 9.Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245–266. doi: 10.1111/j.1365-2559.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- 10.DeMatteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PW, Demetri GD, Blackstein ME, Blanke CD, von Mehren M, Brennan MF. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet. 2009;373:1097–1104. doi: 10.1016/S0140-6736(09)60500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasota J, Miettinen M. KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs) Semin Diagn Pathol. 2006;23:91–102. doi: 10.1053/j.semdp.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Tan CB, Zhi W, Shahzad G, Mustacchia P. Gastrointestinal stromal tumors: a review of case reports, diagnosis, treatment, and future directions. ISRN Gastroenterol. 2012;2012:595968. doi: 10.5402/2012/595968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janeway KA, Kim SY, Lodish M, Nosé V, Rustin P, Gaal J, Dahia PL, Liegl B, Ball ER, Raygada M. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci. 2011;108:314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa I, Lee CH, Kim MK, Rouse BT, Subramanian S, Montgomery K, Varma S, Corless CL, Heinrich MC, Smith KS. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surgi Pathol. 2008;32:210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto H, Kojima A, Nagata S, Tomita Y, Takahashi S, Oda Y. KIT-negative gastrointestinal stromal tumor of the abdominal soft tissue: a clinicopathologic and genetic study of 10 cases. Am J Surg Pathol. 2011;35:1287–1295. doi: 10.1097/PAS.0b013e3182206f15. [DOI] [PubMed] [Google Scholar]
- 16.Agaram NP, Baren A, Arkun K, DeMatteo RP, Besmer P, Antonescu CR. Comparative ultrastructural analysis and KIT/PDGFRA genotype in 125 gastrointestinal stromal tumors. Ultrastruct Pathol. 2006;30:443–452. doi: 10.1080/01913120600854186. [DOI] [PubMed] [Google Scholar]
- 17.Braconi C, Bracci R, Bearzi I, Bianchi F, Costagliola A, Catalani R, Mandolesi A, Ranaldi R, Galizia E, Cascinu S. KIT and PDGFRα mutations in 104 patients with gastrointestinal stromal tumors (GISTs): a population-based study. Ann Oncol. 2008;19:706–710. doi: 10.1093/annonc/mdm503. [DOI] [PubMed] [Google Scholar]
- 18.Miettinen M, Makhlouf H, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical, and molecular genetic study of 906 cases before imatinib with long-term follow-up. Am J Surg Pathol. 2006;30:477–489. doi: 10.1097/00000478-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Lasota J, Dansonka-Mieszkowska A, Sobin LH, Miettinen M. A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Labor Invest. 2004;84:874–883. doi: 10.1038/labinvest.3700122. [DOI] [PubMed] [Google Scholar]
- 20.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J. Clin. Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 21.Demetri GD, von Mehren M, Antonescu CR, DeMatteo RP, Ganjoo KN, Maki RG, Pisters PW, Raut CP, Riedel RF, Schuetze S, Sundar HM, Trent JC, Wayne JD. NCCN Task Force report: update on the management of patients with gastrointestinal stromal tumors. J Natl Compr Canc Netw. 2010;8(Suppl 2):S1–41. doi: 10.6004/jnccn.2010.0116. quiz S42-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 23.West RB, Corless CL, Chen X, Rubin BP, Subramanian S, Montgomery K, Zhu S, Ball CA, Nielsen TO, Patel R. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am J Pathol. 2004;165:107–113. doi: 10.1016/S0002-9440(10)63279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabah M, Cummins R, Leader M, Kay E. Loss of heterozygosity of chromosome 9p and loss of p16INK4A expression are associated with malignant gastrointestinal stromal tumors. Mod Pathol. 2004;17:1364–1371. doi: 10.1038/modpathol.3800199. [DOI] [PubMed] [Google Scholar]
- 25.Miwa S, Nakajima T, Murai Y, Takano Y, Sugiyama T. Mutation assay of the novel gene DOG1 in gastrointestinal stromal tumors (GISTs) J Gastroenterol. 2008;43:531–537. doi: 10.1007/s00535-008-2195-4. [DOI] [PubMed] [Google Scholar]


