Abstract
The myeloid and lymphoid neoplasms with eosinophilia and PDGFRA gene rearrangements usually show a good response to Imatinib and are typically associated with a normal karyotype, occasionally exhibiting a secondary chromosomal abnormality associated with clonal evolution. Five variant translocations involving PDGFRA have been reported. Here, we report a rare case of therapy-related acute myeloid leukemia with PDGFRA rearrangement after chemotherapy for prior B lymphoblastic leukemia (B-ALL). The patient had a history of BCR-ABL negative, hypodiploid B-ALL in complete remission after chemotherapy. However, 15 months later the patient developed acute myeloid leukemia with rapidly increasing eosinophilia, basophilia and a complex karyotype that included a novel t(4;14)(q12;q24). FIP1L1 was not associated with the PDGFRA rearrangement. The patient had a very aggressive clinical course, and died from the disease shortly after diagnosis. This is the first case of a primary therapy-related myeloid neoplasm with secondary PDGFRA rearrangement. The t(4:14)(q12;q24) is joining the growing list of the variant translocations involving PDGFRA.
Keywords: Acute myeloid leukemia (AML), PDGFRA, eosinophilia, basophilia, therapy-related myeloid neoplasm
Introduction
Myeloproliferative and lymphoid neoplasms associated with rearrangement of PDGFRA is a newly recognized entity in the 2008 World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues, and most commonly manifest as de novo chronic eosinophilic leukemia, but also less frequently as acute myeloid leukemia or T-lymphoblastic leukemia [1]. The most common PDGFRA rearrangement is FIP1L1-PDGFRA fusion, which results from an approximate 800 kb interstitial chromosomal deletion that includes the cysteine-rich hydrophobic domain 2 (CHIC2) locus at 4q12 [2,3]. These cases typically show a normal karyotype by conventional cytogenetic study, and the FIP1L1-PDGFRA fusion is inferred by fluoroscent in situ hybridization (FISH) study for CHIC2 deletion. There are five non-FIP1LI associated balanced translocations with PDGFRA gene identified so far: t(2;4)(p22;q12) [4], ins(9;4)(q33;q12q25) [5], t(4;10)(q12;p11) [6], t(4;12)(q12;p13) [4,7] and t(4;22)(q12;q11.2) [8]. The FIP1L1-PDGFRA and other variant translocations of PDGFRA result in a constitutively activated tyrosine kinase that transforms hematopoietic cells, and therefore it is a therapeutic target for tyrosine kinase inhibitors [9]. The small molecule tyrosine kinase inhibitors from the 2-phenylaminopyrimidine class of compounds, e.g. Imatinib mesylate (Gleevec), have been shown to be effective in the treatment of neoplasms associated with PDGFRA rearrangement [4,7,10-12].
The myeloid neoplasm with FIP1L1-PDGFRA translocation often show atypical mast cell proliferation in the marrow. The mast cells are usualy spindle-shaped with aberrant expression of CD25. But clusters of mast cells characteristic of systemic mastocytosis are absent [13-15]. The basophils in the peripheral blood and bone marrow are usually within normal limits. Basophilia can be seen rarely in acute myelogenous leukemia, typically with t(6;9)(p23;q34) (DEK-NUP214), which has distinct clinicopathological features of marrow basophilia, myelodysplasia and high prevalence of FLT3-ITD mutations with poor prognosis [16]. A case of myelodysplastic syndrome accompanied by basophilia and eosinophilia with t(5;12)(q31;p13) has also been reported [17]. However, the association of PDGFRA gene rearrangement with basophilia has not been reported.
Here, we report a unique case of therapy-related acute myeloid leukemia after intensive chemotherapy for prior B-ALL with eosinophilia, basophilia, PDGFRA gene rearrangement, a complex karyotype including t(4;14) and a very poor clinical outcome.
Case report
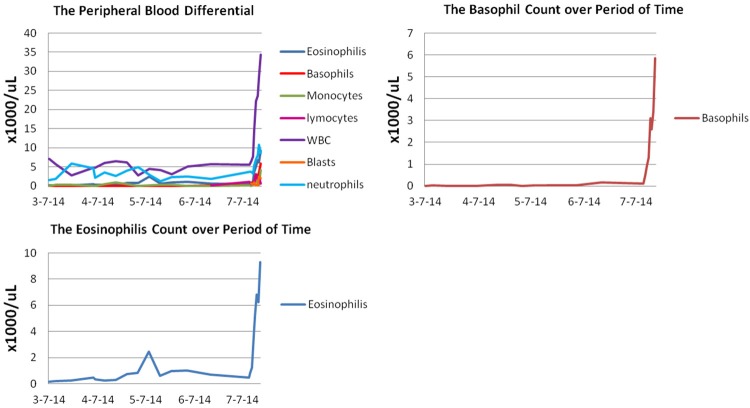
A 77-year-old male presented with weakness, dyspnea on exertion, soaking sweats, low grade fever, loss of appetite, 20-30 pound weight loss, and lower gastrointestinal bleeding while on coumadin. A chest CT with contrast showed mediastinal lympoadenopathy (subcarinal lymph node measuring 3.5 × 2.3 cm and right hilar lymph node measuring 2 × 2 cm). Laboratory tests revealed a white blood cell count of 2.4 × 109/L, hemoglobin of 98 g/L, and platelets of 101 × 109/L. Chemistry profile and liver function tests were normal. Bone marrow biopsy demonstrated hypercellular marrow (95% cellularity) with 95% of blasts (Figure 1), which were positive for TdT, CD34, CD20 and negative for CD3, CD5, CD10, CD23, CD117, MUM1, TRAP, Cyclin D1, and BCL6 by immunohistochemical stains (IHC). Flow cytometry detected a population of blasts positive for TdT, CD34, CD45, CD19, CD20, CD22, and CD38, and negative for CD5, CD10, CD23 and CD117. Cytogentic study showed a hypodiploid karyotype: 39,XY,-3,-7,-13,-14,-15,-16,-17[3]/78,idemx2,-10,+13[9]/74,idemx2,t(2;5)(p21;p14),-4[3]/46,XY[5]. FISH was negative for BCR-ABL gene fusion. He was diagnosed with B lymphoblastic leukemia (B-ALL) and treated with Rituxan plus HyperCVAD (cyclophosphamide, vincristine, doxorubicin, and dexamethasone; methotrexate and cytarabine) and POMP (Purinethol, Oncovin, methotrexate, and prednisone) plus Rituxan; and achieved completely remission. The subsequent restaging marrow demonstrated no evidence of residual B-ALL by IHC and flow cytometry. Cytogenetic study showed normal male karyotype of 46, XY [20]. FISH study was negative for -7, and molecular studies were negative for clonal IgH and light chain gene arrangements. Thirteen months later, the patient presented with mild anemia (hemoglobin: 10.7 g/dL), normal white blood cell and platelet counts. A restaging bone marrow biopsy showed no evidence of residual B-ALL, but multileage dysplasia and focally increased eosinophils (Figure 2). Cytogenetic study demonstrated a complex karyotype of 45,XY,t(4;14)(q12;q24),der(5;21)(p10;q10),add(10)(q22),der(17)t(11;17)(q13;p11.2) [cp7]/46,XY[13]. FISH analysis showed -5q [15/200], but negative for -7/+8/-20q/-p53. These chromosomal abnormalities were unrelated to the previously diagnosed hypodiploid B lymphoblastic leukemia, and suggestive of a myeloid clone. A therapy-related myeloid neoplasm was suspected. Meanwhile the patient started showing fluctuating eosinophilia, with an absolute eosinophil count ranging between 0.20 to 2.44 × 109/L, associated with neutropenia and lymphocytopenia (Figure 3). Two months later, the patient was admitted with fever, weakness, fatigue, decreased activity, cough and diffuse abdominal pain. Peripheral blood showed markedly increased white cell count with rapidly elevated eosinophilia (up to 9.3 × 109/L) and basophilia (up to 5.85 × 109/L) in the course of one week. The rise of white cell count was accompanied by left-shift and circulating blasts (Figure 4A). Bone marrow evaluation performed at this time demonstrated hypercellular marrow with average 75% cellularity, myeloid hyperplasia and prominent dysplasia in all three lineages. In contrast to the previous biopsy, marked eosinophilia (21%), basophilia (10%) with increased blasts (22%) were noted in this sample (Figure 4B-E). The monocytes and mast cells are within normal limits in the marrow. Immunostain for tryptase on the core biopsy showed no mast cell proliferation or abnormal morphology (Figure 4F). Cytogenetic analysis showed a complex karyotype: 45,XY,del(3)(p12),t(4;14)(q12;q24),der(5;21)(p10;q10),add(10)(q22), and 50-51,XY,t(4;14)(q12;q24),+5,der(5;21)(p10;q10),+6,+8,+10,add(10)(q22)x2,+11,der(17)t(11;17)(q13;p11.2),+1-2mar[cp2] (Figure 5A-C). The abrupt presence of prominent eosinophilia in the peripheral blood and bone marrow and 4q12 translocation prompted us to evaluate for PDGFRA rearrangement. FISH analysis performed using myeloproliferative disease probe set targeted at PDGFRA, PDGFRB and FGFR gene rearrangement (Abbott Molecular) revealed deletion of the CHIC2 gene along with deletion of the FIP1LI in 61% of the cells, consistent with a non-FIP1L1 associated PDGFRA rearrangement. FISH study also showed deletion of one PDGFRB gene locus in 78% of the cells. Flow cytometry demonstrated approximately 11% myeloblasts with aberrant expression of CD7 and no phenotypic evidence of B lymphoblastic leukemia. Given the intensive chemotherapy received for B-ALL and complex karyotype, a diagnosis of therapy-related acute myeloid leukema with eosinophilia, basophilia and PDGFRA rearrangement was rendered. Unfortunately the patient’s condition rapidly deteriorated and he expired shortly after the diagnosis, before initiating tyrosine kinase inhibitor treatment.
Figure 1.
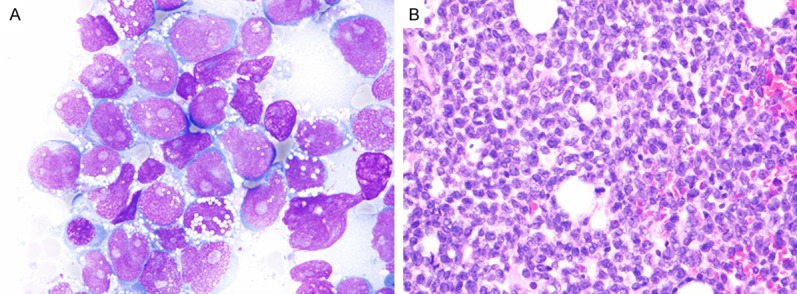
B-lymphoblastic leukemia at initial diagnosis. A. Predominance of large lymphoblasts with round to indented nuclei, fine chromatin, and basophilic cytoplasm with cytoplasmic vacuoles (Aspirate smears, Wright stain, 1000 ×). B. Hypercellular marrow with sheets of blasts (Bone marrow core biopsy, H & E stain, 100 ×).
Figure 2.
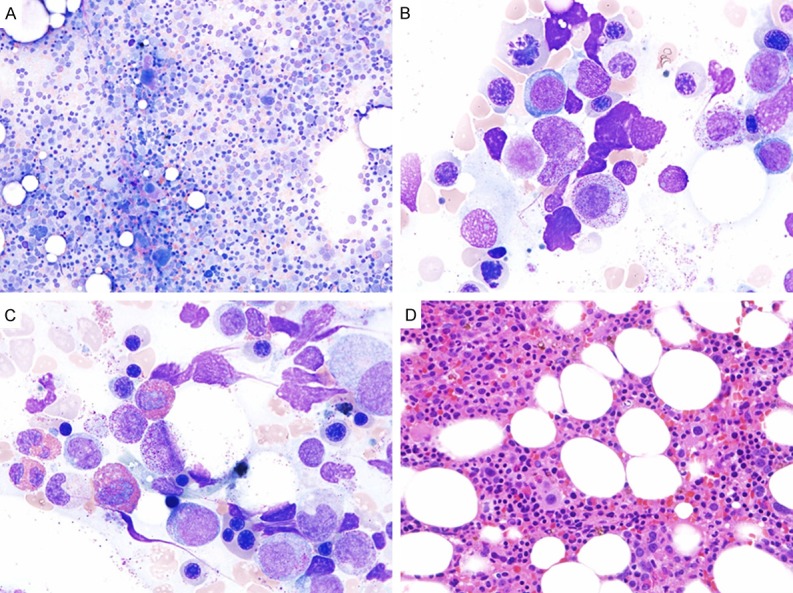
Restaging bone marrow biopsy after chemotherapy for B-ALL showed multilineage dysplasia and focal eosinophilia. A. Megakaryocytic dysplasia with hypolobulated nuclei (Aspirate smears, Wright stain, 100×); B. Erythroid dysplasia with prominent nuclear budding (Aspirate smears, Wright stain, 1000×); C. Focally increased eosinophils and precursors as well as erythroid dysplasia (Aspirate smears, Wright stain, 1000×); D. Hypercellular marrow with dysplastic megakaryocytes (Bone marrow core biopsy, H & E stain, 100×).
Figure 3.
The absolute counts of peripheral blood with differential demonstrated marked leukocytosis with a rapid increase of eosinophils and basophils, along with circulating blasts.
Figure 4.
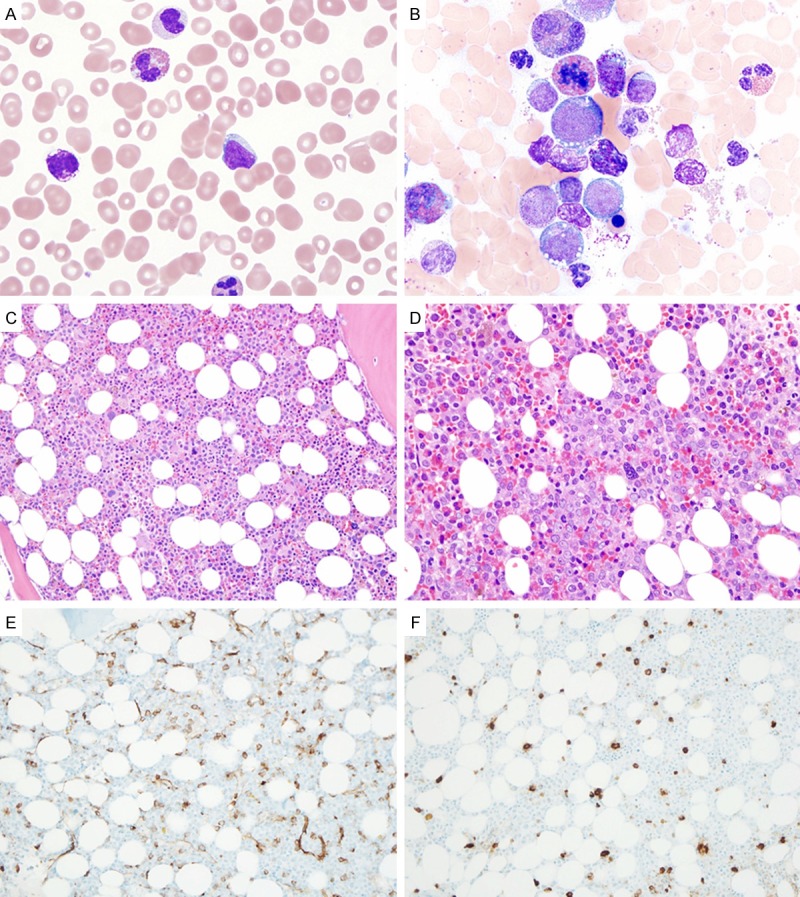
Acute myeloid leukemia with eosinophilia, basophilia and FIP1L1-PDGFRA. A. Eosinophila, basophilia and circulating blasts (Peripheral blood smear, Wright-giemsa stain, 1000×); B. Eosinophilia, basophilia and increased myeloblasts (Aspirate smears, Wright stain, 1000×); C. Hypercellular marrow with hypolobulated dysplastic megakaryocytes (Bone marrow core biopsy, H & E stain, 100×); D. Markedly increased blasts (ALIP) as well as eosinophils and precursors (Bone marrow core biopsy, H & E stain, 200×); E. Approximately 20% CD34-positive blasts (Bone marrow core biopsy, immunostain, 100×); F. Normal number and morphology of tryptase-positive mast cells (Bone marrow core biopsy, immunostain, 100×).
Figure 5.
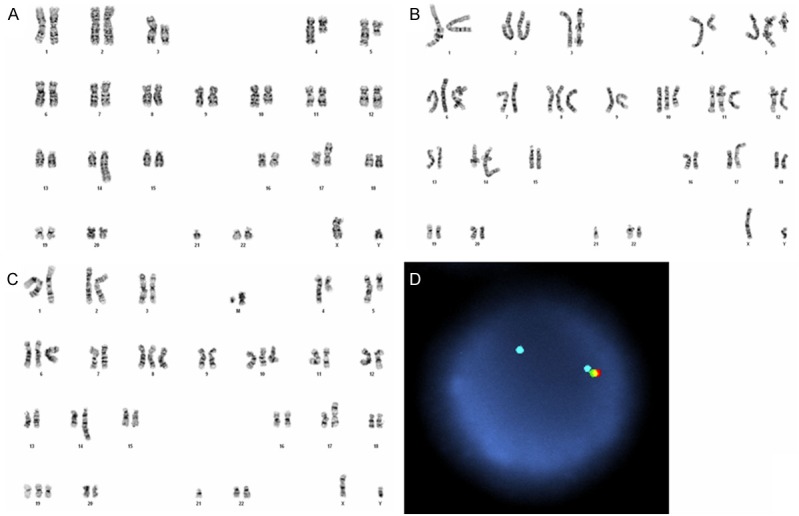
Cytogenetic and FISH findings in acute myeloid leukemia with eosinophilia, basophilia and FIP1L1-PDGFRA. A. Complex karyotype with 45,XY,del(3)(p12), t(4;14)(q12;q24),der(5;21)(p10;q10),add(10)(q22). B and C. Complex karyotype with 50-51,XY,t(4;14)(q12;q24.3),+5,der(5;21)(p10;q10),+6,+8,+10,add(10)(q22)x2,+11,der(17)t(11;17)(q13;p11.2),+1-2mar. D. Interphase fluorescence in situ hybridization (FISH) tricolor 4q12 linked gene analysis: positive for loss of one FIP1L1 signal (green) and one CHIC2 signal (red) from the isolated PDGFRA signal (aqua).
Discussion
The PDGFRA is a member of the class III receptor tyrosine kinase family that also includes c-KIT, and FLT3 [18]. PDGFRA is located on chromosome 4q12 [3] and with a small interstitial deletion of 4q12, leads to juxtaposition of FIP1L1 and PDGFRA resulting in a gain of function fusion protein with signal independent kinase activity and therefore increased cell proliferation and survival [11]. This interstitial deletion is generally cryptic and not detectable using standard cytogenetic banding techniques. A small subset of cases, usually as single case reports showed variant translocation involving PDGFRA. Until now, 5 variant translocations have been reported in the literature: ins(9;4)(q33;q12q25) CDK5RAP2/PDGFRA; t(2;4)(p22;q12) STRN/PDGFRA; t(4;10)(q12;p11) KIF5B/PDGFRA; t(4;12)(q12;p13) PDGFRA/ETV6; and t(4;22)(q12;q11.2) BCR/PDGFRA [3-5,7,8]. The t(4;14)(q12;q24) reported in our case is a new variant translocation involving PDGFRA. Myeloid neoplasms with esoinophilia and PDGFRA rearrangement typically show a normal karyotype with occasional unrelated simple chromosomal abnormality (e.g., trisomy 8, del (20q), del(17p)) [1]. Complex karyotype is exceedingly rare in the setting of PDGFRA rearrangement. To our knowledge, other than our case, only one previous case has been reported in the literature by Védy et al [19].
The myeloid neoplasms associated with PDGFRA rearrangement include predominantly chronic eosinophilic leukemia, and small subset cases of acute myeloid leukemia. They share the similar features of marked peripheral blood eosinophilia and rearrangement of a receptor tyrosine kinase with myeloid and lymphoid neoplasms with abnormalities of PDGFRB and FGFR. Acute myeloid leukemia with inv(16)(p13.1q22) or t(16;16)(p13.1;q22), CBFB-MYH11 also presents with marked eosinophilia. Among these neoplasms, the diagnosis of myeloid neoplasm with PDGFRA rearrangement is particularly problematic as the cytogenetic change is cryptic. FISH study for CHIC2 deletion is the only clinically available test to confirm the PDGFRA rearrangement, which can only be ordered when there is a clinical suspicion for it. Variable eosinophilia can be seen occasionally in MDS and AML without PDGFRA, PDGFRB, FGFR or CBFB rearrangement, and likely represents a reactive response. The peripheral blood eosinophilia is almost always marked (>1.5 × 109/L) in the myeloid neoplasm with PDGFRA rearrangement. However, in our case, the 13 month staging marrow showed the presence of t(4;14)(q12;q24), but no significant peripheral or bone marrow eosinophilia, although focally increased eosinophils and precursors are identified in the bone marrow aspirate. Because of the absence of peripheral blood eosinophilia, a PDGFRA rearrangement was not suspected despite the presence of 4q12 translocation. At this time, there is no clinical guideline for FISH test for PDGFRA rearrangement as to the threshold of eosinophilia. Based on our experience, it appears reasonable to recommend FISH test for PDGFRA rearrangement on cases with either marked eosinophilia (>1.5 × 109/L) or a balanced translocation of chromosome 4q12 as revealed by conventional cytogenetics.
Basophilia is typically absent in myeloid neoplasms associated with PDGFRA, PDGFRB and FGFR rearrangements. Basophilia is usually seen in chronic myeloid leukemia (CML) and AML with t(6;9)(p23;q34) or chromosome 12p abnormalities [16] and is an important diagnostic feature of these neoplasms. The morphologically and functionally closely related mast cells, on the other hands, usually show proliferation in the FIP1L1-PDGFRA associated MPN [13]. This mast cell proliferation is associated with abnormal spindle-shaped morphology and aberrant expression of CD25 [14]. Similar mast cell proliferation was not reported in the cases with variant translocation of PDGFRA. The markedly and rapidly elevated basophilia along with eosinophilia in our case is unique and has not been previously reported. Similar to cases with variant translocations of PDGFRA, the mast cells in our case were within normal limits.
Patients with PDGFRA rearrangements tend to have a more severe disease, associated with extensive end-organ damage [11,20,21]. The identification of the FIP1L1- PDGFRA fusion gene is significant since these patients have excellent response to tyrosine kinase inhibitor Imatinib, including the only previously reported case with complex karyotype [10,11]. The rare patients with variant translocations involving PDGFRA are also responsive to Imatinib [4]. Compared with Védy’s case [19] with complex karyotype which was a de novo AML without prior cytotoxic chemotherapy, the extremely poor clinical course in our case is due to different clinical scenario. Our patient had a history of B-ALL and status post intense cytotoxic chemotherapy, and the AML with PDGFRA rearrangement developed within two years after chemotherapy for B-ALL. Given the presence of a complex karyotype and multi-lineage dysplasia, this should be classified as therapy-related myeloid neoplasm/AML according to the 2008 WHO classification, which typically is associated with poor prognosis. The t(4;14)(q12;q24) and PDGFRA rearrangement is believed to arise along the way of chromosomal evolution to AML.
Transdifferentiation of hematopoietic neoplasms from one lineage to another lineage has been well documented. However, in our case, it is very unlikely that the AML with PDGFRA rearrangement and the prior B-ALL are clonally related. They showed completely different karyotypes including different balanced translocations. In addition, peripheral blood and bone marrow eosinophilia was not present at the time of diagnosis of B-ALL.
In summary, this is a unique case of therapy-related AML with eosinophilia, basophilia, non-FIP1L1 associated PDGFRA rearrangement, and complex karyotype with a new variant translocation t(4;14)(q12;q24) involving the PDGFRA gene locus. The extremely poor prognosis is directly related to chromosomal damage secondary to prior intensive chemotherapy. The variant PDGFRA rearrangement is derived as part of chromosomal change along the evolution of the malignant myeloid clone.
Disclosure of conflict of interest
None.
References
- 1.Bain B, Gilliland D, Horny H, Vardiman J. Myelod and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. In: Swerdlow SHC, Elias , Harris NL, Jaffe ES, editors. WHO Classification of Tumours of haematopoetic and Lymphoid Tissues. 69008 Lyon, France: International Agency for Research on Cancer(IARC); 2008. [Google Scholar]
- 2.Iwasaki J, Kondo T, Darmanin S, Ibata M, Onozawa M, Hashimoto D, Sakamoto N, Teshima T. FIP1L1 presence in FIP1L1-RARA or FIP1L1-PDGFRA differentially contributes to the pathogenesis of distinct types of leukemia. Ann Hematol. 2014;93:1473–1481. doi: 10.1007/s00277-014-2085-1. [DOI] [PubMed] [Google Scholar]
- 3.Bain BJ. Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Haematologica. 2010;95:696–698. doi: 10.3324/haematol.2009.021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis CE, Grand FH, Musto P, Clark A, Murphy J, Perla G, Minervini MM, Stewart J, Reiter A, Cross NC. Two novel imatinib-responsive PDGFRA fusion genes in chronic eosinophilic leukaemia. Br J Haematol. 2007;138:77–81. doi: 10.1111/j.1365-2141.2007.06628.x. [DOI] [PubMed] [Google Scholar]
- 5.Walz C, Curtis C, Schnittger S, Schultheis B, Metzgeroth G, Schoch C, Lengfelder E, Erben P, Müller MC, Haferlach T, Hochhaus A, Hehlmann R, Cross NC, Reiter A. Transient response to imatinib in a chronic eosinophilic leukemia associated with ins(9;4)(q33;q12q25) and a CDK5RAP2-PDGFRA fusion gene. Genes Chromosomes Cancer. 2006;45:950–956. doi: 10.1002/gcc.20359. [DOI] [PubMed] [Google Scholar]
- 6.Tashiro H, Shirasaki R, Noguchi M, Gotoh M, Kawasugi K, Shirafuji N. Molecular analysis of chronic eosinophilic leukemia with t(4;10) showing good response to imatinib mesylate. Int J Hematol. 2006;83:433–438. doi: 10.1532/IJH97.05180. [DOI] [PubMed] [Google Scholar]
- 7.Curtis CE, Grand FH, Waghorn K, Sahoo TP, George J, Cross NC. A novel ETV6-PDGFRB fusion transcript missed by standard screening in a patient with an imatinib responsive chronic myeloproliferative disease. Leukemia. 2007;21:1839–1841. doi: 10.1038/sj.leu.2404728. [DOI] [PubMed] [Google Scholar]
- 8.Baxter EJ, Hochhaus A, Bolufer P, Reiter A, Fernandez JM, Senent L, Cervera J, Moscardo F, Sanz MA, Cross NC. The t(4;22)(q12;q11) in atypical chronic myeloid leukaemia fuses BCR to PDGFRA. Hum Mol Genet. 2002;11:1391–1397. doi: 10.1093/hmg/11.12.1391. [DOI] [PubMed] [Google Scholar]
- 9.Verdu J, de Paz F. Chronic eosinophilic leukemia with FIP1L1-PDGFRA. Blood. 2013;121:1254. doi: 10.1182/blood-2012-09-458224. [DOI] [PubMed] [Google Scholar]
- 10.Barraco D, Carobolante F, Candoni A, Simeone E, Piccaluga P, Tabanelli V, Fanin R. Complete and long-lasting cytologic and molecular remission of FIP1L1-PDGFRA-positive acute eosinophil myeloid leukaemia, treated with low-dose imatinib monotherapy. Eur J Haematol. 2014;92:541–545. doi: 10.1111/ejh.12272. [DOI] [PubMed] [Google Scholar]
- 11.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 12.Kumar AN, Sathyanarayanan V, Devi VL, Rajkumar NN, Das U, Dutt S, Chinnagiriyappa LK. FIP1L1-PDGFRA-Positive Chronic Eosinophilic Leukemia: A Low-Burden Disease with Dramatic Response to Imatinib - A Report of 5 Cases from South India. Turk J Haematol. 2014;31:56–60. doi: 10.4274/Tjh.2013.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadrzadeh H, Abdel-Wahab O, Fathi AT. Molecular alterations underlying eosinophilic and mast cell malignancies. Discov Med. 2011;12:481–493. [PubMed] [Google Scholar]
- 14.Stoecker MM, Wang E. Systemic mastocytosis with associated clonal hematologic nonmast cell lineage disease: a clinicopathologic review. Arch Pathol Lab Med. 2012;136:832–838. doi: 10.5858/arpa.2011-0325-RS. [DOI] [PubMed] [Google Scholar]
- 15.Maric I, Robyn J, Metcalfe DD, Fay MP, Carter M, Wilson T, Fu W, Stoddard J, Scott L, Hartsell M, Kirshenbaum A, Akin C, Nutman TB, Noel P, Klion AD. KIT D816V-associated systemic mastocytosis with eosinophilia and FIP1L1/PDGFRA-associated chronic eosinophilic leukemia are distinct entities. J Allergy Clin Immunol. 2007;120:680–687. doi: 10.1016/j.jaci.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Chi Y, Lindgren V, Quigley S, Gaitonde S. Acute myelogenous leukemia with t(6;9)(p23;q34) and marrow basophilia: an overview. Arch Pathol Lab Med. 2008;132:1835–1837. doi: 10.5858/132.11.1835. [DOI] [PubMed] [Google Scholar]
- 17.Katsura Y, Suzukawa K, Nanmoku T, Nemoto N, Machino T, Obara N, Okoshi Y, Mukai HY, Hasegawa Y, Kojima H, Kawakami Y, Nagasawa T. Myelodysplastic syndrome accompanied by basophilia and eosinophilia with t(5;12)(q31;p13) Cancer Genet Cytogenet. 2007;178:85–88. doi: 10.1016/j.cancergencyto.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Reilly JT. Class III receptor tyrosine kinases: role in leukaemogenesis. Br J Haematol. 2002;116:744–757. doi: 10.1046/j.0007-1048.2001.03294.x. [DOI] [PubMed] [Google Scholar]
- 19.Védy D, Muehlematter D, Rausch T, Stalder M, Jotterand M, Spertini O. Acute myeloid leukemia with myeloid sarcoma and eosinophilia: prolonged remission and molecular response to imatinib. J. Clin. Oncol. 2010;28:e33–35. doi: 10.1200/JCO.2009.23.6976. [DOI] [PubMed] [Google Scholar]
- 20.Vandenberghe P, Wlodarska I, Michaux L, Zachée P, Boogaerts M, Vanstraelen D, Herregods MC, Van Hoof A, Selleslag D, Roufosse F, Maerevoet M, Verhoef G, Cools J, Gilliland DG, Hagemeijer A, Marynen P. Clinical and molecular features of FIP1L1-PDFGRA (+) chronic eosinophilic leukemias. Leukemia. 2004;18:734–742. doi: 10.1038/sj.leu.2403313. [DOI] [PubMed] [Google Scholar]
- 21.Loules G, Kalala F, Giannakoulas N, Papadakis E, Matsouka P, Speletas M. FIP1L1-PDGFRA molecular analysis in the differential diagnosis of eosinophilia. BMC Blood Disord. 2009;9:1. doi: 10.1186/1471-2326-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]