Abstract
Aims: This study was to investigate the expression of microRNA (miR)-144 in malignant solitary pulmonary nodule (SPN) tissues and peripheral blood, as well as the biological function of miR-144 in the occurrence and development of lung cancer. Methods: In this study, 39 malignant and 30 benign SPN patients were included. The expression of miR-144 was examined using quantitative real-time polymerase chain reaction. Receiver operating characteristic (ROC) curve was used to identify the clinical value of miR-144 in the early diagnosis of malignant SPN. MTT assay was performed to determine A549 cell proliferation and Transwell assay was used to detect changes in A549 cell invasion and migration ability. Flow cytometry was performed to monitor cell apoptosis, while Western blotting assay was used to measure protein expression levels. At last, dual-luciferase reporter assay was employed to test whether miR-144 regulates zinc finger E-box-binding homeobox 1 (ZEB1) gene expression. Results: Expression of miR-144 was reduced in patients with malignant SPN. miR-144 had diagnostic value for malignant SPN. Proliferation of A549 cells was inhibited by miR-144. Invasion ability of A549 cells was reduced by miR-144. Apoptosis of A549 cells was promoted by miR-144. miR-144 induced A549 cell apoptosis by targeting ZEB1 protein. miR-144 regulated the expression of ZEB1 by interacting with its 3’-UTR region. Conclusions: Expression of miR-144 is reduced in malignant SPN tissues and peripheral blood, being of clinical value in the diagnosis of malignant SPN. miR-144 promotes the apoptosis of lung cancer cells, and inhibits the proliferation, invasion and migration of lung cancer by regulating ZEB1 gene.
Keywords: Solitary pulmonary nodule, microRNA-144, zinc finger E-box-binding homeobox 1, dual-luciferase reporter assay
Introduction
Solitary pulmonary nodule (SPN) is a singular mass surrounded by lung parenchyma in the lung, with clear edge, opaque image, and a diameter less than 3 cm, but without associated pneumonia, atelectasis or lymphadenopathies. Due to its small size, SPN is not easy to be diagnosed, leading to the misdiagnosis of malignant SPN that severely affects the prognosis of patients [1]. In recent years, development of computed tomography increases the diagnosis rate of SPN. A study shows that the average diagnosis rate of SPN is 20%, with the highest diagnosis rate being less than 50%. In addition, most SPNs has a diameter of less than 1 cm, with about 12% SPNs being malignant [2]. Therefore, determination of SPN malignancy is of clinical importance for the early diagnosis and treatment of lung cancer. Currently, the malignancy of SPN is usually determined by imaging examinations, patients’ disease history, and the growth rate of SPN, but these methods have high possibility for misdiagnosis. Sometimes, SPN can be diagnosed by lung biopsies, but this will cause trauma that may induce infection or tumor spreading. Recent research shows that molecular markers for malignant SPN may exist in the blood [3], indicating that screening of specific molecular marker can be of clinical importance for enhancing the diagnosis rate of malignant SPN.
MicroRNA (miRNA or miR) is a type of endogenous small non-coding RNA molecules (18-22 nucleotides) that target 3’-UTR region of mRNA and regulate target gene expression at post-transcriptional level [4,5]. Studies show that miRNA expression spectrum is significantly changed in colon cancer, breast cancer, esophageal cancer, liver cancer and lung cancer [6-11]. In vitro experiments demonstrate that miRNA has important regulatory effect in tumor proliferation, invasion, and metastasis, as well as angiogenesis [12-14]. In addition, miRNA widely and stably exists in tissues, blood, saliva, and urine, being a natural biomarker candidate [15-17]. These facts indicate that miRNA might be of clinical value in the diagnosis of malignant SPN. In this study, we identify which miRNA can be a potential biomarker of malignant SPN, and investigate its mechanism of action.
Materials and methods
Patients
Between October 2012 and October 2014, tissues and peripheral blood were obtained from 69 patients with SPN. After examination of the excised tissues by the Department of Pathology, 39 cases of lung cancer (malignant SPN), 11 cases of tuberculoma, and 19 cases of inflammatory pseudotumor were diagnosed. The 39 cases of malignant SPN included 31 cases of adenocarcinoma, 6 cases of squamous cell carcinoma, and 2 cases of adenosquamous carcinoma. The remaining 30 patients were benign SPN patients. Before surgery, none of the patients received chemoradiotherapy or any other anti-tumor therapy, or had history of other tumors. The age of the patients ranged from 27.5 to 72 years, with an average age of 48 years. In addition, 30 healthy volunteers with matched ages were included into the control group. All procedures were approved by the Ethics Committee of General Hospital of Chinese People’s Liberation Army. Written informed consents were obtained from all patients or their families.
Cells
Cells were cultured in RPMI-1640 medium complemented with 10% fetal bovine serum. When reaching 70-80% confluence, the cells were transfected using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA). The cells were grouped into normal control group, negative control (NC) group, and miR-144 mimics group.
One day before transfection, log-phase A549 cells (3×105) were seeded onto 24-well plate. Two vials of Opti Memi medium (50 μl) were mixed with 1.5 μl miRNA mimics (25 nM) and 1 μl Lipofectamine 2000, respectively, before st-anding for 5 min. Then, the two vials were combined before another standing at room temperature for 20 min, followed by addition of the mixture into each well for incubation. Six hours later, the medium was changed to fresh RPMI-1640 medium complemented with 10% fetal bovine serum, followed by incubation for 72 h. After incubation, the cells were collected for the determination of zinc finger E-box-binding homeobox 1 (ZEB1) and Caspase-3 protein expression.
Expression profiling of miRNAs in lung cancer tissues
Using GEO2R of PubMed (http://www.ncbi.nlm.nih.gov/geo/geo2r/), we analyzed changes of miRNA expression spectrum from Gene Expression Omnibus datasets of lung cancer tissues (GSE accession No. GSE51853). The search results showed that has-miR-144 levels were significantly reduced in adenocarcinoma, large cell lung cancer, adenosquamous carcinoma, and squamous cell carcinoma tissues.
Quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol reagent (Life Technologies, Grand Island, NY, USA) following manufacturer’s protocol. The integrity of RNA was tested using gel electrophoresis. The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry (Nanodrop ND1000, Thermo Scientific, Waltham, MA, USA). Then, cDNA was obtained by reverse transcription (PrimeScript RT Regent Kit, Takara, Dalian, China) from 1 μg RNA and stored at -20°C. QRT-PCR was performed using SYBR PrimeScript RT-PCR Kit (Takara, Dalian, China). PCR reaction system was composed of 10 μl qRT-PCR-Mix, 0.5 μl upstream primer (TACA-GTATAGATGATGTA), 0.5 μl downstream primer (general sequence in the kit; Takara, Dalian, China), 2 μl cDNA template, and 7 μl ddH2O. PCR amplification conditions were as follows: initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 1 min, and annealing at 60°C for 30 sec.
The levels of miR-144 in peripheral blood and tissues of SPN patients were used to plot receiver operating characteristic (ROC) curve to analyze miR-144 sensitivity and specificity, which could reflect the value of miR-144 in the diagnosis of SPN malignancy.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Three groups of cells were seeded onto 96-well plates with a density of 2×103/well in triplicate. Then, 20 μl MTT (5 g/L) was added at 0, 24, 48 and 72 h for reaction. On the last day of culture, 150 μl dimethyl sulfoxide was added in each well to dissolve purple crystals. After incubation at 37°C for 4 h, optical density (OD) was measured using microplate reader (Model AD340; Beckman, Brea, CA, USA) under 490 nm for plotting cell proliferation curve.
Transwell assay
Transwell assay was performed using Transwell chambers (Corning, Tewksbury, MA, USA). Matrigel was thawed at 4°C overnight and diluted with serum-free RPMI-1640 medium (dilution 1:1). The mixture (50 μL) was evenly smeared into the upper chamber and incubated at 37°C for 60 min. After solidification, 1×105 cells from each group were seeded into the upper chamber containing 200 μl serum-free RPMI-1640 medium. In addition, 500 μl RPMI-1640 medium supplemented with 10% fetal bovine serum was added into the lower chamber. After 24 h, the chamber was removed and the cells in the upper chamber were wiped off. After being fixed with 4% formaldehyde for 10 min, the membrane was stained using Giemsa method for microscopic observation of 5 random fields (200×). The number of transwell cells was calculated for the evaluation of cell invasion and migration ability. All procedures were carried out on ice with pipetting tips being cooled at 4°C.
Flow cytometry
At 48 h after transfection with miR-144 mimics, cells of each group were collected and subject to flow cytometry using ANXN V FITC APOPTOSIS DTEC kit I (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s manual. Single-positive staining by ANNEXIN V indicated early apoptotic cells, single-positive staining by propidium iodide indicated necrotic cells, while double positive staining by both indicated late apoptotic cells.
Western blotting
At 72 h after transfection, A549 cells were washed with precooled phosphate-buffered saline twice, before addition of Radio-Immunoprecipitation Assay (RIPA) lysis buffer for total protein extraction. Protein samples (50 μg) were then mixed with 5× sodium dodecyl sulfate loading buffer (4:1) before denaturation in boiling water bath for 10 min. Then, samples were subject to sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 60 V for 30 min and 100 V for 1 h. The resolved proteins were transferred to polyvinylidene difluoride membranes on ice (100 V, 1 h) and blocked with 5% skimmed milk at room temperature for 1 h. Then, the membranes were incubated with first antibodies (ZEB1, 1:1000; Caspase-3, 1:1000; GAPDH, 1:10000) (Abcam, Cambridge, MA, USA) at 4°C overnight. On the next day, after extensive washing with phosphate-buffered saline with Tween 20 for 3 times of 15 min, the membranes were incubated with second antibodies (goat anti-mouse, 1:5000; Caspase-3 goat anti-rabbit, 1:2000) (Abcam, Cambridge, MA, USA) labeled with horseradish peroxidase (Abcam, Cambridge, MA, USA) for 1 h at room temperature before washing with phosphate-buffered saline with Tween 20 for 3 times of 15 min. Then, the membrane was developed with enhanced chemiluminescence detection kit (Sigma-Aldrich, St. Louis, MO, USA) for imaging. Image lab (Bio-Rad, Hercules, CA, USA) software was used to acquire and analyze imaging signals. The relative content of proteins was expressed as protein/GAPDH ratio. Inhibition rate = (1-relative content in treatment group/relative content in control group) ×100%.
Dual-luciferase reporter assay
Plasmids (0.8 μg) with 3’-UTR and mutant 3’-UTR DNA sequences of ZEB1 were transfected into 293T cells, followed by transfection with miR-144 mimics (100 nM) before incubation for 24 h. Cells were lysed for the determination of fluorescence intensity using GloMax 20/20 luminometer (Promega, Madison, WI, USA). Using Renilla fluorescence as internal reference, dual-luciferase reporter assay was performed by Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s manual.
Statistical analysis
The results were analyzed using SPSS 11.0 software (StatSoft Inc., Chicago, USA). The data were given in means ± standard deviation. Two groups of mean values were compared using Student’s t-test. P value less than 0.05 was considered statistically significant.
Results
Expression of miR-144 is reduced in patients with malignant SPN
To measure the levels of miR-144, SYBR Green qRT-PCR was performed. The data showed that the levels of miR-144 in the tumor tissue (0.17 ± 0.10) and blood (0.23 ± 0.08) of malignant SPN patients were significantly lower than those in benign SPN patients (P < 0.05) (Figure 1). In addition, miR-144 levels in the blood of benign SPN patients were not significantly different from that of healthy subjects (P > 0.05; data not shown). These results suggest that the expression of miR-144 is reduced in patients with malignant SPN.
Figure 1.
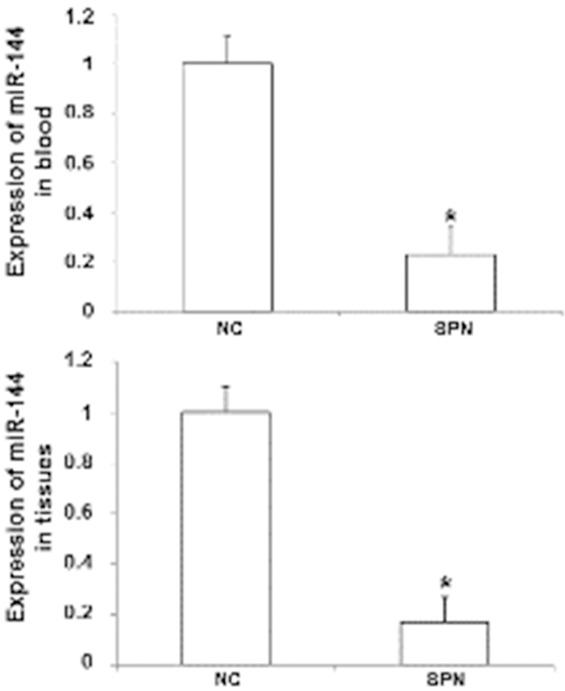
Expression of miR-144 in tissues and peripheral blood of malignant SPN patients. Total RNA was extracted using Trizol reagent and the integrity of RNA was tested using gel electrophoresis. The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry. Then, cDNA was obtained by reverse transcription. QRT-PCR was performed using SYBR PrimeScript RT-PCR kit. *, P < 0.05 compared with negative control group.
miR-144 has diagnostic value for malignant SPN
To evaluate whether miR-144 is of value for the diagnosis of malignant SPN, ROC curve was plotted using preoperative miR-144 levels in the blood of malignant SPN patients. The data showed that the area under the ROC curve for malignant SPN patients was 0.792 (P = 0.003) (Figure 2). This result indicates that miR-144 has diagnostic value for malignant SPN.
Figure 2.
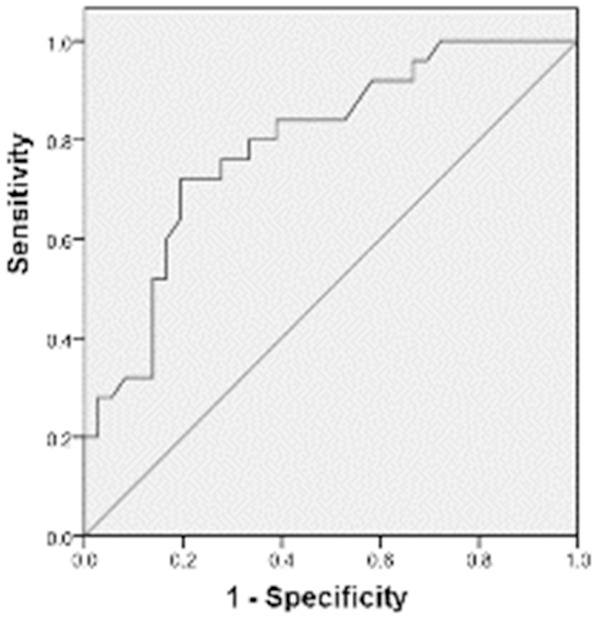
Receiver operating characteristic (ROC) curve for sensitivity and specificity of miR-144. The curve was plotted using preoperative miR-144 levels in the blood of malignant SPN patients. ROC curve is used to evaluate the diagnostic value of miR-144 for malignant SPN.
Proliferation of A549 cells is inhibited by miR-144
To determine the relationship between miR-144 and A549 cell proliferation, MTT assay was performed. The data showed that OD values of cells transfected with miR-144 mimics were significantly lower than those of either normal control or negative control at 24, 48, and 72 h time points (P < 0.05). However, no significant difference was observed between normal control and negative control (P > 0.05) (Figure 3). These results suggest that proliferation of A549 cells is inhibited by miR-144.
Figure 3.
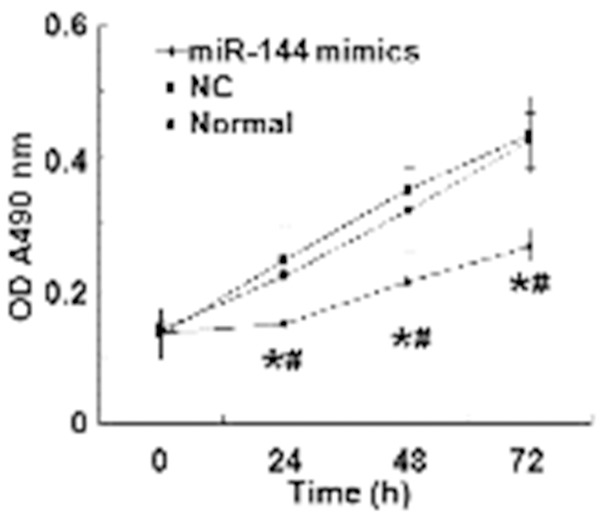
Optical density of A549 cells at 24, 48, and 72 h after transfection. The proliferation of A549 cells in normal control, negative control and miR-144 mimics groups is reflected by optical density values. *, P < 0.05 compared with normal control group; #, P < 0.05 compared with negative control group.
Invasion and migration ability of A549 cells is reduced by miR-144
To study how miR-144 alters the invasion and migration ability of A549 cells, Transwell assay was employed. The data showed that the number of cells that passed through chamber membrane in miR-144 mimics group (28.3 ± 3.2) was significantly lower than those in normal control group (98.5 ± 8.4) and negative control group (87.5 ± 5.8) (P < 0.05), while the invasion cell number of normal control group was not different from that of negative control group (P > 0.05) (Figure 4). These results indicate that the invasion and migration ability of A549 cells is reduced by miR-144.
Figure 4.
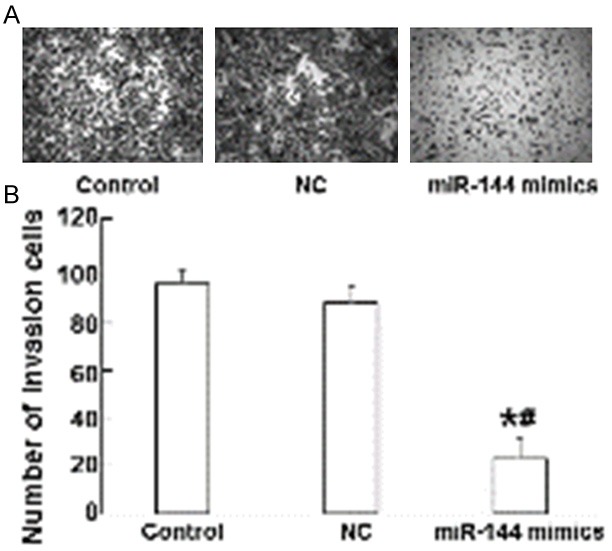
The effect of miR-144 on the invasion and migration ability of A549 cells. Transwell analysis was used to measure cell invasion and migration ability. A. Giemsa staining was used to identify lung cancer cells (magnification ×20). B. The number of invasion cells in normal control, negative control, and miR-144 mimics groups. *, P < 0.05 compared with normal control group; #, P < 0.05 compared with negative control group.
Apoptosis of A549 cells is promoted by miR-144
To test the effect of miR-144 on A549 cell apoptosis, we performed flow cytometry. At 48 h after transfection, cells in miR-144 mimics group showed significantly increased apoptotic rate compared with both control groups (P < 0.05), while the apoptotic rates were not different between normal control and negative control groups (P > 0.05) (Figure 5). These results suggest that the apoptosis of A549 cells is promoted by miR-144.
Figure 5.
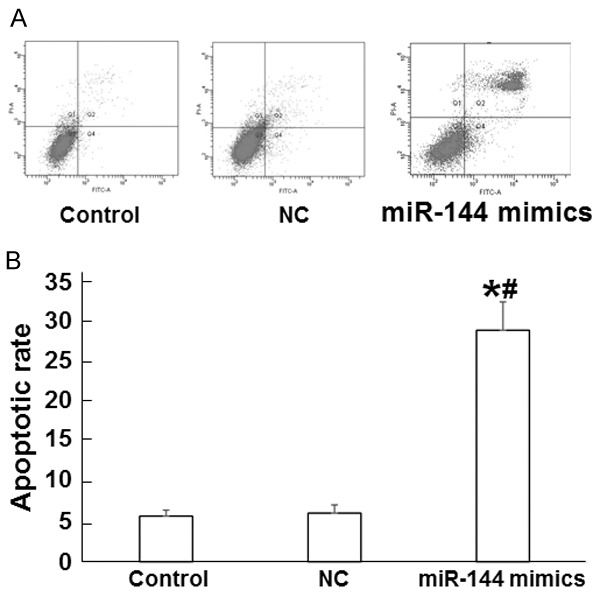
The effect of miR-144 on A549 cell apoptosis. A. Flow cytometry of A549 cells in normal control, negative control, and miR-144 mimics groups. B. Apoptotic rate of A549 cells in normal control, negative control, and miR-144 mimics groups. *, P < 0.05 compared with normal control group; #, P < 0.05 compared with negative control group.
miR-144 induces A549 cell apoptosis by targeting ZEB1 protein
To measure protein expression in A549 cells, we used Western blotting assay. The data showed that ZEB1 protein expression was significantly reduced in miR-144 mimics group compared with both control groups (P < 0.05), but no significant difference was observed between normal control and negative control groups (P > 0.05) (Figure 6A). By contrast, Caspase-3 protein expression in miR-144 mimics group was significantly elevated by the up-regulation of miR-144 (P < 0.05), but normal control was not significantly different from negative control in Caspase-3 protein expression (P > 0.05) (Figure 6B). These results indicate that miR-144 might induce A549 cell apoptosis by targeting ZEB1 protein.
Figure 6.
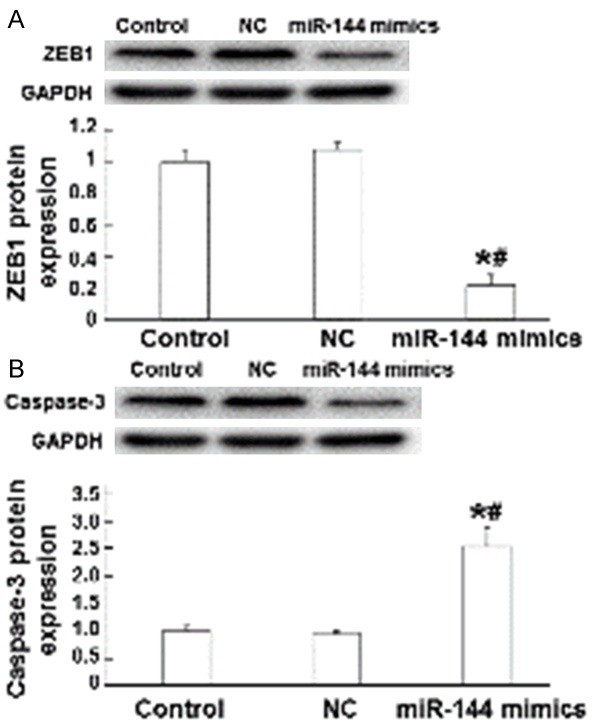
The effect of miR-144 on the expression of ZEB1 and Caspase-3 proteins. (A) ZEB1 and (B) Caspase-3 protein expression in normal control, negative control, and miR-144 mimics groups. Western blotting was used to measure protein expression. *, P < 0.05 compared with normal control group; #, P < 0.05 compared with negative control group.
miR-144 regulates the expression of ZEB1 by interacting with its 3’-UTR region
To confirm the interaction between miR-144 and ZEB1, dual-luciferase reporter assay was employed. The data showed that co-transfection of miR-144 mimics and pMIR-REPORT luciferase reporter plasmids led to significantly decrease relative luciferase activity compared with negative control group (P < 0.05). However, co-transfection with miR-144 mutant and pMIR-REPORT luciferase reporter plasmids did not alter relative luciferase activity (P > 0.05) (Figure 7). These results suggest that miR-144 regulates the expression of ZEB1 by interacting with its 3’-UTR region.
Figure 7.
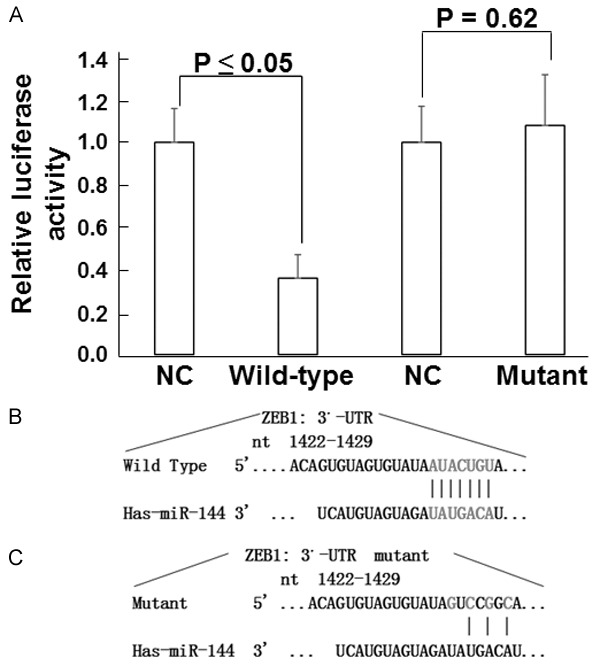
Interaction between miR-144 and ZEB1. A. Relative luciferase activity in cells co-transfected with wild-type or mutant 3’-UTR of ZEB1 and miR-144 mimics. B. Diagram of interaction between wild-type 3’-UTR and miR-144. C. Diagram of interaction between mutant 3’-UTR and miR-144.
Discussion
Studies prove that miRNAs play im-portant roles in the occurrence and development of tumors, especially in the early stage of tumor formation [18-20]. This suggests that miRNA is of clinical value in the early diagnosis of tumors. In order to screen the kind of miRNA that has clinical value in the peripheral blood of malignant SPN patients, we analyzed miRNA chip data and found that miRNA expression spectrum was significantly al-tered in adenocarcinoma, large cell lung cancer, adenosquamous carcinoma, and squamous cell carcinoma tissues. Of note, the expression of miR-144 was significantly down-regulated in all cancer tissues. It is reported that miR-144 regulates AKT3 activity to inhibit tumor cell mass formation and growth [21]. Akiyoshi et al. also report that miR-144 is closely related to the bone metastasis of gastric cancer [22]. However, there are few reports on the expression and biological function of miR-144 in lung cancer. Our qRT-PCR results showed that the expression of miR-144 was significantly reduced in malignant SPN tissue and blood. In addition, ROC curve of miR-144 expression demonstrated that miR-144 was of clinical value in the early diagnosis of malignant SPN.
To better understand the biological function of miR-144 in lung cancer, we studied the effect of miR-144 expression on the proliferation, invasion, migration and apoptosis of A549 cells, using MTT assay, Transwell assay, and flow cytometry. These results show that up-regulation of miR-144 inhibits A549 cell proliferation, reduces its invasion and migration, and promotes apoptosis, suggesting that miR-144 might be a tumor suppressor gene in lung cancer.
To understand the mechanism by which miR-144 exerts its effect on A549 cells, we used bioinformatics method to analyze its downstream target genes, and found possible binding sites of miR-144 in the 3’-UTR region of ZEB1. In addition, studies show that ZEB1 is highly expressed in several tumors, participating tumor formation and development by facilitating their proliferation, invasion and migration [23]. In order to test whether miR-144 directly participates in the regulation of ZEB1 gene expression, we constructed dual-luciferase reporter gene and mutant vectors that were co-transfected with miR-144 mimics into 293T cell. By measuring changes in luciferase activity, we demonstrated the regulation of ZEB1 gene by miR-144. Furthermore, Western blotting assay showed that endogenous ZEB1 protein expression was altered by miR-144 up-regulation. These data suggest that miR-144 directly regulates ZEB1 expression that inhibits the proliferation, invasion and migration of lung cancer cells.
Of note, Western blotting data also showed that Caspase-3 protein expression was up-regulated by miR-144 expression, indicating that miR-144 might have other target genes that activate apoptotic pathway. However, this needs further studies in the future. In conclusion, the present study demonstrates that the expression of miR-144 is significantly reduced in malignant SPN tissues and peripheral blood, exhibiting its diagnostic value for malignant SPN. In vitro experiments show that miR-144 inhibits the proliferation, invasion and migration, and promotes the apoptosis of lung cancer cells by regulating ZEB1 expression. miR-144 is a potential novel target for the treatment of lung cancers.
Acknowledgements
This work was supported by the General Hospital of Chinese People’s Liberation Army and Navy General Hospital of People’s Libe-ration Army.
Disclosure of conflict of interest
None.
References
- 1.Truong MT, Ko JP, Rossi SE, Rossi I, Viswanathan C, Bruzzi JF, Marom EM, Erasmus JJ. Update in the evaluation of the solitary pulmonary nodule. Radiographics. 2014;34:1658–1679. doi: 10.1148/rg.346130092. [DOI] [PubMed] [Google Scholar]
- 2.Rezaeetalab F, Aryana K, Attaran D, Bagheri R, Nattagh F, Lari SM. The role of octreotate scan in discrimination of solitary pulmonary nodule. World J Nucl Med. 2014;13:46–49. doi: 10.4103/1450-1147.138574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni LF, Liu XM. Diagnostic value of serum tumor markers in differentiating malignant from benign solitary pulmonary nodules. Beijing Da Xue Xue Bao. 2014;46:707–710. [PubMed] [Google Scholar]
- 4.Stepanowsky P, Levy E, Kim J, Jiang X, Ohno-Machado L. Prediction of MicroRNA Precursors Using Parsimonious Feature Sets. Cancer Inform. 2014;13:95–102. doi: 10.4137/CIN.S13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim W, Song G. Discovery of prognostic factors for diagnosis and treatment of epithelial-derived ovarian cancer from laying hens. J Cancer Prev. 2013;18:209–220. doi: 10.15430/JCP.2013.18.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi P, Middleton J, Jeon YJ, Garofalo M. MicroRNAs in lung cancer. World J Methodol. 2014;4:59–72. doi: 10.5662/wjm.v4.i2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rokavec M, Li H, Jiang L, Hermeking H. The p53/microRNA connection in gastrointestinal cancer. Clin Exp Gastroenterol. 2014;7:395–413. doi: 10.2147/CEG.S43738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao B, Azmi AS, Ali S, Zaiem F, Sarkar FH. Metformin may function as anti-cancer agent via targeting cancer stem cells: the potential biological significance of tumor-associated miRNAs in breast and pancreatic cancers. Ann Transl Med. 2014;2:59. doi: 10.3978/j.issn.2305-5839.2014.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai NS, Samia-Aly E, Barbera M, Fitzgerald RC. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin Cancer Biol. 2013;23:512–521. doi: 10.1016/j.semcancer.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JN, Brewer HM, Nicora CD, Weitz KK, Morris MJ, Skabelund AJ, Adkins JN, Smith RD, Cho JH, Gelinas R. Protein and microRNA biomarkers from lavage, urine, and serum in military personnel evaluated for dyspnea. BMC Med Genomics. 2014;7:58. doi: 10.1186/1755-8794-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva SS, Lopes C, Teixeira AL, Carneiro de Sousa MJ, Medeiros R. Forensic miRNA: potential biomarker for body fluids? Forensic Sci Int Genet. 2015;14:1–10. doi: 10.1016/j.fsigen.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y, Liu Q. MicroRNA-130b promotes cell aggressiveness by inhibiting peroxisome proliferator-activated receptor gamma in human hepatocellular carcinoma. Int J Mol Sci. 2014;15:20486–20499. doi: 10.3390/ijms151120486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao D, Jia P, Wang W, Zhang G. VEGF-mediated suppression of cell proliferation and invasion by miR-410 in osteosarcoma. Mol Cell Biochem. 2015;400:87–95. doi: 10.1007/s11010-014-2265-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Li G, Zhou J, Han N, Liu Z, Yin J. miR-107 activates ATR/Chk1 pathway and suppress cervical cancer invasion by targeting MCL1. PLoS One. 2014;9:e111860. doi: 10.1371/journal.pone.0111860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degagne E, Pandurangan A, Bandhuvula P, Kumar A, Eltanawy A, Zhang M, Yoshinaga Y, Nefedov M, de Jong PJ, Fong LG, Young SG, Bittman R, Ahmedi Y, Saba JD. Sphingosine-1-phosphate lyase downregulation promotes colon carcinogenesis through STAT3-activated microRNAs. J Clin Invest. 2014;124:5368–5384. doi: 10.1172/JCI74188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitao Mda C, Coimbra EC, Lima Rde C, Guimaraes Mde L, Heraclio Sde A, Silva Neto Jda C, de Freitas AC. Quantifying mRNA and microRNA with qPCR in cervical carcinogenesis: a validation of reference genes to ensure accurate data. PLoS One. 2014;9:e111021. doi: 10.1371/journal.pone.0111021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao ZG, Jin JY, Zhang AM, Zhang LP, Wang XX, Sun JG, Chen ZT. MicroRNA profile of tumorigenic cells during carcinogenesis of lung adenocarcinoma. J Cell Biochem. 2015;116:458–466. doi: 10.1002/jcb.25206. [DOI] [PubMed] [Google Scholar]
- 21.Liu BB, Luo L, Liu XL, Geng D, Liu Q, Yi LT. 7-Chlorokynurenic acid (7-CTKA) produces rapid antidepressant-like effects: through regulating hippocampal microRNA expressions involved in TrkB-ERK/Akt signaling pathways in mice exposed to chronic unpredictable mild stress. Psychopharmacology. 2014:1–10. doi: 10.1007/s00213-014-3690-3. [DOI] [PubMed] [Google Scholar]
- 22.Akiyoshi S, Fukagawa T, Ueo H, Ishibashi M, Takahashi Y, Fabbri M, Sasako M, Maehara Y, Mimori K, Mori M. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br J Cancer. 2012;107:1345–1353. doi: 10.1038/bjc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlert U, Suwala A, Raabe E, Siebzehnrubl F, Suarez M, Orr B, Bar E, Maciaczyk J, Eberhart C. ZEB1 Promotes invasion in human fetal neural stem cells and hypoxic glioma neurospheres. Brain Pathol. 2014 doi: 10.1111/bpa.12240. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


