Abstract
Extracellular vesicles (EVs), including exosomes and shed vesicles, have been implicated in intercellular communication; however, their biomarker potential is less clear. Therefore, EVs derived from MCF7 and MCF10A cells were analyzed to identify unique microRNA (miR) profiles that distinguish their origin. One characteristic common to the miR profiles of MCF7 EVs and their parent cells is the high abundance of miR-21, let-7a, miR-100, and miR-125b, and low levels of miR-205. A second characteristic is the high abundance of “microRNA-like” transfer RNA (tRNA) fragments, which is unique to the MCF7 EVs, and is not found in comparing the cellular profiles. In addition, correlations were examined in the MCF7 cellular expression levels of these five miRs and two tRNA-derived miRNAs, miR-720 and miR-1274b, and compared to the correlations in MCF7 EV levels. Interestingly, correlations in the cellular expression of miR-125b, miR-100, and let-7a are mirrored in the EVs. In contrast, correlations in tRNA-derived miRNA levels are found only in the EVs. The findings suggest that EV miR clusters can be defined based on functional miR interactions related to correlated cellular expression levels or physical miR interactions – e.g., aggregation due to comparable binding affinities to common targets. Implications: These results point to using high levels of tRNA-derived small RNA fragments in combination with known miR signatures of tumors to distinguish tumor-derived EVs in circulation from EVs derived from other cell sources. Such biomarkers would be unique to the EVs where high abundances of tRNA fragments are amplified with respect to their cellular levels.
INTRODUCTION
There is growing recognition that microRNAs (miRNAs) are important epigenetic regulators of gene expression, and that abnormalities in miRNA expression play a causal role in cancer pathogenesis [1–4]. Changes in miRNA expression levels in cancer were first identified for miR-15 and miR-16-1 in B-cell chronic lymphocytic leukemia [5]. Since then, dysregulation of miRNA expression has been observed in a myriad of solid and hematopoietic tumors. The differential expression of miRNAs in tumors relative to normal tissues has led to exploring miRNA expression levels as biomarkers for cancer. To date, the vast majority of cancer-specific miRNA expression profiles have been derived from tumors or cancer cell lines. However, cancer cells also release extracellular vesicles (EVs) containing miRNAs that can be retrieved from peripheral blood, urine, saliva, or other body fluids. Cell-secreted EVs consist primarily of two populations, exosomes and shed vesicles, released from all cell types by entirely different mechanisms [6–8]. Exosomes are released by exocytosis of multivesicular bodies in response to specific stimuli, while shed vesicles are released by the budding of small cytoplasmic protrusions that then detach from the cell surface. Until recently, EVs were largely regarded as cellular debris without biological function. However, it is now recognized that they can mediate intercellular communication [8].
A recent, exciting development in studies of EVs has been the discovery of mRNA and miRNA encapsulated in and/or associated with the vesicles, which has been confirmed for a variety of cell types [9–18]. The significance of these findings to the pathogenesis of cancer is to suggest that EV-mediated transfer of miRNAs in particular can be a mechanism for epigenetic reprogramming cells in general, and cells in the tumor microenvironment, specifically. Most recent studies of EV-mediated miRNA transfer in tumor progression have focused on tumor-derived exosomes. Exosomes are generally defined by their spherical, unilamellar morphology, their size – average diameters less than ~100 nm [19, 20] – and the expression of specific biomarkers, including tetraspanins [8], while shed vesicles are morphologically more heterogeneous and typically larger in size with characteristic lengths up to one micron. Both exosomes and shed vesicles can contain miRNAs, although at different compositions [8].
Identification of cancer-specific miRNA signatures in circulating EVs released from tumors offers the possibility of developing minimally invasive biomarkers for cancer. The existence of unique cancer-specific miRNA profiles in peripheral blood is based on the hypothesis that if miRNAs are present in circulation, they must be protected from endogenous RNase activity [21]. Encapsulation of miRNAs within EVs and/or their association with ribonuclear proteins, such as Ago2 [22], affords this protection. However, miRNAs originating from many other cell types are also present in circulation, creating a high background of non-cancer cell-derived species [23, 24]. Since cancer-related inflammation can alter miRNA expression in these cells, tumor-specific changes in blood-based miRNA profiles will also be confounded with systemic changes. In addition, large scale miRNA profiling studies comparing normal and tumor tissues have shown that the vast majority of miRNA species in a particular cell type are expressed both in normal and diseased states, with only abundances changing between cell states [25, 26]. A more effective approach to detecting tumor-induced changes in circulating EV miRNAs would be to correlate cancer-specific miRNA profiles with molecular signatures that identify those EVs in circulation originating exclusively from the tumor cells. Recognizing that cell-secreted EVs have miRNA profiles that are distinct from their parent cells, we postulate that the EV miRNA profiles themselves have unique features that lead to this identification.
In this study, we analyze the miRNA profiles of EVs secreted from MCF7 and MCF10A cells to determine whether unique EV miRNA signatures can be identified that distinguish their cell source. We find that the EV miRNA profiles are significantly different from the miRNA profiles of their parent cells, although the most abundant miRNAs detected in the cells are also detected in the secreted EVs. Moreover, the EV miRNA profiles show striking differences in composition, thereby enabling a MCF7 cell-specific EV miRNA signature to be defined based on two distinguishing features. One feature common to both the MCF7 cellular and EV miRNA profiles is high levels of miR-21, miR-100, miR-125b, and let-7a, and a low level of miR-205. The second distinguishing feature is high levels of “microRNA-like” transfer RNA (tRNA) fragments in the MCF7 EVs that are not seen in the MCF10A EVs. The distinguishing, high abundances of tRNA-derived miRNAs are unique to the EV miRNA profiles, and are not found in comparing the cellular profiles. We also find correlations in MCF7 cellular expression levels of miR-100, miR-125b, and let-7a that are mirrored in the MCF7 EVs, suggesting that this miRNA cluster in the EVs is defined by functional miRNA interactions related to the correlated cellular expression levels. In contrast, correlations in abundances of the tRNA-derived miRNAs, miR-720 and miR-1274b, are found only in the EVs, suggesting that this EV miRNA cluster is defined based on purely physical miRNA interactions that may result, e.g., from aggregation due to comparable binding affinities to common targets.
MATERIALS AND METHODS
Cell culture
MCF10A cells were grown in DMEM/F12 (Life Technologies) containing 5% fetal bovine serum (FBS), as described previously [27]. MCF7 cells were grown in RPMI media (Life Technologies) containing 10% FBS, 1% 100x NEAA and 10 µg/ml Gentamicin. Cells were allowed to reach 70–80% confluence in complete culture media containing FBS at 37°C in a 5% CO2 humidified incubator. These subconfluent cultures were then washed twice in 1x phosphate buffered saline (PBS) and either analyzed at that time, or further incubated in serum-free media. Incubation in serum-free media avoids cross-contamination of the MCF10A and MCF7 miRNAs from serum-derived EV miRNAs. Total cell numbers were determined using a hemocytometer after harvesting the cells.
Extracellular Vesicle Isolation
EVs were isolated from the serum-free media following 72 hrs of serum deprivation. To obtain 100 ng of total RNA required for the NanoString miRNA assays, a minimum of 350 million cells were cultured for each EV RNA sample. EVs were isolated from approximately 200 million cells for each qRT-PCR miRNA assay. Sequential centrifugation/ultracentrifugation was used to isolate the EVs, as described in detail elsewhere [28].
RNA extraction
Total RNA was extracted from cells and the EVs using TRIzol reagent. The RNA samples were purified further by ethanol precipitation. For the NanoString assays, samples were standardized to a concentration of 33 ng/µl total RNA before storage at −80°C. Total RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific,). RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). For the cellular RNA samples, the A260/A280 absorption ratio was >1.9, while the A260/A230 absorption ratio was >2.0. Bioanalyzer results for cellular RNA showed negligible degradation, confirming the high quality of these samples. These absorption ratios for the EV RNA samples were considerably lower: 1.6 and 0.7, respectively. The lower absorption ratios do not necessarily reflect a lower quality of the RNA, but simply a lack of ribosomal RNA, which was confirmed in the bioanalysis of the RNA in the EV samples.
NanoString nCounter™ miRNA Assay
Sample preparation and hybridization reactions were carried out according to the manufacturer’s protocol. After hybridization, the solution was placed directly in the nCounter Prep Station for analysis. The complexes consisting of the miRNA and the capture and reporter probes were purified, aligned, and counted using a CCD camera and a microscope objective lens. The nCounter Digital Analyzer acquired images of each field of view using four different excitation wavelengths (480, 545, 580 and 622 nm). The processed images and raw barcode counts were tabulated and reported in a CSV format. Subsequent data analysis was carried out in an Excel worksheet. Twelve RNA samples are analyzed in a single NanoString assay, and 654 human miRNAs were profiled in each sample.
Nonspecific hybridization for the miRNA capture and reporter probes used in our assays was also determined by running a no-template control assay. Correction factors for 66 of the 654 human miRNAs were then applied to the raw counts obtained for these specific miRNAs in our subsequent assays. The correction factors supplied by NanoString were specific to the lot of our reagents.
NanoString nCounter™ miRNA Data Analysis
The miRNA counts in each sample were first normalized to account for sample-to-sample variations in a single NanoString assay. The normalization factors were within 1.00 ± 0.22, and were computed by summing the counts for the six positive controls in each sample and dividing by the average of this sum for all 12 samples. Normalized miRNA counts were then converted to counts above a background threshold defined as two standard deviations above background for an assay, which was computed by averaging the normalized counts for the eight negative controls in each sample. Counts below the background threshold, which varied between 50 and 100 counts, were considered “not detectable.”
Normalized, background-adjusted counts for each miRNA were converted to miRNA copy numbers using a standard curve generated from the dilution series of the six positive controls in each sample. The miRNA copy numbers obtained from this standard curve are based on 10 ng of total RNA in a sample. For the cellular miRNA, these copy numbers are converted to miRNA copy numbers per cell using the total RNA measured in a sample after TRIzol extraction and the total number of cells from which this RNA was derived. An estimated efficiency of 75% for the TRIzol extraction of cellular RNA was taken from the literature [29] and applied to carry out this conversion. An efficiency of 100% was assumed for the ethanol purification following TRIzol extraction, since the measured total RNA concentration did not change significantly before and after this purification step. EV miRNA copy numbers per cell were similarly obtained from the total amount of RNA measured after TRIzol extraction and the total number of cells from which the EVs were derived. For this conversion, we assumed an efficiency of 15% in isolating EVs from the cells using sequential centrifugation/ ultracentrifugation based on previous work that reported this efficiency to be between 5 and 25% [29]. An efficiency of 10% for EV RNA recovery by TRIzol extraction was estimated by comparing the total EV RNA concentration of measured after hypotonic lysis of the EVs to that measured after TRIzol extraction. EV RNA recovery in the ethanol purification was estimated to be 19% from direct measurements of the total RNA concentration before and after this step. We note that an alternative “spike in” method of normalization using the mean of several stable RNAs added after the ultracentrifugation steps and before the TRIzol extraction could account for the overall loss of miRNA during the TRIzol extraction. This “spike in” method using synthetic miR-39 has been applied to determine PCR amplification efficiencies for miRNA derived from exosome samples compared to miRNA derived from supernatant fractions [30].
The NanoString assays were also evaluated by computing a sampling efficiency, a lower detection limit, and an inter-assay reproducibility. An average sampling efficiency of ~1% was obtained by taking the difference between the counts observed in the assay and the actual theoretical number of molecules for each of the six positive controls in each sample. The lower detection limit was defined as the concentration above which the normalized counts for the dilution series of positive controls from 128 fM to 0.125 fM were not significantly different from the average background. This detection limit was determined to be 0.125 fM or 25 to 50 counts. Reproducibility was assessed by the linearity of log-log plots of miRNA copy numbers derived from independent assays.
Quantitative real-time PCR (qRT-PCR)
The qRT-PCR assays were performed using Taqman Assay kits (Applied Biosystems) according to the manufacturer’s protocol [31]. All reactions were performed in triplicate. Reverse transcription (RT) was carried out using the Taqman miRNA RT kit with 10 ng of total RNA per 15 µl RT reaction. The reactants were incubated in a thermal cycler for 30 min at 16°C, followed by 30 min at 42°C and 5 min at 85°C, and then held at 4°C. No-template controls and RT minus controls were also performed. PCR was carried out in a 20-µl reaction volume containing 3 µl of RT product and 1 µl of Taqman probes specific for each miRNA. The reaction mixture was incubated at 95°C for 10 minutes, followed by 40 cycles of denaturation (95°C for 15 seconds) and extension (60°C for 1 minute).
Measured Ct values were converted to miRNA copy numbers using standard curves obtained by reverse transcription and amplification of synthetic miR-21 and miR-720 (Integrated DNA Technologies, Coralville, IA). These miRNA copy numbers were converted to copy numbers per cell, as described above in the analysis of the NanoString miRNA data, using an efficiency of 75% (10%) for the TRIzol extraction of cellular (EV) RNA, 100% (19%) recovery of cellular (EV) RNA in the ethanol purification, and an efficiency of 15% for isolating EVs from the cells by sequential centrifugation/ultracentrifugation.
Dynamic Light Scattering (DLS)
Relative EV size distributions were measured using a BI-200SM Laser Light Scattering Goniometer (Brookhaven Instruments). Samples consisted of 1 ml of EV suspension equilibrated at room temperature. The samples were diluted to the required range of 10–200 kilocounts per second (kcps). All measurements were made in triplicate.
Cryo-Transmission Electron Microscopy
Samples were prepared within a Controlled Environment Vitrification System (CEVS) at 25°C and 100% relative humidity. A 10-µl suspension of EVs was applied onto glow discharged Lacey Formvar/Carbon 200 Mesh copper grids (Ted Pella, Inc.) and blotted before plunging into liquid ethane. Vitrified grids were transferred to a Gatan cryo-sample holder and visualized in a FEI Tecnai G2 Spirit electron microscope. The microscope was operated at 120 kV and under low dose conditions to minimize radiation damage to the samples. The total electron dose was between 10 and 100 e−/Å2. Images were captured on a 4k×4k Gatan Ultrascan CCD camera at magnifications of 4,800x, 18,000x and 30,000x.
Anti-CD63 antibody microarray assay
Microarrays were fabricated by printing a solution of the anti-CD63 antibody on polyacrylamide film-coated glass slides using a PerkinElmer Piezorray noncontact microarrayer, as described in detail elsewhere [32]. Unlabeled and DiI-labeled EVs were contacted with the antibody microarray. The unbound, unlabeled EVs were collected for DLS measurements of the size distribution, which was compared to the measured size distribution of unlabeled EVs in solution before contacting the microarray. Microarray images were acquired using a ProScanArray fluorescence scanner (PerkinElmer). Image quantification and data analysis were performed using ScanArray Express 3.0 (PerkinElmer) and JMP 6.0 (SAS) software.
RESULTS
Cellular Responses to Serum Deprivation
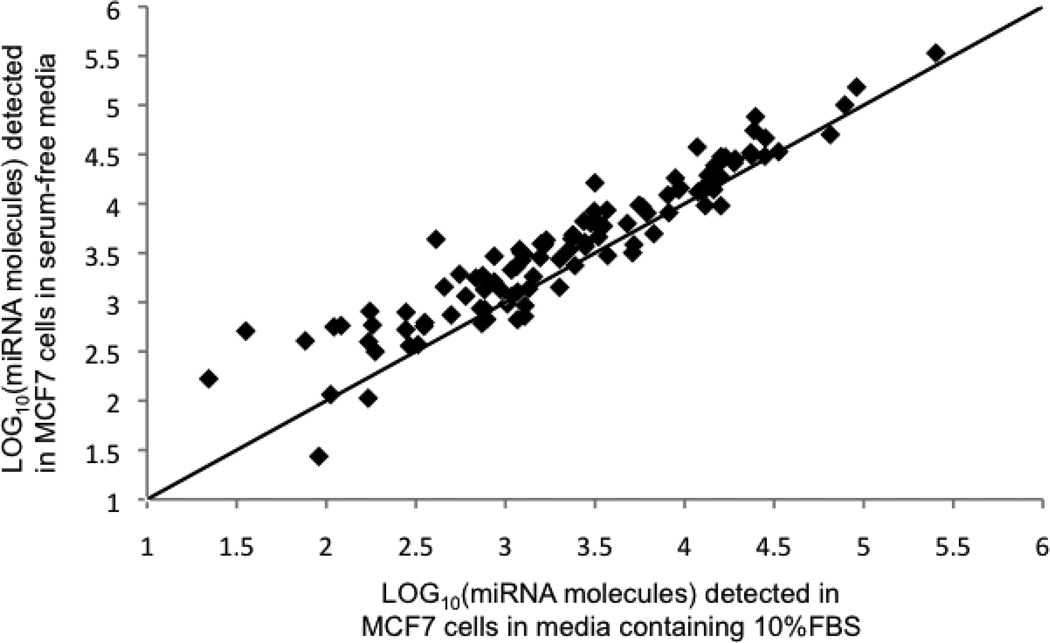
We determined that MCF7 and MCF10A cells grow in serum-free media over 72 hrs, but at slower rates than cells grown in media supplemented with 10% FBS. The increase in MCF7 (MCF10A) cell numbers was ~10% (40%) of that for cells grown in FBS-supplemented media over the same period of time. Visual inspection for detached cells confirmed the absence of significant cell death during this period. Temporal responses to serum deprivation were tracked by measuring total RNA concentration and miRNA expression levels for miR-21, miR-720, miR-1274a, and miR-1274b over 72 hrs of serum deprivation. These miRNAs were chosen as the most abundant miRNA species in MCF7 cells (miR-21), while miR-720, miR-1274a and miR-1274b were chosen to represent the tRNA-derived miRNAs. Although the temporal responses were noticeably different for the two cell lines at early times (<10 hrs), the total RNA concentration and the cellular expression levels for these miRNAs stabilized near their original levels between 24 and 72 hrs (Figure S1). The expression levels of miR-21 and the tRNA-derived miRNAs for each cell line show remarkably similar temporal responses over the 72 hrs of serum deprivation in contrast to the different responses observed between the cell lines. Moreover, no dramatic shifts in relative expression levels of the individual miRNA species were observed for either cell line; the most abundant miRNA species prior to serum removal and at 72 hrs after removal were largely unaffected by the change in culture conditions as shown in Figure 1 for MCF7 cells grown in media supplemented with 10% FBS and after 72 hrs in serum-free media. A similar plot (not shown) was obtained for MCF10A cells.
Figure 1.
Comparison of miRNA copy numbers detected in MCF7 cells grown in media supplemented with 10% FBS and after 72 hs in serum-free media (0% FBS). The 45° diagonal line indicating a 1:1 correlation is drawn to guide the eye.
Extracellular Vesicle Biophysical Characterization
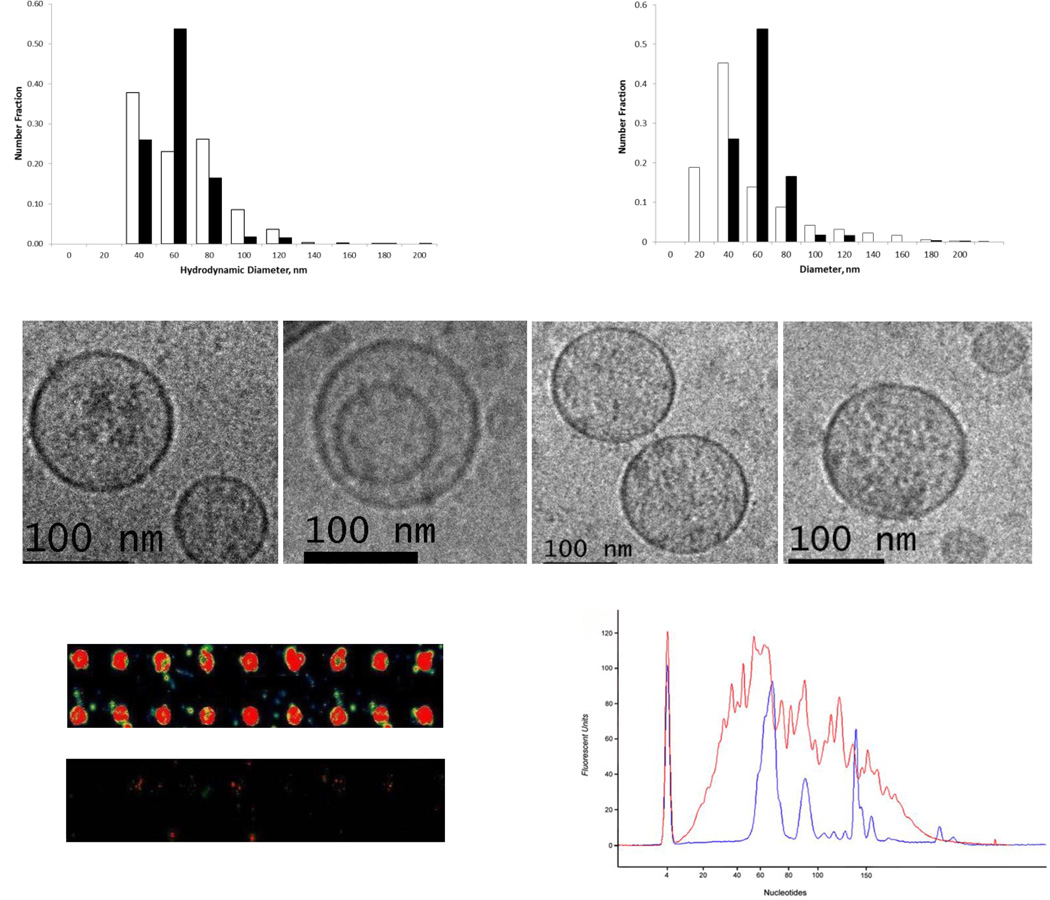
MCF7 and MCF10A EV size distributions characterized by DLS were found to be unimodal with hydrodynamic diameters ranging from 40 to 120 nm (Figure 2). A comparable unimodal distribution of MCF7 EV geometric diameters shifted to lower diameters by ~20 nm was derived from cryo-TEM images (Figure 2). Slightly larger EV diameters measured by DLS are expected from the bias in light scattering to detect larger particles, as well as the difference between hydrodynamic diameters (measured by DLS) and geometric diameters (measured in cryo-TEM images). Vesicle diameters in this range are considered to be characteristic of the exosome subpopulation of EVs released from cells by the exocytosis of multivesicular bodies [19], although spherical vesicles ~100 nm in diameter can also bud from the plasma membrane [33].
Figure 2.
Top (right): Number-weighted distributions of EV hydrodynamic diameters measured by dynamic light scattering (DLS) for EVs secreted by MCF10A cells (open bars) and MCF7 cells (filled bars). Top (left): MCF7 EV size distributions obtained directly from cryoTEM images by measuring the diameters of >1000 EVs (open bars) compared with the distribution of EV diameters obtained by DLS (filled bars). Middle: Representative cryo-TEM images of MCF7 cell-secreted EVs. An internal or nested vesicle within a larger EV is shown in the second image from the left. Scale bar in each image is 100 nm. Bottom (left): Representative image of 9×2 subarrays of anti-CD63 antibody spots on Nexterion slides contacted with DiI-labeled EVs from MCF10A cells (top); the control (bottom) is the supernatant following EV isolation by ultracentrifugation. EV size distributions were also measured DLS. A unimodal size distribution with an average hydrodynamic diameter of 190 nm was obtained for the unbound EV fraction, compared to a bimodal distribution with average hydrodynamic diameters of 50 nm (60%) and 260 nm (40%) for the EVs in solution prior to contacting the microarray. Bottom (right): Bioanalyzer small RNA profiles measured for equivalent amounts of total RNA isolated from MCF7 cells after 72 hrs in serum-free media and from their secreted EVs. The cellular RNA profile is shown in blue and the EV RNA profile is shown in red. The prominent peak on the left in both profiles represents an internal standard.
EV morphologies were characterized using cryo-TEM (Figure 2). The vast majority of EVs from both cell lines were found to be unilamellar and spherical in shape. Some EVs with diameters >100 nm were also observed to contain internal or nested vesicles, which has previously been reported for cell-secreted EVs of comparable diameters and confirmed by three-dimensional reconstruction of cryo-TEM tilt series images [34]. Electron dense material within the EVs was also clearly visible in the cryo-TEM images (Figure 2). This material could be uniformly distributed or concentrated in specific regions within the vesicles. It is unclear whether the non-uniform distributions reflect physical aggregation of electron dense material or specific attachments of this material to molecules anchored to the internal surface of the lipid bilayer. Many EVs, however, appeared to be devoid of encapsulated material as determined by comparing the cryo-TEM images of these EVs with those obtained for synthetic liposomes (100 nm in diameter) prepared with and without encapsulated protein.
The presence of the tetraspanin CD63 on the surface of the EVs was determined by capturing DiI-labeled EVs isolated from both cell lines on anti-CD63 antibody microarrays, as shown for the MCF10A EVs in Figure 2. This protein is a known surface marker for exosomes [9, 12, 16] in general, and has been used as a surface marker for MCF7 cell-secreted exosomes, [14, 35]. DLS measurements of EV size distributions in the sample solutions before and after contact with the microarray confirmed that the smaller vesicles, corresponding exosomes, were preferentially captured on the microarray.
Small RNA profiles for MCF7 cells (Figure 2) show three distinct peaks at 75, 90, and 140 nucleotides for the cellular RNA profile, compared to a broad distribution of nucleotides with a maximum at ~50–75 nucleotides for MCF7 EVs. The fractional abundance of mature miRNAs, estimated from the area under the curve between 15 and 25 nucleotides divided by the total area, is ~0.5%, indicating that mature miRNAs represent only a small fraction of the small RNAs in MCF7 EVs. We also confirmed (data not shown) that the EVs contained no intact ribosomal RNA (rRNA) compared to the donor cells, and no small RNA oligomers greater than ~800 nucleotides in length, as has been reported elsewhere for a variety of cell lines [9, 11, 15, 16].
miRNA profiles for MCF7 and MCF10A cells and extracellular vesicles
For both cell lines, about one quarter of the 654 human mature miRNAs analyzed in the NanoString assay were detected above a threshold set at two standard deviations above the average background (described in Materials and Methods). The number of miRNA species detected in the EVs compared to the parent cells was much lower: 48 miRNAs were detected in the MCF10A EVs compared to 185 miRNAs in MCF10A cells, and 96 miRNAs were detected in the MCF7 EVs compared to 163 miRNAs in MCF7 cells. A few miRNAs detected in high abundance in the EVs were not detected above threshold in the parent cells; e.g., miR-630 in MCF7 EVs and cells. To determine whether these miRNAs were secreted during early stages of serum deprivation and not replenished in the cells at 72 hrs, we assayed for these miRNAs in the cells immediately before removing the serum from the culture media, and detected none of them at this initial time point, suggesting that these miRNAs partition into the EVs with high selectivity.
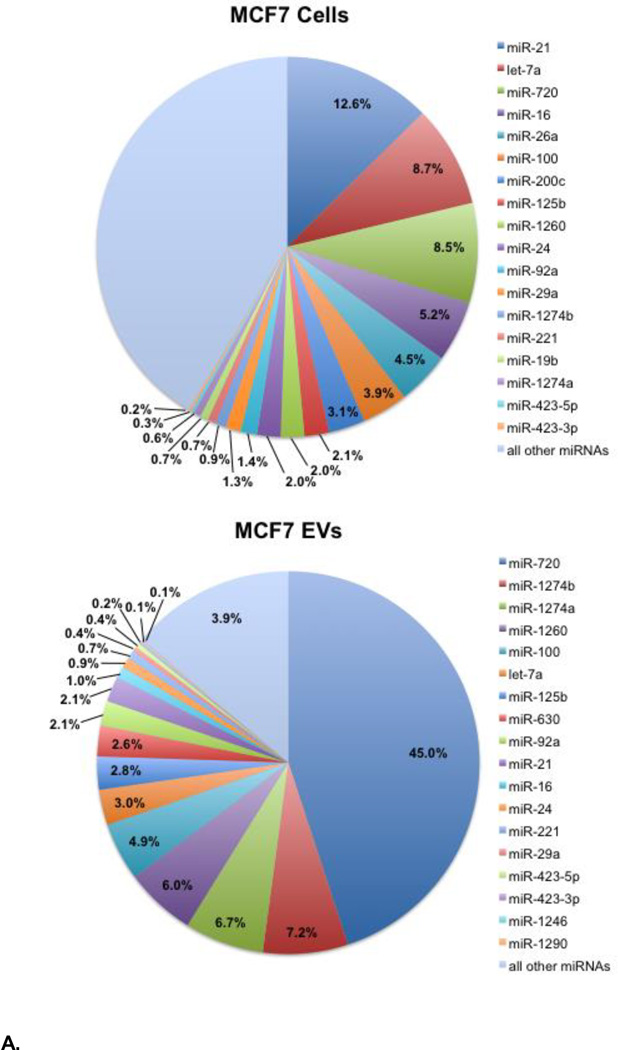
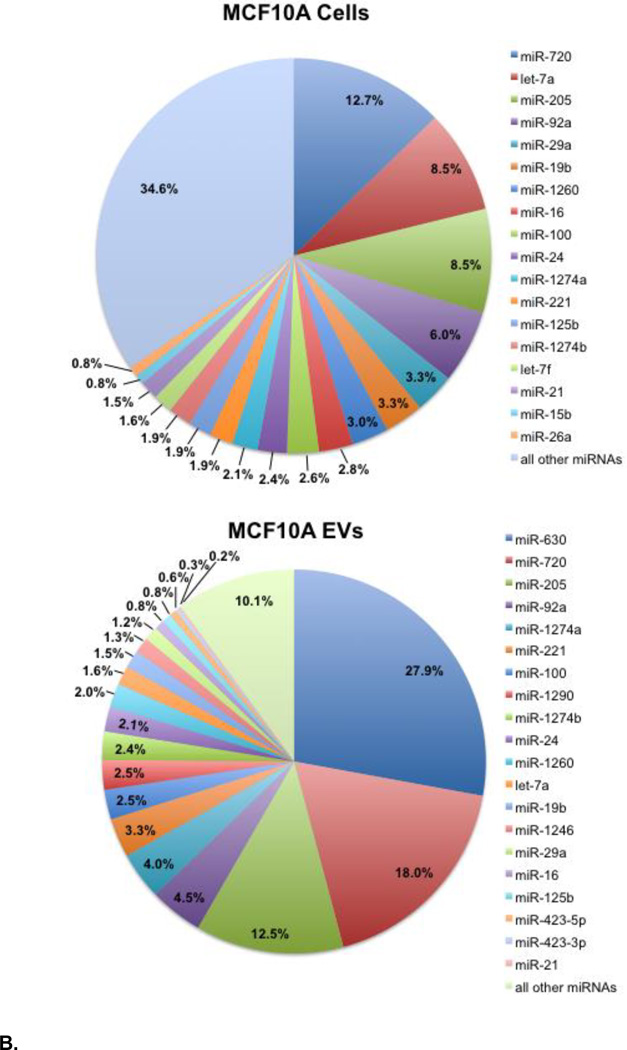
Compositions of the most abundant miRNAs detected in MCF7 and MCF10A cells and their EVs identified in the NanoString assay are shown in Figure 3. For both cell lines, 10 miRNAs comprise more than half of the total number of miRNA transcripts detected, with six of the ten common to both cell lines: miR-720, let-7a, miR-16, miR-100, miR-1260, and miR-24. We note that let-7a, miR-16, and miR-100 have been implicated in cell survival pathways and suppressing cell growth and proliferation [36–39], which may reflect common cellular responses to serum deprivation. Indeed, lower expression levels of let-7a and miR-100 have been measured for MCF7 cells grown in the same culture medium, but supplemented with 10% EV-depleted FBS [14]. The remaining four miRNAs are specific to one cell line: miR-21, miR-26a, miR-200c, and miR-125b (MCF7 cells) and miR-205, miR-92a, miR-29a, and miR-19b (MCF10A cells).
Figure 3.
A. Composition of the most abundant miRNAs detected in MCF7 cells (top) and in the EVs derived from MCF7 cells (bottom) at 72 hrs of serum depletion. Selected miRNAs with copy numbers that differ significantly between the donor cells and secreted EVs (e.g., miR-1260), or between the two cell lines are also included. Percentages are based on the specific miRNA copy numbers/cell relative to the total copy numbers/cell for all miRNAs detected. Copy numbers/cell for these miRNA are given in Table S1. 3B. Composition of the most abundant miRNAs detected in MCF10A cells (top) and in the EVs derived from MCF10A cells (bottom) at 72 hrs of serum depletion. Selected miRNAs with copy numbers that differ significantly between the cells and secreted EVs (e.g., miR-630), or between the two cell lines are also included. Percentages are based on the specific miRNA copy numbers/cell relative to the total copy number/cell for all miRNAs detected. Copy numbers/cell for these miRNA are given in Table S1.
The EV miRNA compositions are dominated by even smaller numbers of miRNA species with five miRNAs accounting for two thirds of the total number of transcripts in each case (Figure 3). The four most abundant MCF7 EV miRNAs are tRNA-derived miRNAs: miR-720, miR-1274a, miR-1274b, and miR-1260 [40, 41], which comprise 65% of the total number of MCF7 EV miRNA transcripts; they are also abundant in MCF7 cells, but are present at considerably lower compositions. The three most abundant MCF10A EV miRNAs are miR-630, miR-720, and miR-205 (Figure 3). A high expression level of miR-205 is a distinguishing characteristic of MCF10A cells. Interestingly, miR-630 is not among the most abundant miRNAs detected in MCF10A cells. As noted above, miR-630 is abundant in MCF7 EVs, but is not detected above threshold in MCF7 cells. These observations indicate that miR-630, in particular, is selectively targeted for secretion from both cell types. The three tRNA-derived miRNA – miR-1274a, miR-1274b, and miR-1260 – in addition to miR-720 are likewise among the most abundant miRNA in the MCF10A EVs, but at notably lower compositions compared to MCF7 EVs. The tRNA-derived miRNA are at least an order of magnitude more abundant in MCF7 EVs if we consider the total copy numbers/cell for the four tRNA-derived miRNA relative to a ‘signature’ miRNA in each case – miR-21 for MCF7 EVs and miR-205 for MCF10A EVs.
Over-expression of tRNAs encoded for charged or polar amino acids, such as lysine and threonine, has been reported specifically for MCF7 cells relative to MCF10A cells [42]. The high abundance of the tRNA-derived miRNAs in MCF7 EVs may reflect the over-expression of the intact tRNAs in MCF7 cells, coupled with tRNA cleavage pathways that effectively compete with miRNA biogenesis [43–45], and an export process that selects for the tRNA fragments. Although starvation can induce tRNA cleavage to produce miRNA-like fragments [46], tRNA cleavage induced by serum deprivation is not considered to be a factor here for miR-720, since this miRNA was also found to be the most abundant cellular species and the second most abundant, cell-secreted EV species when MCF7 cells are grown in media supplemented with EV-depleted FBS [14].
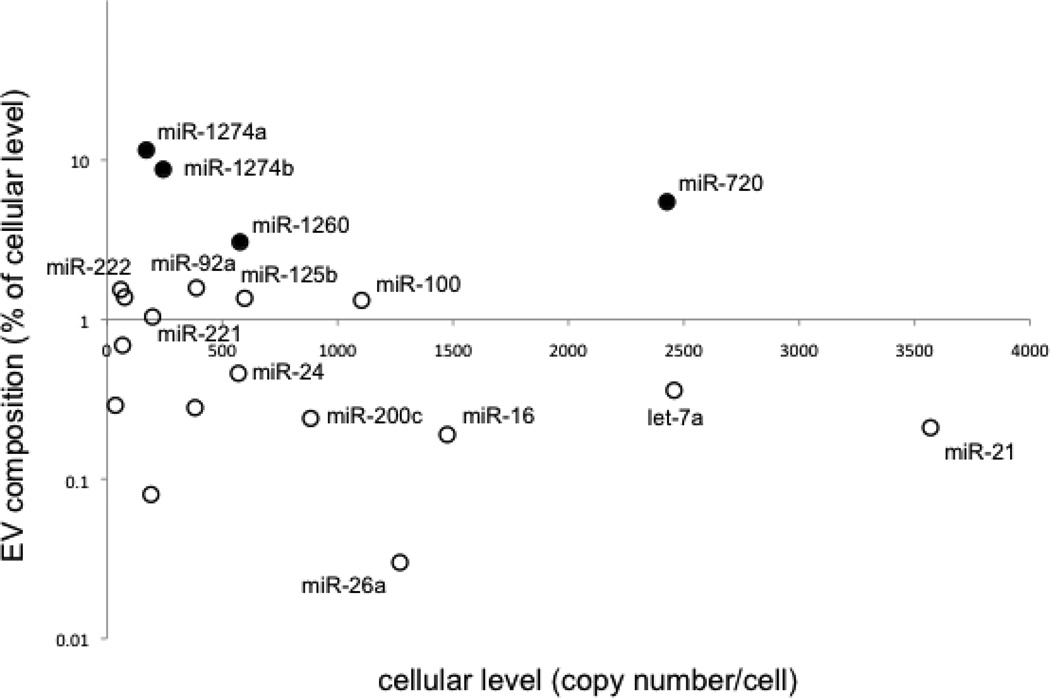
Comparing miRNA copy numbers/cell in the MCF7 cells and EVs (Table S1) reveals that the net secretion of miRNA transcripts as a whole over 72 hrs of serum deprivation represents only a small fraction of the total pool of cellular miRNA. This fraction is ~1% of the total number of miRNA transcripts per MCF7 cell. However, the EV composition of individual miRNA species as a fraction of their cellular levels (copy numbers/cell) can be as much as an order of magnitude higher depending on the miRNA. The tRNA-derived miRNAs in MCF7 EVs, in particular, stand out with EV compositions that are much higher than those for other miRNAs (Figure 4). In contrast, miR-21 and let-7a, the two most abundant miRNAs in MCF7 cells, have MCF7 EV compositions that are more than an order of magnitude lower. The uniformly higher levels of the tRNA-derived miRNA in the MCF7 EVs relative to their cellular levels suggest that these species are targeted for secretion from MCF7 cells.
Figure 4.
MCF7 EV miRNA composition as a percentage of miRNA cellular level of selected miRNAs as a function of their MCF7 cellular level measured at 72 hrs of serum deprivation. The tRNA-derived miRNAs are shown as filled circles. Percent of cellular level is defined as the EV miRNA copy numbers/cell as a percentage of the copy number per cell of that miRNA in MCF7 cells (Table S1).
Seven miRNAs were further analyzed by qRT-PCR. Six of these miRNAs, including two tRNA-derived miRNAs – let-7a, miR-21, miR-100, miR-125b, miR-720, miR-1274b – were selected based on two criteria: (1) high copy numbers/cell in MCF7 EVs, and (2) greatest fold change in miRNA copy numbers/cell for MCF7 EVs relative to MCF10A EVs (Table S1). The seventh miRNA, miR-205, was selected based on these criteria applied to MCF10A EVs. We note that the MCF10A cellular expression level for miR-205 and the MCF7 cellular expression levels for let-7a, miR-21, miR-100, and miR-125b are greater than 240 copies/cell, which we estimate as the threshold level required to elicit a measurable cellular response [16]. This estimate is also more than twice the lower threshold of 100 copies of a miRNA transcript required for the measurable repression of mRNAs within a single cell [47]. Thus, the selected miRNAs are considered to be present in MCF10A cells (miR-205) or MCF7 cells (let-7a, miR-21, miR-100, miR-125b) at functionally significant levels. The copy numbers/cell derived from the qRT-PCR assays for these seven miRNAs in MCF10A and MCF7 cells and EVs are given in Table S2.
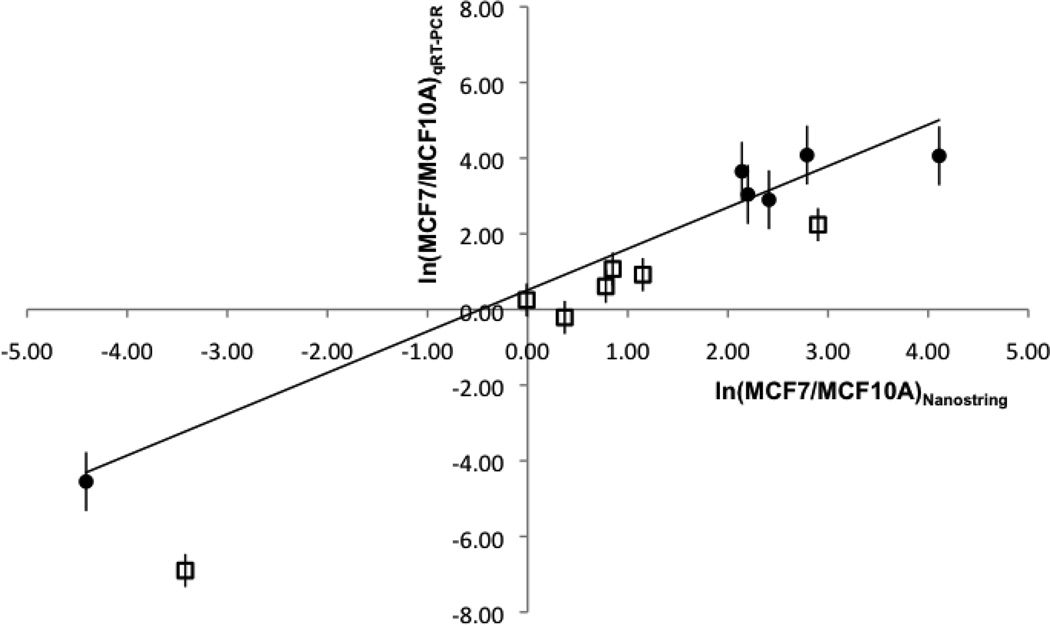
MCF7:MCF10A fold changes in cellular and EV miRNA copy numbers/cell obtained from the NanoString and qRT-PCR assays are compared in Figure 5, and show essentially quantitative agreement between the two assays. Greater fold changes for the EV miRNAs are also evident in this plot, with a majority of the cellular MCF7:MCF10A fold changes clustered nearer the origin; the fold changes for cellular miR-21 and for cellular miR-205 are the two exceptions. The quantitatively similar relative miRNA abundances derived from the two assays stands in contrast to the measured absolute abundances (miRNA copy numbers/cell), which are significantly higher for the qRT-PCR assay (Table S2) compared to the NanoString assay (Table S1) for both cellular and EV miRNA. We note that the qRT-PCR assay results for the cellular abundances of let-7a and miR-21 are consistent with cellular abundances on the order of a few thousand copy numbers/cell for let-7a and up to roughly an order of magnitude higher for miR-21 that have been measured by others [31, 48].
Figure 5.
Natural logarithm of the fold change in MCF7:MCF10A EV miRNA copy numbers/cell (filled circles) and for MCF7:MCF10A cellular miRNA copy numbers/cell (open squares) derived from NanoString and qRT-PCR assays. Linear fit with a correlation coefficient of 0.963 is shown for the EV miRNA copy numbers/cell. Error bars represent one standard deviation for qRT-PCR measurements (n=7 for MCF7 EVs and cells; n=3 for MCF10A EVs and cells). The miRNA are: let-7a, miR-21, miR-100, miR-125b, miR-205, and miR-720 (miR-1274b for cells only). miRNA copy numbers/cell obtained by qRT-PCR are given in Table S2.
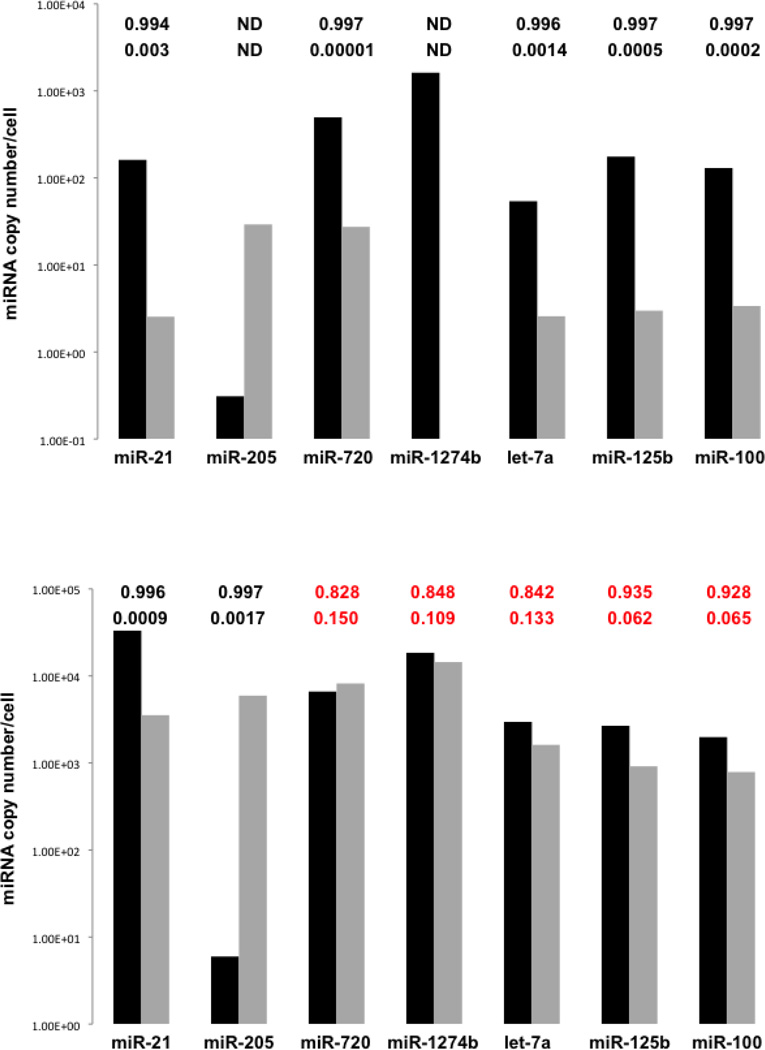
Average miRNA copy numbers/cell for the seven selected miRNAs in MCF7 and MCF10A cells and in their EVs derived from the qRT-PCR assays are compared in Figure 6. Statistically significant, higher expression levels of miR-21 in MCF7 cells and miR-205 in MCF10A cells are the only characteristics of the cellular miRNA profiles that distinguish the two cell lines. Higher miR-21 expression in MCF7 cells is expected since this miRNA is over-expressed in many solid tumors, including breast cancer [25, 26], as well as in MCF7 cells specifically [2, 14, 49]. Lower levels of miR-205 in MCF7 cells are also consistent with the differential expression of this miRNA observed in human breast cancer and MCF7 cells [2, 14, 49, 50]. Conversely, differences in the mean copy numbers/cell for let-7a, miR-100, and miR-125b, and tRNA-derived miR-720 and miR-1274b in MCF7 cells relative to MCF10A cells are not statistically significant. The p-value in each case is 0.062 or higher (Figure 6), corresponding to odds of only 5:1 that the mean copy numbers/cell for these miRNA are different between the two cell lines compared to odds of 249:1 that the mean copy number/cell of miR-21 is higher in MCF7 cells.
Figure 6.
Average miRNA copy numbers/cell in MCF7 (black) and MCF10A (gray) EVs (top) over 72 hrs of serum deprivation, and cells (bottom) at 72 hrs of serum deprivation derived from qRT-PCR assays (n=7 for MCF7 cells and EVs; n=3 for MCF10A cells and EVs; Table S2) for seven miRNAs selected for distinguishing MCF7 cells from MCF10A cells based on miRNA compositions of their EVs. The upper row of numbers above each pair of average miRNA copy numbers/cell are Bayesian probabilities that the higher population mean in each case is higher than the lower population mean. An explanation of the Bayesian probability calculations is given as supplemental material. The numbers in lower row above each pair are p-values for the null hypothesis that the means are the same. “ND” for miR-1274b indicates that this miRNA was not detected in the MCF10A EVs in the qRT-PCR assay; for miR-205, only two measurements of the miRNA copy numbers/cell were obtained for this miRNA in MCF10A EVs. The p-values and Bayesian probabilities in red indicate mean miRNA copy numbers/cell that are not considered statistically different between the two cell lines.
High levels of miR-21 in MCF7 EVs and miR-205 in MCF10A EVs likewise distinguish the EV miRNA profiles, but additional distinguishing characteristics are also found exclusively in the EV profiles (Figure 6). Specifically, the mean copy numbers/cell for let-7a, miR-100, and miR-125b are significantly higher in the MCF7 EVs: p-values ≤ 0.0014 (odds > 249:1). Another prominent characteristic not found in the cellular profiles is the statistically significant higher mean copy number/cell of tRNA-derived miR-720 in the MCF7 EVs: p-value of 0.00001 (odds of 332:1). In comparison, the difference between MCF7 and MCF10A mean cellular levels of miR-720 is not statistically significant: p-value of 0.150 (odds of 5:1). Collectively, these results show that this repertoire of miRNAs and tRNA-derived miRNAs exhibit a wider array of distinguishing features in the EVs compared to their parent cells, as noted above.
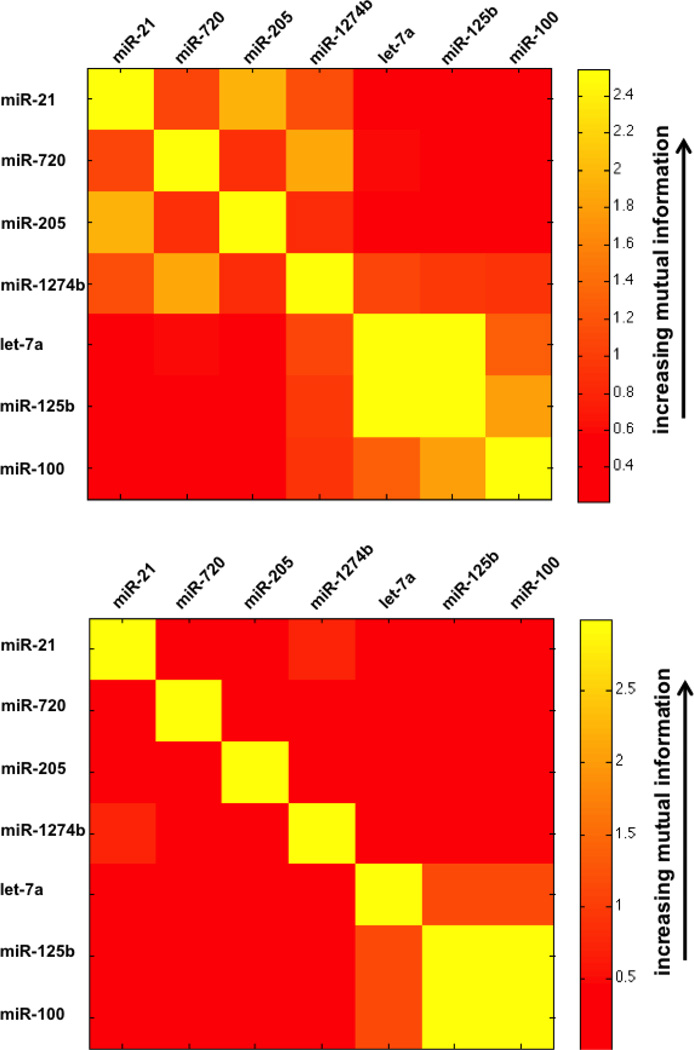
An interesting feature of the MCF7 cellular miRNA expression revealed in the qRT-PCR assays is the correlations in the cellular expression levels of let-7a, miR-125b, and miR-100, which we determined extends to the MCF7 EV miRNA as well. These correlations are shown in Figure 7 where the correlations are expressed as mutual information (MI) in units of bits for each miRNA pair. Note that MI = 0 if the cellular expression levels or the EV miRNA copy numbers/cell for two miRNAs are uncorrelated and MI > 0 if they are either correlated or anti-correlated. See supplemental material for MI defined in terms of the correlation coefficient. High MI implies functional and/or physical miRNA interactions. MCF7 cellular expression levels (Figure 7) are largely uncorrelated, except for miR-125b and miR-100, with let-7a, miR-125b, and miR-100 forming a cluster of interacting miRNAs in MCF7 cells. This miRNA cluster is even more apparent in the MCF7 EVs (Figure 7). Additional MCF7 EV miRNA clusters are also found for miR-21 and miR-205, and for the tRNA-derived miRNAs, miR-720 and miR-1274b (Figure 7). The miR-21/miR-205 miRNA cluster is likely an artifact of the exceptionally low copy numbers/cell for miR-205 in MCF7 EVs (Table S2). This is clearly not the case for the miR-720/miR-1274b miRNA cluster, however. The positive MI in the miR-720 and miR-1274b copy numbers/cell in the EVs in contrast to essentially no MI in their cellular expression levels suggests that these miRNA interactions may be physical rather than functional in nature. We hypothesize that these physical interactions are reflected in the high sequence similarities of miR-720 and miR-1274b – that is, if these tRNA-derived miRNAs bind with similar affinity to other species associated with the EVs, then their sequences would have significantly more matching nucleotides than miRNA pairs that are primarily functionally interactive, such as miR-125b and let-7a or miR-125b and miR-100. Indeed, 10 of 18 nucleotides in the aligned sequences of miR-720 and miR-1274b are identical, whereas only 5 and 6 of 22 nucleotides are the same in the aligned sequences of miR-125b and let-7a and miR-125b and miR-100, respectively (sequence alignments are shown in Figure S2).
Figure 7.
Matrices of mutual information for MCF7 EV miRNA copy numbers/cell (top) and MCF7 cellular miRNA expression levels (bottom) of seven miRNAs selected for distinguishing MCF7 cells from MCF10A cells based on the miRNA profiles of their EVs derived from qRT-PCR assays for multiple (n=7) biologically distinct samples. Calculations of mutual information are described in the supplemental material.
DISCUSSION
Elevated cellular levels of tRNA are a hallmark of proliferative diseases, such as cancer [43, 44], and breast cancer specifically [42]. A growing body of evidence also points to the precise cleavage of mature tRNAs by specific cellular processes, rather than random degradation, to produce small RNA fragments with miRNA-like silencing activity [43, 44, 45]. A manifestation of tRNA over-expression in cancer cells is therefore likely to be high levels of “miRNA-like” tRNA fragments, or tRNA-derived miRNAs, as we have found here for miR-720 and miR-1274b in MCF7 cells. We also found that these tRNA-derived miRNAs are secreted from MCF7 cells at significantly higher fractions of their cellular expression levels compared to other miRNAs – specifically, miR-21, let-7a, miR-100, and miR-125b. Consequently, the cellular levels of these tRNA-derived miRNAs are amplified in the secreted EVs. Further, we found this amplification to be much greater in the MCF7 EVs. The high abundance of these tRNA-derived miRNAs in the MCF7 EVs clearly distinguishes this EV population from MCF10A EVs, and complements the high abundance of miR-21, let-7a, miR-100, and miR-125b, and the low level of miR-205 in identifying MCF7 cells as the source. These findings point to the appealing prospect that it may be possible to identify tumor cell-specific EVs in circulation by high levels of tRNA-derived small RNA fragments in combination with miRNA signatures derived from known miRNA profiles of cancer tumors. We stress that this distinguishing feature of high abundance of miRNA-like tRNA fragments is likely to be most apparent in the EV miRNA profiles rather than the cellular profiles.
Our comparisons of the correlations in cellular miRNA expression levels and correlations in EV miRNA levels show that miRNA clusters in the EVs and their parent cells can be defined by functional miRNA interactions – i.e., correlations in cellular miRNA expression levels that are mirrored in the EVs, such as those found for let-7a, miR-125b, and miR-100. However, other miRNA clusters with uncorrelated cellular expression levels were identified in the EVs based on correlations in their EV miRNA copy numbers/cell. We propose that these miRNA clusters are defined by purely physical miRNA interactions – e.g., aggregation due to comparable binding affinities to common targets. It is also evident that intracellular regulatory functions alone do not drive the partitioning of specific miRNAs into the EVs. For example, miR-26a and miR-100 have similar regulatory functions in suppressing MCF7 cell growth and proliferation, which is consistent with their comparable, high cellular levels under culture conditions of serum deprivation. However, miR-100 and miR-26a levels in MCF7 EVs differ by more than an order of magnitude, which does not match their similar functional roles. The difference may be related instead to different binding affinities as suggested here, as well as different stabilities and/or processing efficiencies. An important implication of these observations is that more informative cancer-specific miRNA profiles may be obtained by developing new clustering algorithms that consider physical miRNA interactions in addition to functional miRNA interactions. Moreover, physically interactive miRNA clusters that represent distinguishing features of cancer-specific miRNA signatures are likely to be amplified in secreted vesicles, thus highlighting EV miRNA signatures specifically as biomarkers for cancer.
We estimate the total number of miRNAs secreted per cell over 72 hrs of serum deprivation to be much smaller than the total number of EVs secreted per cell over this time interval, even when considering the higher EV miRNA copy numbers/cell measured by qRT-PCR. It follows therefore that most EVs do not contain miRNA, a conclusion also reached in a recent stoichiometric analysis of miRNA content in EVs from diverse sources, including cancer patient plasma, healthy donor seminal fluid, and in vitro sources: dendritic cells, mast cells, and ovarian cancer cells [30]. A low probability of miRNA-containing EVs does not preclude the EV-mediated functional delivery of the miRNAs to target cells, however. The functional delivery of miRNA to target cells would depend instead on the extent to which miRNA are concentrated in a small number of vesicles. In this context, measurements of miRNA copy numbers in single vesicles over a population of EVs will be more informative than measuring the average miRNA copy number/vesicle for specific transcripts of interest. In addition, the low miRNA copy numbers/cell in the EVs reported here reflect only the net secretion of miRNA-containing EVs that are not targeted for re-internalization by the parent cells. Since this re-internalization will depend on the cellular microenvironment, we expect the net secretion of miRNA-containing EVs to depend on the cellular microenvironment. Exploring this dependence is an important future research direction.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Stefano Volinia (Ohio State University Comprehensive Cancer Center) for helpful discussions on NanoString miRNA data analysis, and Professor Ishi Talmon (Technion) for sharing his expertise and providing valuable training in cryo-TEM sample preparation and imaging. This work was supported by grants from the NIH (P01 CA124570 and 1RC2AG036559-01 and 1UH2TR000914-01) and NSF (EEC-0425626 and EEC-0914790). N.G. was partially supported by a NASA Ohio Space Grant Consortium Fellowship. N.G. performed EV isolations/characterizations and RNA purifications, and carried out the NanoString miRNA assays and analyzed the data with L. Y. K.A. performed the EV isolations/characterizations and RNA purifications, and carried out the qRT-PCR assays and the antibody microarray assays. D.A. carried out the statistical analyses of the qRT-PCR assays, M.S. maintained the cell cultures and designed the EV isolation and RNA purification experiments, and the qRT-PCR assays. M.R. and M.P. designed the research, analyzed the experimental results, and wrote the manuscript.
NanoString miRNA assay data reported in this publication have been deposited in NCBI's Gene Expression Omnibus (Guzman et al., 2015) and are accessible through GEO Series accession number GSE66165 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66165).
Footnotes
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Calin GA, et al. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Farazi TA, et al. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong YW, et al. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 4.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocucci E, et al. Shedding microvesicles: artifacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Akers JC, et al. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptic bodies. J Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Deregibus MC, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 11.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbings DJ, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 13.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2(282):1–10. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pigati L, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE. 2010;5(10):e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor DD, C Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 16.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte-'t Hoen EN, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellingham SA, et al. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracellular Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arraud N, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thrombosis Haemostasis. 2014;12:614–627. doi: 10.1111/jth.12554. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard CC, et al. Blood Cell Origin of Circulating MicroRNAs: A Cautionary Note for Cancer Biomarker Studies. Cancer Prev Res (Phila) 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debnath J, et al. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 28.Théry C, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3. [DOI] [PubMed] [Google Scholar]
- 29.Lamparski HG, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 30.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111(41):14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucl Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue C, et al. Novel cellular microarray assay for profiling T-cell peptide antigen specificities. J. Proteome Res. 2010;9:5629–5637. doi: 10.1021/pr100447b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman BM, et al. Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 2012;26:4160–4173. doi: 10.1096/fj.11-202077. [DOI] [PubMed] [Google Scholar]
- 35.Palma J, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobert S, et al. Regulation of β-Tubulin Isotypes by Micro-RNA 100 in MCF7 Breast Cancer Cells. Cytoskeleton. 2011;68:355–362. doi: 10.1002/cm.20517. [DOI] [PubMed] [Google Scholar]
- 37.Sampson VB, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67(20):9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36(16):5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Hoon MJ, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20(2):257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schopman NC, et al. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010;7(5):573–576. doi: 10.4161/rna.7.5.13141. [DOI] [PubMed] [Google Scholar]
- 42.Pavon-Eternod M, et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuck AC, Tollervey D. RNA in pieces. Trends in Genetics. 2011;27(10):422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SR, Collins K. Starvation-induced Cleavage of the tRNA Anticodon Loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280(52):427–449. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 47.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25(12):1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 48.Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes & Development. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fix LN, et al. MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer Genomics Proteomics. 2010;7(5):261–277. [PubMed] [Google Scholar]
- 50.Iorio MV, et al. microRNA-205 Regulates HER3 in Human Breast Cancer. Cancer Res. 2009;69(6):2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.