SUMMARY
SGI-110 is a second-generation hypomethylating prodrug whose active metabolite is the well-characterized drug decitabine. This novel compound is an oligonucleotide consisting of decitabine linked through a phosphodiester bond to the endogenous nucleoside deoxyguanosine. The dinucleotide configuration provides protection from drug clearance by deamination, while maintaining at least equivalent effects on gene-specific and global hypomethylation both in vitro and in animal model systems. This agent is currently being tested in phase I and II clinical trials in humans and has been demonstrated to be safe and well tolerated as a single agent, with evidence of promising activity in heavily pretreated (including currently FDA approved hypomethylating drugs) myelodysplastic syndrome and acute myeloid leukemia patients. Ongoing trials are also open in platinum-resistant ovarian cancer and hepatocellular carcinoma.
Keywords: Hypomethylating prodrug, Myelodysplastic syndrome, Acute myeloid leukemia, ovarian cancer, Hepatocellular carcinoma, SGI-110
Graphical abstract
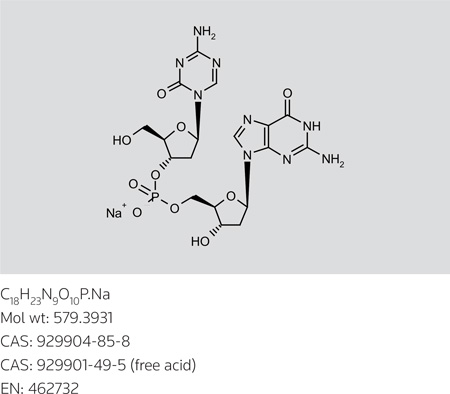
S-110
2'-Deoxy-5'-O-[(2'-deoxy-5-azacytidin-3'-O-yl)(hydroxy)phosphoryl]guanosine sodium salt
InChI: 1S/C18H24n9o10P.na/c19-16-22-6-27(18(31)25-16)12-2-8(9(3-28)35-12)37-38(32,33)34-4-10-7(29)1-11(36-10)26-5-21-13-14(26)23-17(20)24-15(13)30;/h5-12,28-29H,1-4H2,(H,32,33)(H2,19,25,31)(H3,20,23,24,30);/q;+1/p-1/t7-,8-,9+,10+,11+,12+;/m0./s1
SYNTHESIS*
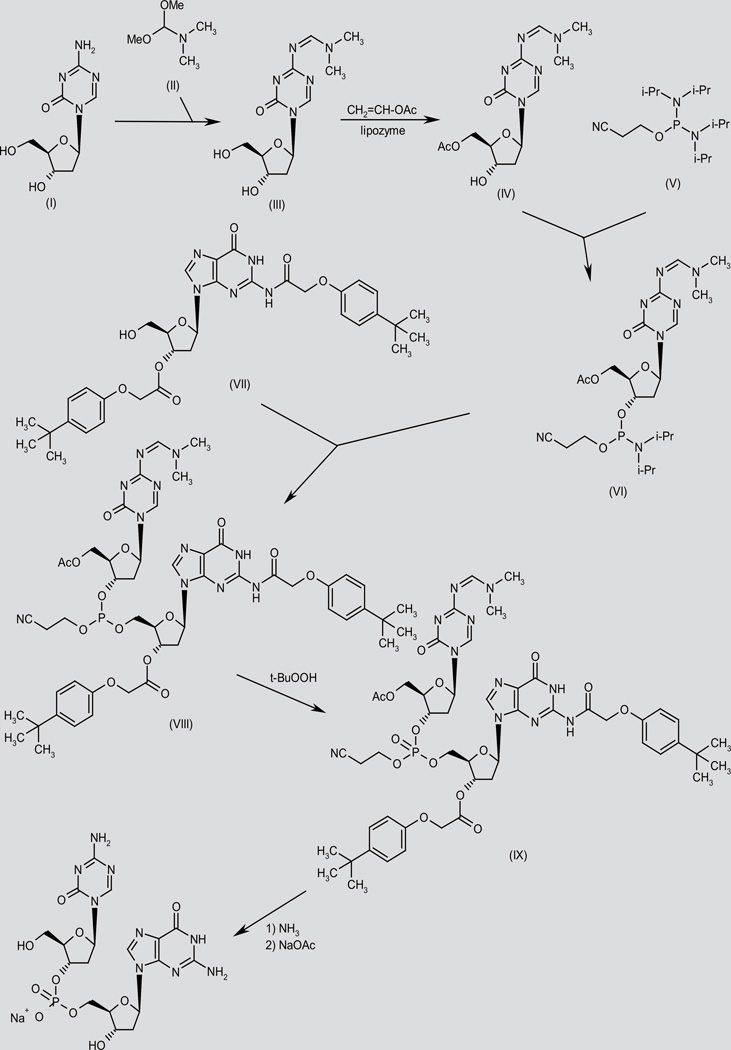
SGI-110 has been prepared by two different methods.
In one method, condensation of decitabine (I) with N,N-dimethylform-amide dimethylacetal (II) gives the formamidine derivative (III), which by enzymatic acetylation with vinyl acetate in the presence of immobilized lipozyme in acetonitrile/1,4-dioxane yields the primary acetate (IV). Condensation of the deoxyribofuranose derivative (IV) with 2-cyanoethyl N,N,N’,N’-tetraisopropylphosphorodiamidate (V) in CH2Cl2 provides the phosphorodiamidate (VI), which then condenses with the deoxyguanosine derivative (VII) in CH2Cl2 to afford the trisubstituted phosphite (VIII) (1). Oxidation of phosphite (VIII) with t-BuOOH provides the corresponding phosphate (IX), which is fully deprotected by means of NH3 in MeOH, and finally salified using NaOAc in H2O/EtOH (1). Scheme 1.
Scheme 1.
Synthesis of SGI-110
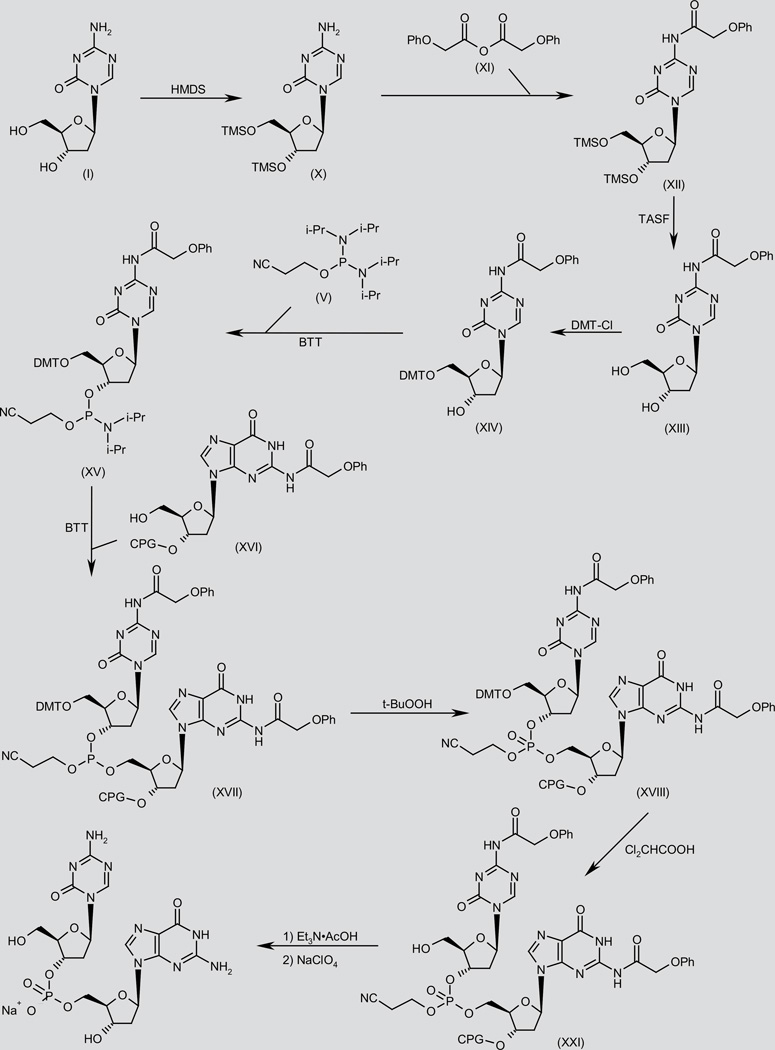
In an alternative, solid-phase method, O-protection of decitabine (I) by means of HMDS in DMF, followed by N-acylation of the obtained bis-TMS ether (X) with phenoxyacetic anhydride (XI) in pyridine gives amide (XII). Desilylation of compound (XII) using TASF in DMF, followed by selective protection of the primary alcohol group in the resulting diol (XIII) with 4,4’-dimethoxytrityl chloride (DMT-Cl) in pyridine furnishes ether (XIV). Condensation of the free secondary alcohol of (XIV) with 2-cyanoethyl N,N,N’,N’-tetraisopropylphosphorodiamidate (V) using benzylthiotetrazole (BTT) in acetonitrile provides phosphoramidate (XV), which is then condensed with the 3’-deoxyguanosine derivative (XVI) linked onto controlled pore glass (CPG) support in the presence of BTT in MeOH to afford trisubstituted phosphite (XVII) (2). Oxidation of phosphite (XVII) with t-BuOOH in acetonitrile provides the corresponding phosphate (XVIII), which is then O-deprotected by means of Cl2CHCOOH in toluene to give the primary alcohol (XIX). Finally, CPG-bound dinucleotide (XIX) is submitted to deprotection and cleavage from the solid support with aqueous Et3N·AcOH, followed by precipitation of the resulting triethylammonium salt in the presence of NaClO4 in H2O/acetone (2). Scheme 2.
Scheme 2.
Synthesis of SGI-110
BACKGROUND
Methylation of cytosine-phospho-guanine (CpG) residues within DNA has been identified to produce a so-called “fifth base” methylcytosine within DNA. DNA methylation of this kind is important in mammalian cells for the maintenance of normal gene expression; it is used for the regulation of imprinted genes and is critical to the process of lyonization of the X-chromosome in women (3–5). The pattern of methylated cytosine residues within DNA is carefully maintained by enzymes called DNA methyltransferases (DNMTs), whose function is to copy the methylation signature from parent to daughter DNA strand (6). In general, methylated cytosines are associated with intergenic DNA regions, including peri-centromeric chromatin and repetitive DNA elements, such as long interspersed nucleotide elements (LINEs) and Alu repeats (short repetitive ~300-base pair stretches of DNA characterized by susceptibility to the Arthrobacter luteus restriction endonuclease; the most abundant transposable elements in the human genome), while CpG rich regions associated with gene promoters (CpG islands) are remarkably absent of methylation (7). The function of these enzymes is critical for normal embryonic development, and indeed, their absence during embryogenesis in the mouse is lethal (8, 9).
In general, cancers are associated with a disruption in the normal cellular methylation signature, demonstrating an overall global hypomethylation, particularly of repetitive DNA elements and LINEs, associated frequently with hypermethylation of CpG island sequences (10). These hypermethylation events have been demonstrated to induce loss of gene expression in a variety of cancers, most notably tumor suppressor genes (TSGs), in a manner analogous to mutation-induced loss of gene expression. However, unlike mutation-induced gene silencing, methylation events preserve the DNA signature and are potentially reversible through inhibition of cellular methylation machinery.
Two drugs, azacitidine and decitabine, classified as DNA methylation inhibitors (hypomethylating agents; HMAs), are approved by the U.S. FDA for the treatment of myelodysplastic syndrome (MDS) and low blast count (< 30%) acute myeloid leukemia (AML). Both agents are prodrugs activated by phosphorylation, decitabine directly by deoxycytidine kinase and azacitidine through the serial action of uridine/cytidine kinase and ribonucleotide reductase. It is important to note that cancer cells which lack these enzymes are inherently resistant to decitabine and azacitidine respectively (11, 12). Both drugs are relatively unstable in alkaline and acid solution, and undergo spontaneous decomposition under these conditions, resulting in an in vitro half-life in these solutions of between 3 and 4 hours (13–15). They are also unstable in vitro in neutral aqueous solution at 37 °C, but with slightly longer half-lives of 7 hours for azacitidine and 12–20 hours for decitabine (16, 17). Currently, azacitabine is available for either subcutaneous (s.c.) or intravenous (i.v.) injection, while decitabine is given only i.v. Both drugs have very short in vivo half-lives following clinical administration; patients treated with i.v. decitabine demonstrate > 90% clearance of drug within an hour following its infusion (18), while those treated with s.c. azacitidine have similarly rapid clearance (19, 20). Elimination of these drugs occurs both by hydrolysis and through deamination by the action of cytidine deaminase.
Both decitabine and azacitidine were approved for MDS in the U.S. based upon the demonstration of improvements in transfusion requirements and quality of life (21–24). Subsequently, azacitidine, but not decitabine, was demonstrated to improve overall survival in landmark studies involving particularly high-risk MDS patients (25, 26).
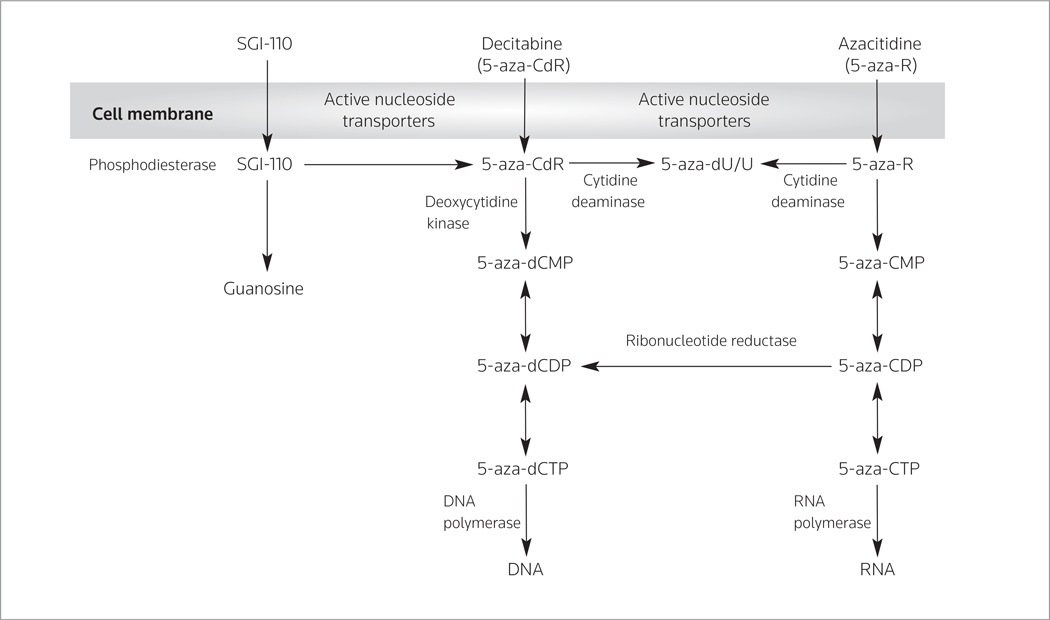
The HMAs are hypothesized to exert their effects through the re-expression of TSGs, as described above. Both agents are nucleoside analogues which, following intracellular uptake mediated by nucleoside-specific transport by means of the transporters human solute carrier family 29 member 1 (ENT1, SLC29A1) and 2 (ENT2, SLC29A2), are phosphorylated into mono di-and tri phosphate forms and then incorporated into the DNA of replicating cells in place of the base cytidine (18, 27). In the case of both drugs, the metabolite incorporated into DNA is decitabine. While decitabine is incorporated entirely into DNA, azacitidine is incorporated 60–80% into RNA and only ~20% into DNA (Figure 1). The importance of this differential incorporation is not well understood, although a majority of the data on their mechanisms suggests that DNA methylation changes are likely central to their activity.
Figure 1.
Uptake and serial steps for the incorporation of SGI-110, azacitidine and decitabine into RNA and DNA.
The substitution of nitrogen for carbon at the 5’ position of the carbon ring in decitabine results in an inability to accept a methyl group through the action of DNMTs. DNMTs are therefore trapped on the aza-nucleotide, depleting the cell of active DNMTs and allowing subsequent cycles of DNA replication to go forward without maintenance of the parent DNA methylation signature. Low-dose exposure to these drugs in the nanomolar range (200–500 nM) for decitabine and the micromolar range (0.5–3 µM) for azacitidine, both in vitro and in human studies, has been repeatedly demonstrated to induce both global and gene-specific DNA hypomethylation in this fashion (28–31). The action of these drugs is S-phase-specific. In addition to the action of these drugs to induce alterations in DNA methylation, trapping of DNMTs on the parent DNA strand produces a bulky DNA adduct, resulting in double-strand DNA breaks (32). At higher doses, both azacitidine (> 5 µM) and decitabine (> 1 nM) induce cell cycle arrest and their effects on DNA methylation plateau (28, 30).
SGI-110 is a novel DNA methylation inhibitor whose active metabolite is the FDA-approved drug decitabine. This novel drug was synthesized by Astex Pharmaceuticals, Inc. and was rationally designed to be more stable than decitabine in order to provide enhanced pharmacokinetic and pharmacodynamic properties. The decitabine moiety is linked through a phosphodiester bond to deoxyguanosine, and this linkage results in reduced susceptibility to immediate inactivation by cytidine deaminases and a prolonged plasma half-life compared to the parent drug. Following the cleavage of the phosphodiester bond, free decitabine undergoes inactivation by cytidine deaminase in the standard fashion. Due to its chemical structure, the molar equivalent of a 1-mg dose of decitabine is approximately 2.5 mg of SGI-110. Preclinical studies both in vitro and in animal models with SGI-110 demonstrated that exposure to this drug induces similar effects on both gene-specific (mir29b, HgF) and global (LINE-1) methylation when compared with azacitidine and decitabine (33–35). Furthermore, early pharmacokinetic results suggest an improved exposure window with similar AUC exposure profiles between i.v. decitabine and s.c. SGI-110, albeit with lower maximal concentrations (Cmax) based on molar equivalence in dose. Presently, SGI-110 is being tested against both myeloid malignancies and solid tumors in phase I and II clinical trials. Results from a portion of the phase I trial in MDS/AML have been published in abstract form and suggest meaningful clinical activity, notably in some patients who have failed prior HMAs (36). As yet, data from the phase I trials in solid tumors have not yet been presented. These data, as well as the published preclinical data, will be reviewed in detail below.
PRECLINICAL PHARMACOLOGY
A majority of the preclinical studies undertaken with SGI-110 have compared its effect on methylation and gene re-expression to those seen with the parent drug decitabine. The following paragraphs will attempt to synthesize the results from a large number of these preclinical studies, which largely rely on evaluation of changes in DNA methylation using bisulfite converted DNA. Exposure to sodium bisulfite in vitro deaminates only unmethylated cytosines; methylated CpG sequences will remain cytosines, while unmethylated cytosine will be converted to uracil and will be sequenced as thymidine. Direct sequencing or sequence-specific polymerase chain reaction (PCR) can then be used to determine what effects the drug of interest has had on the methylation pattern within a cell. Bisulfite pyrosequencing, in which bisulfite converted DNA is amplified outside a CpG-rich region and specific CpG sites are analyzed for a sequence change, can be used both to assess gene-specific changes in methylation, as well as an effective surrogate for genome-wide changes in methylation using an assay specific for LINE-1.
Initial studies of SGI-110 were undertaken in order to demonstrate superior stability and support the hypothesis that protection from deamination by cytidine deaminase would prolong drug exposure relative to decitabine. Indeed, Yoo et al. demonstrated that, in comparison with decitabine, SGI-110 showed significantly better stability (~80% intact drug vs. < 5% over 100 minutes) when incubated with recombinant cytidine deaminase at 37 °C in vitro (37). Similar results were obtained by incubating SGI-110 with human plasma over 40 minutes. Unfortunately, hydrolytic cleavage of the drug did not appear to be significantly better. These authors further demonstrated that SGI-110 produced LINE-1 and p16 (a TSG) hypomethylation in a concentration-dependent fashion, associated with robust re-expression of p16 mRNA and protein in both the human bladder carcinoma T24 cell line, as well as the human colon carcinoma HCT 116 cell line. Furthermore, exposure to both decitabine and SGI-110 induced significant depletion of DNMT protein by Western blot at equimolar concentrations. These results suggest an enhanced potency for the decitabine derived from SGI-110 over parent decitabine, since SGI-110 delivers only half as much decitabine at equimolar doses of the drug.
An analogous study in a mouse model was undertaken by the same group to assess the tolerability and efficacy of SGI-110 in comparison with decitabine in athymic nude mice. Non-tumor-bearing mice were injected i.v. with either decitabine or SGI-110 at equimolar doses of 15 and 36 mg/kg divided over one of three different schedules: whole dose once per week, 1 of 2 doses twice per week or 1 of 3 doses 3 times per week over a period of 3 weeks. These mice were then assessed for drug tolerability using weight and morbidity. SGI-110 was better tolerated at all dose levels than decitabine, although the latter schedule (3 doses per week) was not tolerated, as mice receiving both drugs on the 3-dose per week schedule all died prior to the end of the experiment. This group further treated human bladder carcinoma EJ6 bearing nude mice with either SGI-110 (10 or 12.5 mg/kg/day) or decitabine (5 mg/kg/day) × 6 days, injected either intraperitoneally (i.p.) or s.c. to determine relative effects on tumor growth, hypomethylation and gene re-expression of p16. Both drugs effectively slowed tumor growth (evaluated for both delivery methods), while effectively hypomethylating the p16 gene promoter and re-expressing the gene product (only the i.p. method). Mice treated with SGI-110 lost a similar amount of weight compared with decitabine-treated mice, but appeared healthier and more active than their decitabine-treated counterparts. S.c. injection was deemed superior to i.p. injection in terms of toxicity and the daily schedule was better tolerated than the 3-dose per week schedule.
There is significant interest in the possibility that re-expression of epigenetically silent TSGs could re-sensitize resistant cancer cells to chemotherapy-induced death. This concept has been demonstrated previously with decitabine in combination with carboplatin in ovarian cancer, and given the potentially superior pharmacokinetic/pharmacodynamic (longer half-life/demethylation effect) and administration profile (s.c.) of SGI-110, preclinical demonstration of a similar profile was necessary in order to facilitate clinical development of this concept (38–40). Daniela Matei et al. recently published in abstract form that the platinum-resistant human ovarian carcinoma A2780 cell line could be re-sensitized to carboplatin following a 48-hour pre-incubation with 5 µM SGI-110 in vitro (41).
These authors also demonstrated that the combination of SGI-110 (2–5 mg/kg/day) and carboplatin (2 mg/kg/day) was well tolerated in tumor-bearing mouse xenografts, and that the combination resulted in both global and gene-specific hypomethylation. These as well as previously published data have been sufficient to prompt a phase I/II study of SGI-110 in combination with conventional therapies in relapsed/refractory ovarian cancer patients (see below).
Interest in the ability of HMAs to re-sensitize cancer cells to conventional therapy is not limited to ovarian cancer. Another group has looked in vitro at the activity of SGI-110 in hepatocellular carcinoma (HCC). At the 2012 EORTC meeting, Simone Jueliger et al. presented data showing that p53 wild-type HCC cell lines were sensitive to SGI-110 at relatively low concentrations (< 100 nM) (42). They further showed that SGI-110 demonstrated some activity alone and prior to sorafenib (the current standard of care for patients with inoperable HCC) and suggested evidence of synergy both in vitro and in a mouse xenograft model of HCC. Based upon these data, a phase I trial in HCC is under way.
In addition to TSG hypomethylation, there has been increasing interest in the effect of HMAs on modulation of the immune response to cancer. Data for this hypothesis stem from the recognition that members of the cancer testis antigen (CTA) family, a group of genes whose normal expression is restricted to the adult testis and embryonic ovary, are silenced by dense promoter methylation in adult somatic tissues (43). These genes can be aberrantly expressed (due to hypomethylation of the gene promoter) in some cancers, and in this context they are recognized to be immunogenic; based on these data, we and others have developed the hypothesis that induced anti-CTA-based immunity following HMA exposure may be clinically relevant (33, 44). In addition to their ability to re-express CTAs, HMAs, including SGI-110, have been shown to upregulate the expression of a variety of co-stimulatory molecules in cancer cell lines (44).
Michele Maio’s group has published both in abstract and manuscript form data demonstrating that solid tumor cell lines exposed to doses of SGI-110 ranging from 1–10 µM in vitro will re-induce the expression of a plethora of CTAs, including nY-ESo-1 and MAGEA1 (44). These genes are of particular interest, since clinically relevant spontaneous immune responses have been demonstrated in patients with a variety of tumor subtypes and relevant vaccines are currently under clinical development. Dr. Maio’s group treated a series of cancer cell lines, including cutaneous melanoma, mesothelioma, renal cell carcinoma and sarcoma, in vitro with either decitabine or SGI-110. All the treated cell lines demonstrated induced expression of CTAs by real-time PCR, including MAGEA1, MAGEA2, MAGEA3, MAGEA4, MAGEA10, GAGE-12, GAGE-1-6, NY-ESO-1 and SSX 1–5. Furthermore, the NY-ESO-1 and MAGEA1 promoters demonstrated hypomethylation (assessed by methylation-specific PCR) following drug exposure. The authors demonstrate that exposure to both HMAs induced enhanced expression of HLA class I, HLA-A2 and ICAM-1 in human melanoma Mel 275 HLA-A2-restricted cells, which correlate with enhanced recognition by HLA-A2-restricted gp100 specific T cell clones. In addition to these data from solid tumor cell lines, our group has demonstrated that similar re-expression of the CTAs MAGEA1 and NY-ESO-1 are inducible in a variety of AML cell lines following exposure to HMAs, including SGI-110 (33). These phenomena are true both in vitro and in mouse xenograft models.
Overall, these studies suggest that SGI-110 is at least as potent as decitabine in inducing epigenetically silenced genes, including TSGs and CTAs, and furthermore that the SGI-110 formulation appears to be better tolerated in mice and more potent at equimolar equivalent doses than decitabine.
PHARMACOKINETICS AND METABOLISM
The pharmacokinetics, bioavailability and safety of SGI-110 have been studied in a variety of different animal model systems. Tolerability has been demonstrated in mouse, rat and rabbit models using multiple dose routes and schedules (35). These studies demonstrate that a majority of the SGI-110 doses are converted to decitabine and the relative bioavailability in mice and rats was nearly 100% when the drug was dosed s.c. compared to i.v.
In addition to studies in rodents, two studies of SGI-110 have demonstrated tolerability and bioavailability in non-human primates. The first study was done to assess the ability of SGI-110 to induce the fetal hemoglobin (HgF) gene. The authors first bled two baboons for eight days to a hematocrit of 20%. Animals were given either decitabine 0.5 mg/kg or SGI-110 1 mg/kg for 9 days. The animals were treated with each drug sequentially on the same protocol to allow for intraanimal comparison of drug pharmacokinetics. In both animals, HgF was induced with a latency of about 15 days following drug exposure. The first animal induced HgF levels of 46% following SGI-110 and 56.9% following decitabine, the second animal induced HgF levels of 75.5% with SGI-110 and 80.6% following decitabine; untreated baboons demonstrated an average HgF level of 5.6%. These data suggest that decitabine and SGI-110 are equivalent at the epigenetic endpoint of HgF induction. Induction of HgF in treated baboons was associated with hypomethylation of the gene promoter to 28.9% following SGI-110 and 36.5% following decitabine; untreated baboons demonstrated 75.6% methylation of the HgF promoter. In addition to effects on HgF, the authors compared the effect of both drugs on platelet and neutrophil counts; both animals demonstrated increased platelet counts around day 20 of treatment and a fall in neutrophil counts beginning around day 5 following drug exposure and demonstrating some recovery by day 30. Decitabine given i.v. at a dose of 0.5 mg/kg reached a maximum concentration (Cmax) of 16 ng/mL, whereas SGI-110 administered i.v. at a dose of 0.5 mg/kg produced a Cmax for the prodrug of 6 and 17 ng/mL of decitabine.
Another study of the drug in comparison to decitabine was undertaken in cynomolgus monkeys in which male monkeys were injected for 5 days with either 1.7 mg/kg i.v. of decitabine, 1.7 mg/kg of SGI-110 s.c. or 3.0 mg/kg of SGI-110 s.c. (34). This study was done specifically to demonstrate safety, pharmacokinetics and pharmacodynamics in comparison with decitabine in order to facilitate the phase I first-inhuman study of SGI-110. The dose of decitabine was chosen to mimic the current clinically utilized decitabine schedule of 20 mg/m2/day × 5 days, while the SGI-110 schedules reflect 42% and 75%, respectively, of the molar equivalent decitabine doses. Plasma levels of decitabine and SGI-110 were evaluated every 30 minutes following drug treatment. In the monkeys, SGI-110 administered s.c. was rapidly converted to decitabine, producing a similar exposure window when compared with i.v. decitabine. For the groups treated with s.c. SGI-110, the Cmax ranged from 157–200 (1.7 mg/kg) or 190–378 ng/mL (3 mg/kg) for SGI-110, and 52–82 (1.7 mg/kg) or 60–163 ng/mL (3.0 mg/kg) for decitabine. For i.v. decitabine, the Cmax was 215–525 ng/mL. The AUCs for the SGI-110 groups ranged from 90–101 (1.7 mg/kg) or 123–324 (3 mg/kg) ng*h/mL, while decitabine AUCs ranged from 37–68 (1.7 mg/kg) or 69–155 (3 mg/kg). The AUC for i.v. decitabine was 99–221.
Treated monkeys developed reversible decreases in hemoglobin, neutrophil and total white blood cell counts, which nadired between days 8 and 14 and recovered between days 21 and 28; decitabine-treated monkeys had lower nadirs when compared with either of the two SGI-110-treated groups. The doses of SGI-110 tested produced superimposable changes in LINE-1 and gene-specific methylation (miR29b) when compared with decitabine. A fourth cohort of monkeys was treated with a schedule of 3.0 mg/kg of s.c. SGI-110 once per week for 3 weeks. This schedule produced significantly less impact on the hemogram of the monkeys compared with decitabine and a less robust reduction in LINE-1 and miR29b methylation. The treated monkeys tolerated both drugs well, and based upon these data, the phase I/II trial of SGI-110 in relapsed refractory MDS or AML randomized patients to receive SGI-110 s.c. either on a daily or a weekly schedule every 28 days (36).
CLINICAL STUDIES
The first-in-human phase I/II trial of SGI-110 opened in December of 2010 and was initially designed to have two parts (nCT01261312). The first, a dose-escalation phase, enrolled 78 patients who were randomized to receive either of two regimens, SGI-110 s.c. daily on days 1–5 or s.c. weekly × 3 weeks of a 28-day cycle with a dose-escalation schema guided by both tolerability and pharmacokinetics based on the AUC of decitabine, and pharmacodynamics by observed changes in LINE-1 methylation in mononuclear cells harvested from the peripheral blood. Currently, the dose-escalation component of the study has completed accrual and was presented by Dr. Kantarjian on behalf of the study investigators at the 2012 American Society of Hematology Meeting in Atlanta, Georgia (34). The primary endpoints of the dose-escalation portion of the study were definition of the maximum tolerated dose (MTD) and biologically effective dose (BED), as defined by the lowest dose to achieve maximum hypomethylation or gene re-expression in at least three dose levels. Secondary endpoints included adverse events, pharmacokinetic profiles of SGI-110 and decitabine, and response rates.
overall seven cohorts were enrolled in this portion of the study beginning at doses of 3 mg/m2/day on the daily × 5 schedule and 6 mg/m2/week on the once-weekly × 3 schedule. Dose escalation up to 125 mg/m2 occurred for both dose schedules; the MTD was not reached for the weekly schedule and was designated as 90 mg/m2 daily for 5 days (for MDS patients) and the BED (chosen due to the observation of maximal LINE-1 demethylation plateau) as 60 mg/m2 on the daily × 5 schedule. The once-weekly×3 schedule produced less efficient LINE-1 demethylation. A majority of the patients enrolled in the phase I portion had AML (82%); they were predominantly male (65%), had good performance status (PS 0–1; 87%) and had been previously exposed to decitabine, azacitidine or both (63%). All MDS patients had received prior decitabine, azacitidine, or both.
The pharmacokinetic profile suggested delayed appearance of decitabine after s.c. SGI-110 injection, resulting in a significantly longer exposure window and prolonged effective half-life. Dose-dependent demethylation of LINE-1 elements (~25% relative to baseline) were observed up to the dose of 60 mg/m2 daily × 5 days, with doses of 90 and 125 mg/m2 producing similar changes in LINE-1 methylation. On the weekly regimen, only limited changes in LINE-1 methylation were observed at all dose levels. Despite the fact that clinical responses were not a primary endpoint of the study, 10 patients developed a response to treatment (5 MDS and 5 AML patients). All 5 AML responses occurred in patients who demonstrated a ≥ 10% decrease in LINE-1 methylation. Adverse events judged to be related to SGI-110 on the daily × 5 regimen were as expected from the dosing route: injection-site pain (18% overall; no grade 3 or 4); and from the disease and drug class: thrombocytopenia, neutropenia and febrile neutropenia, all < 20% each. Additional side effects included nausea, fatigue, decreased appetite and diarrhea, none of which were above grade 2. The once per week × 3 regimen was well tolerated, with toxicities consistent with the daily × 5 regimen; there were no dose-limiting toxicities on the weekly regimen.
Based on the data from the phase I portion of the study, the dose-expansion cohort was opened in June 2012 and is currently scheduled to enroll about 200 patients either front-line or with relapsed/refractory AML and MDS. Patients will be randomized to receive either 60 or 90 mg/m2 daily × 5 days; one additional cohort of relapsed/refractory AML patients will also be enrolled to receive 60 mg/m2/day × 10 days. This study is ongoing.
Two other trials of SGI-110 are currently listed on the ClinicalTrials.gov web site; the first is a phase I/II trial of SGI-110 with carboplatin (or physicians’ choice in the phase II portion) in relapsed/refractory ovarian cancer patients (NCT01696032), and the second a single-agent phase II trial in patients with HCC (NCT01752933).
The ovarian cancer trial was opened in September of 2012 and proposes to enroll an estimated 116 patients with platinum-resistant recurrent ovarian cancer in an open-label, randomized study. The initial portion of the study is designed to assess the safety of SGI-110 in combination with carboplatin. In this portion of the study, patients will receive escalating doses of SGI-110 and carboplatin in order to identify an optimum dose based mainly on safety. In the second portion of the study, patients will be randomized to receive the dose of SGI-110 and carboplatin identified in the phase I study, or one of three treatment choices (topotecan, pegylated liposomal doxorubicin or paclitaxel) at the discretion of the investigator. Study endpoints include: objective tumor response rates, changes in the ovarian cancer serum biomarker CA125, progression-free survival and overall survival. This study is projected to complete enrollment in 2014.
The last open trial with SGI-110 is a single-agent, non-randomized phase II study in advanced HCC patients who have failed prior treatment with sorafenib. This trial was designed using a Simon’s 2-stage design in which a minimum number of patients must demonstrate disease control at 16 weeks to proceed to stage 2. At stage 2, a set number of patients must have disease control at 16 weeks to declare that SGI-110 is of interest in the treatment of advanced HCC after failure of prior sorafenib. If both stages are completed the study is projected to include 46 patients. Primary endpoints for the study include disease control rate at 16 weeks, and best overall response rates at 16 weeks; secondary endpoints include safety and tolerability, changes in the serum biomarker of HCC (alpha-fetoprotein) from pretreatment levels, duration of response and progression-free/overall survival. The study is projected to be complete in 2014.
DRUG INTERACTIONS
To date, there have been no studies of drug interactions for SGI-110. Overall, the rapid clearance from plasma, lack of cytochrome P450 inhibition, and the metabolism of the drug both by hydrolysis of the active metabolite, as well as by cytidine deaminase, suggest that, like decitabine and azacitidine, SGI-110 is unlikely to have significant drug interactions.
CONCLUSIONS
The currently available data support the clinical development of SGI-110. Presently, the data from both preclinical and clinical trials suggest that this drug has significant activity in myeloid malignancy. The drug appears to have a favorable safety profile and demonstrates impressive evidence of response, even in heavily pretreated MDS and AML patients who have failed first-generation HMAs. The improved pharmacokinetic profile of SGI-110 relative to decitabine suggests that toxicity with this agent may be considerably less, while providing for a potentially longer exposure period. Given that the putative mechanism for HMAs depends upon DNA incorporation, and furthermore, that the previously studied HMAs have significantly shorter half-lives in humans than SGI-110, it seems reasonable to test this drug in the context of solid tumors that might not grow with sufficient rapidity to be susceptible to first-generation HMAs.
ACKNOWLEDGMENTS
DISCLOSURES
E.A. Griffiths and A.R. Karpf have received research support from Astex Pharmaceuticals, Inc.; E.A. Griffiths is currently a principal investigator on the phase I/II MDS/AML clinical trial of SGI-110 and has served on advisory boards for Alexion Pharmaceuticals and Celgene, Inc. G. Choy, P. Taverna, M. Azab and S. Redkar are employees of Astex Pharmaceuticals, Inc.
Footnotes
Synthesis prepared by J. Bolòs, R. Castañer. Thomson Reuters, Provença 398, 08025 Barcelona, Spain.
SOURCE
Astex Pharmaceuticals, Inc. (US).
REFERENCES
- 1.Redkar S. Scaleup and development of a process for a low-volume subcutaneous formulation of SGI-110, a potent hypomethylating agent. 103rd Annu Meet Am Assoc Cancer Res (AACR); March 31–April 4; Chicago. 2012. Abst. [Google Scholar]
- 2.Phiasivongsa P, Redkar S. CA 2623090, CN 101282986, EP 1943264, JP 2009509531, KR 2008059612, US 2007072796, US 7700567, WO 2007041071. Astex Pharmaceuticals, Inc.; Oligonucleotide analogues incorporating 5-aza-cytosine therein.
- 3.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 5.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153(4):773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylin SB, Esteller M, Rountree MR, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 7.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG. Epigenetic changes in cancer and preneoplasia. Cold Spring Harb Symp Quant Biol. 2005;70:329–333. doi: 10.1101/sqb.2005.70.036. [DOI] [PubMed] [Google Scholar]
- 11.Grant S, Bhalla K, Gleyzer M. Effect of uridine on response of 5-azacytidine-resistant human leukemic cells to inhibitors of de novo pyrimidine synthesis. Cancer Res. 1984;44(12, Pt. 1):5505–5510. [PubMed] [Google Scholar]
- 12.Stegmann AP, Honders MW, Willemze R, Landegent JE. De novo induced mutations in the deoxycytidine kinase (dck) gene in rat leukemic clonal cell lines confer resistance to cytarabine (AraC) and 5-aza-2’-deoxycytidine (DAC) Leukemia. 1995;9(6):1032–1038. [PubMed] [Google Scholar]
- 13.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975;64(7):1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- 14.Lin KT, Momparler RL, Rivard GE. High-performance liquid chromatographic analysis of chemical stability of 5-aza-2’-deoxycytidine. J Pharm Sci. 1981;70(11):1228–1232. doi: 10.1002/jps.2600701112. [DOI] [PubMed] [Google Scholar]
- 15.Tomankova H, Zyka J. Study of the time dependence of the stability of 5-aza-2’deoxycytidine in acid medium. Microchem J. 1980;25(3):281–288. [Google Scholar]
- 16.Momparler RL. Pharmacology of 5-aza-2’-deoxycytidine (decitabine) Semin Hematol. 2005;42(3) Suppl. 2:S9–S16. doi: 10.1053/j.seminhematol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123(1):8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 18.van Groeningen CJ, Leyva A, O’Brien AM, et al. Phase I and pharmacokinetic study of 5aza-2’-deoxycytidine (NSC 127716) in cancer patients. Cancer Res. 1986;46(9):4831–4836. [PubMed] [Google Scholar]
- 19.Chabner BA, Drake JC, Johns DG. Deamination of 5-azacytidine by a human leukemia cell cytidine deaminase. Biochem Pharmacol. 1973;22(21):2763–2765. doi: 10.1016/0006-2952(73)90137-8. [DOI] [PubMed] [Google Scholar]
- 20.Chabot GG, Bouchard J, Momparler RL. Kinetics of deamination of 5-aza-2’-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol. 1983;32(7):1327–1328. doi: 10.1016/0006-2952(83)90293-9. [DOI] [PubMed] [Google Scholar]
- 21.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 22.Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 23.Silverman LR, Holland JF, Demakos EP. Azacitidine (Aza C) in myelodysplastic syndromes (MDS), CALGB studies 8421 and 8921. Ann Hematol. 1994;68:A12. [Google Scholar]
- 24.Silverman LR, Holland JF, Ellison RR. Low dose continuous infusion azacytidine is an effective therapy for patients with myelodysplastic syndromes, a study of Cancer and Leukemia Group B. J Cancer Res Clin Oncol. 1990;116(Suppl.):816. [Google Scholar]
- 25.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubbert M, Wijermans P, Kunzmann R, et al. Cytogenetic responses in high-risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5aza-2’-deoxycytidine. Br J Haematol. 2001;114(2):349–357. doi: 10.1046/j.1365-2141.2001.02933.x. [DOI] [PubMed] [Google Scholar]
- 27.Wiley JS, Jones SP, Sawyer WH, et al. Cytosine arabinoside influx and nucleoside transport sites in acute leukemia. J Clin Invest. 1982;69(2):479–489. doi: 10.1172/JCI110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2’-deoxycytidine. J Biol Chem. 1982;257(4):2041–2048. [PubMed] [Google Scholar]
- 29.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 30.Khan R, Aggerholm A, Hokland P, Hassan M, Hellström-Lindberg E. A pharmacodynamic study of 5-azacytidine in the P39 cell line. Exp Hematol. 2006;34(1):35–43. doi: 10.1016/j.exphem.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Marcucci G, Byrd JC, Grever M, Xiao J, Chan KK. Characterization of decomposition products and preclinical and low dose clinical pharmacokinetics of decitabine (5-aza-2’-deoxycytidine) by a new liquid hromatography/tandem mass spectrometry quantification method. Rapid Commun Mass Spectrom. 2006;20(7):1117–1126. doi: 10.1002/rcm.2423. [DOI] [PubMed] [Google Scholar]
- 32.Kiianitsa K, Maizels N. A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 2013;41(9):e104. doi: 10.1093/nar/gkt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths E, Srivastava P, Collamat-Lai G, et al. SGI-110, a novel 5-aza-2’deoxycytidine containing dinucleotide, induces cancer germline (CG) antigen expression in acute myeloid leukemia cells. 104th Annu Meet Am Soc Cancer Res (AACR); April 6–10; Washington, D.C.. 2013. Abst 681. [Google Scholar]
- 34.Taverna P, Scholl J, Shi C, et al. SGI-110, a novel subcutaneous (SQ) second generation DNA hypomethylating agent achieves improved pharmacodynamics (PD), safety and pharmacokinetics (PK) in comparison to IV decitabine in a non-human primate in vivo study. 103rd Annu Meet Am Soc Cancer Res (AACR); March 31–April 4; Chicago. 2012. Abst 4076. [Google Scholar]
- 35.Scholl J, Vollmer D, Bearss J, et al. In vivo activity of SGI-110, a novel hypomethylating agent for treatment in hematology and solid malignancies. AACI-NCI-EORTC Int Conf Mol Targets Cancer Ther; Nov 16–19; Boston. 2009. Abst A190. [Google Scholar]
- 36.Kantarjian H, Roboz G, Rizzieri D, et al. Results from the dose escalation phase of a randomized phase 1–2 first-in-human (FIH) study of SGI-110, a novel low volume stable subcutaneous (SQ) second generation hypomethylating agent (HMA) in patients with relapsed/refractory MDS or AML. 54th Annu Meet Am Soc Hematol; Dec 8–11; Atlanta. 2012. Abst 414. [Google Scholar]
- 37.Yoo CB, Jeong S, Egger G, et al. Delivery of 5-aza-2’-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67(13):6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 38.Matei D, Fang F, Shen C, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang F, Balch C, Schilder J, et al. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116(17):4043–4053. doi: 10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balch C, Huang TH, Brown R, et al. The epigenetics of ovarian cancer drug resistance and resensitization. Am J obstet Gynecol. 2004;191(5):1552–1572. doi: 10.1016/j.ajog.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Matei D, Taverna P, Miller D, et al. Chemosensitizing effects of the novel, small molecule DNA methylation inhibitor SGI-110 in ovarian cancer. 103rd Annu Meet Am Assoc Cancer Res (AACR); March 31–April 4; Chicago. 2012. Abst 4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jueliger S, Lyons J, Azab M, et al. SGI-110, a novel second generation DNA hypomethylating agent, enhances sorafenib activity and alters the methylation signature of HCC cell lines. Eur J Cancer; 24th EORTC-NCI-AACR Symp Mol Targets Cancer Ther; Nov 6–9; Dublin. 2012. 2012, Abst 465. [Google Scholar]
- 43.Fratta E, Coral S, Covre A, et al. The biology of cancer testis antigens: Putative function, regulation and therapeutic potential. Mol Oncol. 2011;5(2):164–182. doi: 10.1016/j.molonc.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coral S, Parisi G, Nicolay H, et al. Immunomodulatory activity of SGI-110, a 5-aza-2’-deoxycytidine-containing demethylating dinucleotide. Cancer Immunol Immunother. 2013;62(3):605–614. doi: 10.1007/s00262-012-1365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]