Abstract
Recent studies have shown that Th17 cells may be involved in the pathological process of acute myeloid leukemia. This CD4+ cell subgroup secretes highly homologous interleukin (IL)-17A and IL-17F, and also expresses IL-23 receptor (IL-23R) on the cell surface. Our study aims to investigate the relationship of IL-17A, IL-17F, and IL23R with disease susceptibility, and clarify the relationship between gene polymorphism variation and serum IL-17 level. 62 acute myeloid leukemia patients and 125 healthy controls were included in this study. Restriction fragment length polymorphism polymerase chain reaction (PCR-RFLP) was applied to analyze IL-17A (rs2275913; G-197A), IL17F (rs763780; A7488G; His161Arg), and IL-23R (rs11209026, G1142A; Arg381Gln) alleles. At the same time, enzyme-linked immunoassay analysis (ELISA) was used to test serum IL-17 level in patients. Acute myeloid leukemia patients presented higher rate of IL-17F G single mutant (RR = 4.75, P < 0.001) and GG mutation homozygote (RR = 23.01, P < 0.005). While IL-17A, IL-23R A single mutant and purified AA mutation homozygote showed no correlation with acute myeloid leukemia susceptibility. In addition, ELISA showed that serum IL-17 exhibited no significant difference between acute myeloid leukemia patients and healthy controls had (8.8 ± 7.19 pg/ml vs. 1.4 ± 0.2 pg/ml, P > 0.05). IL-17F G single mutant and GG mutation homozygote were correlated with acute myeloid leukemia susceptibility, while IL-17 gene polymorphism and serum IL-17 level were not. Furthermore, IL-17A and IL-23R gene polymorphism were not associated with acute myeloid leukemia susceptibility.
Keywords: Interleukin-17, gene polymorphism, correlation, acute myeloid leukemia
Introduction
As a kind of fatal hematopoietic stem cell tumor, acute myeloid leukemia (AML) is caused by bone marrow invaded by leukemia cells resulting in normal hematopoiesis inhibition [1-3]. AML usually cause lethal infection, bleeding or organ invasion, and may accompany the phenomenon of white blood cell hyperplasia [4,5]. The pathogenesis of AML has the characteristics of heterogeneity and complexity. Current study suggested that environmental and genetic factors play an important role in the process of acute myeloid leukemia occurrence.
In recent years, researches had confirmed that Th17 cells (main cells that secrete IL-17) play an important role in AML patients [6]. It was reported that the number of Th17 cells or the levels of IL17 and its related cytokines have significant differences between in normal cells and malignant AML, indicating that Th17 may influence the pathophysiological process of AML [7,8]. Other studies have found that the number of Th17 cells reduced obviously in the AML patients got complete remission after chemotherapy, which suggested that Th17 cell number has great clinical value in evaluating treatment effect [8].
The present study suggested that IL-17A and IL-17F (with high homology) generation and IL-23 receptor expression on cell surface is important symbol of the Th17 cells subgroup [9]. IL-23 plays an important role in Th17 cells differentiation, and pathological Th17 cells development. IL-17 cytokines produced by pathological Th17 cells can induce the production of multiple proinflammatory cytokines, including tumor necrosis factor-α, IL-6 and chemokines [10-12]. In addition, IL-23 and IL-21 can induce retinoic acid receptor-related orphan nuclear receptor γt (RORγt) synergy STAT3 in promoting IL-17 expression [13].
At present, there is still lack of report about the correlation of IL-17A, IL-17F, and IL-23R gene polymorphism with disease susceptibility, progression, and treatment response. Therefore, we analyzed IL-17A (rs2275913; G-197A), IL17F (rs763780; A7488G; His161Arg), and IL-23R (rs11209026, G1142A; Arg381Gln) alleles in AML patients and healthy controls, to clarify the relationship between IL-17 gene polymorphism and AML patients.
Materials and methods
Patients and healthy control
62 acute myeloid leukemia patients between Jan 2013 and Dec 2013 in our hospital were enrolled. Inclusion criteria: patients with detailed medical history; with no myelodysplastic syndrome (MDS) previously; myeloproliferative disease history; no contact with treatment or drugs that may cause leukemia potentially; older than 16 years old. There were 24 women and 38 men with median age as 52 years old (19-80 years old). In addition, 125 healthy controls were also included. Inclusion criteria: with no blood relationship to the patients; with no tumor history; matched patients in age, sex, and occupational distribution. There were 62 males and 63 females with no significant difference to the patients in age and sex (P > 0.05).
Experimentation protocols were submitted to and approved by the ethics committee of Sichuan Provincial People’s Hospital.
Genome DNA extraction
Peripheral blood was extracted from the normal control, while bone marrow was collected from AML patient. Ficoll separation method was applied to obtain the mononuclear cells. Purelink genomic DNA kit (Life technology, USA) was used for separation and extraction according to the manufacturer specification.
IL-17A, IL-17, and IL-23R genotype identification
IL-17A (rs2275913; G-197A), IL17F (rs763780; A7488G; His161Arg), and IL-23R (rs11209026, G1142A; Arg381Gln) alleles were identified for polymorphism.
Restriction fragment length polymorphism polymerase chain reaction (PCR-RFLP) [14] was applied to analyze IL17F (rs763780; A7488G) gene polymorphism by amplifying the promoter regions. The primer used as follows [15]: forward, 5’-GTT CCC ATC CAG CAA GAG AC-3’; reverse, 5’-AGC TGG GAA TGC AAA CAA AC-3’. The PCR cycling conditions consisted of an initial, single cycle of 3 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. After 2% agarose electrophoresis, the PCR products were ultraviolet imaged by ethidium bromide staining. The PCR products was digested by NlaIII restriction enzyme (NEB Company, USA) and analyzed after 2% agarose electrophoresis. Three forms of products could be found after digestion and electrophoresis: 412 bp single band (homozygous IL17F G allele, no NlaIII loci); 3 bands at 412, 288 and 124 bp in the length (heterozygous); two bands at 288 and 124 bp (IL17F A homozygous allele). Thermal Cycler 2720 PCR (ABI, USA) was used for IL-17F gene polymorphism PCR amplification.
IL-17A (rs2275913; G-197A) and IL-23R (rs11209026, G1142A; Arg381Gln) alleles were amplified by real-time quantitative PCR and analyzed by Roche LightCycler 480. TaqMan SNP Genotyping Assay (rs2275913 and rs11209026) (Life Technology, USA) were applied for IL - 17A and IL-23R alleles analysis.
ELISA analysis of serum IL-17 level
All of the patient’s blood was collected before chemotherapy. Meanwhile, 20 patients were randomly selected from the patients achieved complete remission for analysis. The blood was centrifuged at 3000 g for 30 min to collect the supernatant. IL-17 level was detected by ELISA kit (R & D, USA) according to the manufacture’s specification. The lowest detection concentration of IL-17 was lower than 15 pg/ml. 10 healthy volunteers was enrolled as control.
Data analysis
All statistical analyses were performed using SPSS19.0 software (Chicago, IL). Genotype and allele frequency between groups were compared through chi-square test and Fisher’s exact test. Odds ratio (OR) and risk ratio (RR) were calculated by Haldane correction method, while significant differences was estimated by Fisher’s exact test. All p values were used for the comparison after calibration (pc). Serum level of IL-17 was tested by the Mann-Whitney U test. P < 0.05 was considered to indicate a statistically significant result.
Results
IL-17A and IL-23R allele and genotype distribution in acute myeloid leukemia and control
IL-17A and IL-23R allele and genotype distribution were lack of significant difference. The detection number of GG, GA, and AA genotype in IL-17A (rs2275913) in AML patients was 23 (37.1%), 25 (40.3%), and 14 (22.6%), respectively; while it was 38 (30.4%), 67 (53.6%), and 20 (16%) in the control. IL-17A heterozygote exhibited a slight increased frequency with the RR as 1.73 (P = 0.091). IL - 17A gene A single mutant allele frequency was 0.427 and 0.428 in patients and controls, respectively (Table 1).
Table 1.
IL-17A allele in acute myeloid leukemia and control
| Polymorphism | AML (n, %) | Healthy control (n, %) | P value |
|---|---|---|---|
| IL-17A (G-197A) rs2275913 | |||
| GG homozygotic type | 23 (37.1%) | 38 (30.4%) | > 0.05 |
| GA heterozygous mutant | 25 (40.3%) | 67 (53.6%) | > 0.05 |
| AA homozygous mutant | 14 (22.6%) | 20 (16.0%) | > 0.05 |
| G single genotype | 48 (77.4%) | 105 (84.0%) | > 0.05 |
| A single mutant | 14 (62.9%) | 87 (69.6%) | > 0.05 |
IL-23R gene A single mutant is relatively rare. No IL-23R AA mutations homozygous genotype was detected in this study. 55 patients (88.7%) and 111 healthy controls (88.8%) presented IL-23R G wild type gene homozygote. IL23R GA heterozygous type showed 7 (11.3%) in AML and 14 (11.2%) in healthy controls, respectively (Table 2). In addition, the frequency of A variant allele was both 0.056 in patients and healthy controls.
Table 2.
IL-23R allele in acute myeloid leukemia and control
| Polymorphism | AML (n, %) | Healthy control (n, %) | P value |
|---|---|---|---|
| IL-23R(G1142A)rs11209026 | |||
| GG homozygotic type | 55 (88.7%) | 111 (88.8%) | > 0.05 |
| GA heterozygous mutant | 7 (11.3%) | 14 (11.2%) | > 0.05 |
| AA homozygous mutant | 0 (0) | 0 (0) | > 0.05 |
| G single genotype | 62 (100%) | 125 (100%) | > 0.05 |
| A single mutant | 7 (11.3%) | 14 (11.2%) | > 0.05 |
Correlation of IL-17 gene polymorphism and AML susceptibility
According to the gene polymorphism results, IL-17F (A7488G) polymorphism variation was associated with AML. IL-17F G allele frequency was higher in patients than in healthy controls. In AML patients and healthy controls, the incidence of G allelic mutants were 32.3% (20/62) and 8.8% (11/125), with G single mutation RR = 4.75 (adjusted P value = 0.001). The allele frequency of IL-17 G single mutant in patients and healthy controls were 0.202 and 0.044, respectively (Table 3; Figure 1).
Table 3.
IL-17F allele and genotype in acute myeloid leukemia and control
| Polymorphism | AML (n, %) | Healthy control (n, %) | P value |
|---|---|---|---|
| IL-17F(A7488G)rs763780 | |||
| AA homozygotic type | 42 (67.7%) | 114 (91.2%) | > 0.05 |
| AG homozygous mutant | 15 (24.2%) | 11 (8.8%) | > 0.05 |
| GG heterozygous mutant | 5 (8.1%)b | 0 (0)b | < 0.01 |
| A single genotype | 57 (91.9%) | 125 (100%) | > 0.05 |
| G single mutant | 20 (32.3%)a | 11 (8.8%)a | < 0.05 |
IL-17F (A7488G) polymorphism variation was associated with AML;
RR = 4.75, P = 0.0009, pc = 0.0027;
RR = 23.01, P = 0.0036, pc = 0.0108.
Figure 1.
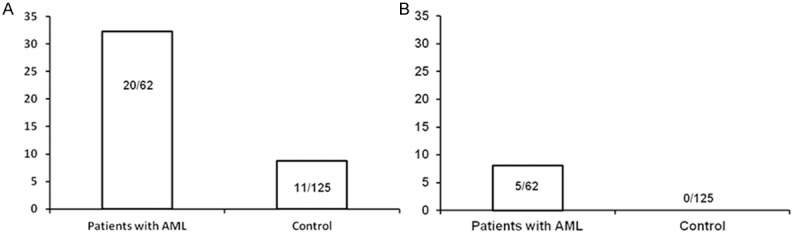
The correlation of IL-17 gene polymorphism and AML susceptibility. A. AML susceptibility was correlated with IL-17 gene G single mutant; B. AML was associated with IL-17 gene GG heterozygous mutant.
IL-17F GG homozygous genotype showed higher correlation with acute myeloid leukemia. The detection number of IL-17F GG homozygote in patients and healthy controls were 8.1% (5/62) and 0 (0/125), with RR = 23.01 (adjusted P value = 0.05).
Correlation of IL-17F gene polymorphism and plasma IL-17 level
IL-17 protein was detected in 9 AML patients (at diagnosis) (9/62), 2 AML patients with complete remission (2/19), and 2 healthy controls (2/10). The IL-17 protein level in 9 AML patients was 8.8 ± 7.19 pg/ml (1.37-19.10 pg/ml), while it was 1.4 ± 0.2 pg/ml in AML patients with complete remission. We did not analyze the correlation among plasma IL-17 level, disease progression, and gene polymorphism.
Discussion
In this study, we analyzed the correlation of AML susceptibility with IL-17A, IL-17F, and IL-23R gene polymorphic variants. In addition, we also calculated the relationship between IL-17 gene polymorphism variations and serum IL-17 level. The results showed that IL-17F gene G single mutant and GG homozygous mutant were associated with AML susceptibility to acute myeloid leukemia, while IL-17A and IL-23R gene polymorphism showed no correlation with it.
After analyzing three pairs of alleles, only IL-17F (A7488G) polymorphism showed association with AML susceptibility. Our results revealed that AML patients exhibited significantly higher frequency of IL-17F G variant and homozygous than that of healthy controls. It was first reported in Chinese patients.
Kawaguchi et al. [16] observed the functional result of IL – 17F polymorphisms and found that the expression and activity of IL-17 would be suppressed during G allele mutation. However, we failed to found the similar effect. According to Kawaguchi et al. [16] results, we expected the patients carrying the allelic variation might express lower levels of IL-17. However, we did not find the significant differences of plasma IL-17 level in patients with different IL-17F genotypes.
For rs2275913 IL17A gene polymorphism, Espinoza et al. [17] confirmed that T cells from healthy control exhibited significant higher level of IL-17 protein after stimulated for 197 A allele in vitro. Similar to IL-17F polymorphism, however, we failed to observe the significant differences between IL-17A genotype and plasma IL-17 level (data not shown). In addition, there was no significant difference between the plasma level of IL-17 in AML patients and healthy controls, which was the same to the previous report [18].
It has been reported that IL-17 concentration is different from the number of cells that can produce IL-17, but they both can cause adverse reactions. Wu et al. [8] revealed that the number of peripheral Th17 cells increased significantly in AML patients comparing with healthy controls. They also found that IL-17 concentration increase may be accompanied by an increase of Th17 cell number in patients, and Th17 cell number decreased in patient with complete response undergoing chemotherapy. Recently, Abousamra et al. [7] suggested that the number of circulating Th17 cells significantly increased in acute lymphoblastic leukemia and AML patients compared with healthy controls. In these patients, Th17 cell number decreased obviously in patients with complete remission after treatment. Tian et al. [19] indicated that compared with normal controls, Th17 cell number upregulated markedly in acute lymphoblastic leukemia patients, while IL-17 level decreased obviously.
It is necessary to point out that the patient’s race in the last three studies [7,8,19] race was not same. Variant results may be due to the difference of patient population, correlation between IL-17F mutants and plasma IL-17 level might be related to the different races of patient population.
Han et al. found that the increase of Th17 cells number and IL-17 level was associated with poor prognosis of AML patients [20]. However, we failed to observe the relationship between IL-17 gene polymorphism and the therapeutic response (data not shown).
To sum up, our study tested the relationship of AML with IL-17A, IL-17F, and IL-23R gene polymorphism in Chinese for the first time. Our results showed that IL-17F gene polymorphism was associated with AML susceptibility.
Disclosure of conflict of interest
None.
References
- 1.O’Donnell MR, Abboud CN, Altman J, Appelbaum FR, Coutre SE, Damon LE, Foran JM, Goorha S, Maness LJ, Marcucci G. Acute myeloid leukemia. J Natl Compr Canc Netw. 2011;9:280–317. doi: 10.6004/jnccn.2011.0027. [DOI] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Pediatr Clin North Am. 2008;55:21–51. doi: 10.1016/j.pcl.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37:649–658. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 5.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 6.Li P, Ji M, Park J, Bunting KD, Ji C, Tse W. Th17 related cytokines in acute myeloid leukemia. Front Biosci (Landmark Ed) 2012;17:2284–2294. doi: 10.2741/4052. [DOI] [PubMed] [Google Scholar]
- 7.Abousamra NK, Salah El-Din M, Helal R. Prognostic value of Th17 cells in acute leukemia. Med Oncol. 2013;30:732. doi: 10.1007/s12032-013-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, Wang S, Wang F, Chen Q, Peng S, Zhang Y, Qian J, Jin J, Xu H. Increased frequencies of T helper type 17 cells in the peripheral blood of patients with acute myeloid leukaemia. Clin Exp Immunol. 2009;158:199–204. doi: 10.1111/j.1365-2249.2009.04011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hot A, Zrioual S, Toh ML, Lenief V, Miossec P. IL-17A- versus IL-17F-induced intracellular signal transduction pathways and modulation by IL-17RA and IL-17RC RNA interference in rheumatoid synoviocytes. Ann Rheum Dis. 2011;70:341–348. doi: 10.1136/ard.2010.132233. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 12.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 14.Aydin-Sayitoglu M, Hatirnaz O, Erensoy N, Ozbek U. Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias. Am J Hematol. 2006;81:162–170. doi: 10.1002/ajh.20434. [DOI] [PubMed] [Google Scholar]
- 15.Resende RG, Correia-Silva Jde F, Silva TA, Salomao UE, Marques-Silva L, Vieira EL, Dutra WO, Gomez RS. IL-17 genetic and immunophenotypic evaluation in chronic graft-versus-host disease. Mediators Inflamm. 2014;2014:571231. doi: 10.1155/2014/571231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi M, Takahashi D, Hizawa N, Suzuki S, Matsukura S, Kokubu F, Maeda Y, Fukui Y, Konno S, Huang SK, Nishimura M, Adachi M. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. J Allergy Clin Immunol. 2006;117:795–801. doi: 10.1016/j.jaci.2005.12.1346. [DOI] [PubMed] [Google Scholar]
- 17.Espinoza JL, Takami A, Nakata K, Onizuka M, Kawase T, Akiyama H, Miyamura K, Morishima Y, Fukuda T, Kodera Y, Nakao S. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS One. 2011;6:e26229. doi: 10.1371/journal.pone.0026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrobel T, Mazur G, Jazwiec B, Kuliczkowski K. Interleukin-17 in acute myeloid leukemia. J Cell Mol Med. 2003;7:472–474. doi: 10.1111/j.1582-4934.2003.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian T, Sun Y, Li M, He N, Yuan C, Yu S, Wang M, Ji C, Ma D. Increased Th22 cells as well as Th17 cells in patients with adult T-cell acute lymphoblastic leukemia. Clin Chim Acta. 2013;426:108–113. doi: 10.1016/j.cca.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Han Y, Ye A, Bi L, Wu J, Yu K, Zhang S. Th17 cells and interleukin-17 increase with poor prognosis in patients with acute myeloid leukemia. Cancer Sci. 2014;105:933–942. doi: 10.1111/cas.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]


