Abstract
Alternative cleavage and polyadenylation (APA) can diversify coding and non-coding regions, but has particular impact on increasing 3′ UTR diversity. Through the gain or loss of regulatory elements such as RNA binding protein and microRNA sites, APA can influence transcript stability, localization, and translational efficiency. Strikingly, the central nervous systems of invertebrate and vertebrate species express a broad range of transcript isoforms bearing extended 3′ UTRs. The molecular mechanism that permits proximal 3′ end bypass in neurons is mysterious, and only beginning to be elucidated. This landscape of neural 3′ UTR extensions, many reaching unprecedented lengths, may help service the unique post-transcriptional regulatory needs of neurons. A combination of approaches, including transcriptome-wide profiling, genetic screening to identify APA factors, biochemical dissection of alternative 3′ end formation, and manipulation of individual neural APA targets, will be necessary to gain fuller perspectives on the mechanism and biology of neural-specific 3′ UTR lengthening.
Introduction
The 3′ untranslated regions (3′ UTRs) of mRNAs coordinate post-transcriptional control via diverse trans-acting factors, including RNA-binding proteins (RBPs) and microRNAs (miRNAs). Genome-wide analyses of mRNA 3′ termini reveal that most genes undergo alternative cleavage and polyadenylation (APA) to yield multiple 3′ UTRs. Notably, 3′ UTR diversity was recently recognized to be particularly widespread in the central nervous system (CNS) of Drosophila and mammals. Neurons are well-recognized to comprise exceptional cell diversity and profound needs for post-transcriptional gene regulation. Thus, the unique architecture and functionality of neurons suggest that neural APA has implications for mRNA transport, mRNA localization, and localized translation. We review current progress and outline future prospects towards elucidating this largely mysterious process that substantially diversifies the neural transcriptome, may influence brain development and neuronal plasticity, and whose deregulation may contribute to neural dysfunction.
Formation of 3′ ends and usage of alternative polyadenylation sites
The 3′ termini of most eukaryotic pre-mRNAs (replication-dependent histone mRNAs being an exception) undergo endonucleolytic cleavage and polyadenylation (CP). The resulting untemplated polyadenosine (pA) tail is required for stability, export and translation of the mature mRNA. CP is mediated by sequence elements such as the polyadenylation signal (PAS), canonically defined as an AAUAAA hexamer located ~10–30 nt upstream of the cleavage site, and the downstream sequence element (DSE), a U/GU-rich region downstream of the cleavage site [1]. The PAS is recognized by the cleavage and polyadenylation specificity factor (CPSF) complex, while the DSE is bound by the cleavage stimulation factor (CstF) complex (Figure 1). The core CP machinery includes many other factors [2,3], such as the 68 kDa subunit of the mammalian cleavage factor I (CFI) that associates with UGUA motifs upstream of the cleavage site [4]. Additional auxiliary elements can influence CP, especially in the context of polyA sites that lack the canonical PAS [5]. CP is executed co-transcriptionally, and connected to other processes mediated by RNA polymerase II [6].
Figure 1. The cleavage and polyadenylation (CP) machinery at the polyA site.
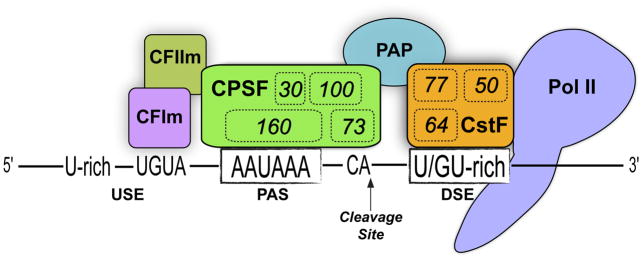
A multiprotein complex cleaves the mRNA 3′ terminus and adds an untemplated polyA tail. RNA polymerase II (Pol II) recruits polyA factors and enhances the cleavage reaction, and is thus considered part of the CP machinery. The cleavage and polyadenylation specificity factor (CPSF) complex recognizes the polyadenylation signal (PAS); CPSF160 binds the AAUAAA signal and CPSF executes endonucleolytic cleavage, preferably at CA dinucleotides. The Cleavage stimulation factor (CstF) complex aids CP by recognizing the downstream sequence element (DSE) via the CstF-64 subunit. Other numbers within complexes correspond to the names of additional factors. Other auxiliary polyA factors have been omitted for clarity, as have other physical connections that the CP machinery has with the transcription complex and splicing factors. USE, upstream sequence element; CFIm, Cleavage factor Im; PAP, polyA polymerase.
The CP machinery has the capacity to cleave and polyadenylate at more than one site at the distal end of a gene, a process referred to as alternative polyadenylation (APA). Depending on the configuration of events, this process yields mRNAs with alternative terminal exons or tandem 3′ UTRs of variable length. Alteration of coding exons by APA can increase protein diversity, analogous to the action of alternative splicing. However, the majority of APA events occur within 3′ UTRs, and thus have consequences for post-transcriptional control.
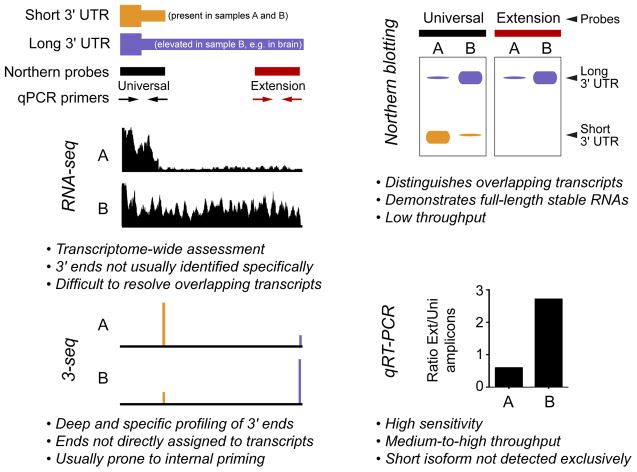
Individual cases of genes with multiple functional polyA sites have been known for some time (i.e., by Northern blotting or RT-PCR, Figure 2), but only more recently has APA been recognized to operate broadly across the transcriptome. Initial cDNA analyses estimated that >50% of human genes are subject to APA [7]. This ballpark figure has held up with deeper and more detailed analyses afforded by RNA-sequencing (RNA-seq) and procedures that specifically capture mRNA 3′ termini (3′-seq, Figure 2). Such methods demonstrate that APA broadly diversifies the transcriptomes of fungi, plants, and diverse animal species [8,9].
Figure 2. Experimental approaches to study alternative polyadenylation.
The schematics at top left depict a model gene that expresses short (orange) and long (purple) 3′ UTR isoforms, of which the latter is preferentially expressed in sample B and not in sample A. This pattern is representative of many neural-restricted 3′ UTR extensions of genes that are expressed more broadly. These APA isoforms can be analyzed at the level of individual genes using Northern probes or RT-PCR primers directed at proximal and distal regions of the 3′ UTR; the former is “universal” since it detects both isoforms, whereas the “extension” reagents specifically recognize the long 3′ UTR. The right panels illustrate model data for Northern and qPCR measurements of APA isoforms. The middle and bottom left panels illustrate genomewide approaches, and how model data appear at specific genomic loci. RNA-seq data provides an overview of the transcriptome as reconstructed from short overlapping reads, whereas 3′-seq methods are designed to specifically capture and sequence the polyadenylated 3′ termini of mRNAs.
The 3′ UTR is a central hub of post-transcriptional regulation
3′ UTRs comprise a dominant binding location for many RNA-binding proteins (RBPs), which collectively influence mRNA stability, localization, and translational efficiency (Figure 3). RBPs can recognize relatively specific motifs, more degenerate sites, and/or secondary structures. Some RBPs contribute positively to gene activity. For example, mRNA translation can be enhanced by Hu proteins binding AU-rich elements [10], and Cytoplasmic Polyadenylation Element Binding (CPEB) protein binding U-rich elements [11]. Other 3′ UTR-interacting RBPs act as negative regulators (Figure 3). For instance, AUF1 [12] and KSRP [13] induce mRNA decay via AU-rich elements, while other RBPs mediate translational inhibition [14,15]. Factors that promote or inhibit mRNA stability and translation can compete for the same sequence elements, resulting in dynamic post-transcriptional control mechanisms [16,17].
Figure 3. Post-transcriptional sequence elements located in 3.
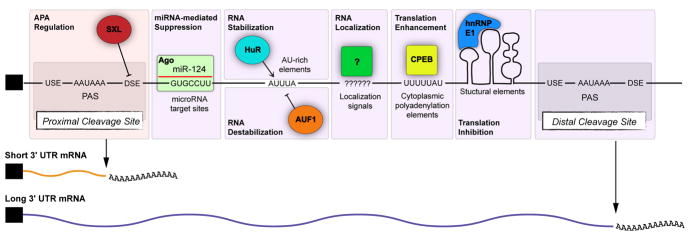
′ UTRs.
Sequence elements located in the 3′ UTR are targets for multiple levels of post-transcriptional control. The RBP SXL binds to GU-rich DSEs to block cleavage at proximal sites, and favors generation of long APA isoforms for the e(r) gene in the Drosophila ovary. miRNAs recruit Argonaute complexes to complementary sites, mostly in 3′ UTRs, leading to decay and/or translational suppression of target transcripts. AU-rich binding proteins such as HuR and AUF1 compete for AU-rich elements to differentially regulate mRNA stability. Diverse sequence elements and structures can direct mRNA localization to subcellular regions, such as dendrites and axons. CPEB binding to targets results in cytoplasmic polyadenylation. In addition to primary sequences, secondary structures are important for binding to certain RBPs; hnRNPE1 binds a structural motif to mediate translation suppression.
Argonaute proteins are a special RBP class whose association with target transcripts is guided by short RNAs known as microRNAs (miRNAs). These ~21–24 nt RNAs recruit Argonaute complexes to binding sites, typically Watson-Crick matches within 3′ UTRs to nucleotides 2–8 of the miRNA [18,19], to induce target degradation and/or translational suppression. Argonaute/miRNA complexes have extensive impact on post-transcriptional networks, as computational and experimental approaches indicate that individual conserved miRNAs frequently have hundreds of functional targets [20].
Altogether, diverse trans-acting factors can mediate complex regulatory transactions via 3′ UTRs. Against this backdrop, APA diversifies 3′ UTR isoforms that can be subject to, or can escape from, these regulatory events.
Tissue-specific APA: The extreme case of the nervous system
Recent years have witnessed a growing appreciation for how 3′ UTR lengths can be globally modulated according to cell state, a process that can render 3′ UTR-mediated regulation conditional. For example, cell proliferation has been associated with 3′ UTR shortening in various cell types [21–23]. Characteristic differences in 3′ UTR lengths are also observed in several tissues. For example, mammalian and invertebrate testes utilize many shorter 3′ UTRs compared to other tissues, whereas the central nervous system is biased towards longer 3′ UTRs [24–28].
In fact, individual neural-specific 3′ UTR lengthening events were documented by Northern blotting over 20 years ago [29,30]. Subsequent analyses of cDNA data revealed that brains were biased to utilize distal PAS, resulting in longer 3′ UTRs in mice and humans [31]. More recently, deep RNA-seq data from multiple human (Illumina BodyMap 2.0) and mouse tissues [32] showed that amongst broadly expressed genes, the predominant isoforms in brain favor distal PAS [26]. This trend holds up in comparisons with other terminally differentiated tissues, such as skeletal and cardiac muscle, supporting the notion that brains exhibit 3′ UTR lengthening beyond what might be a consequence of differentiation status alone.
In the wake of the most recent state-of-the-art annotations of the Drosophila and human genomes [33,34], one might have considered the extent of their mRNA-encoding transcriptomes to be essentially complete. However, recent studies showed that extended 3′ UTR isoforms were vastly underestimated in these species. Using stringent annotation criteria, many hundreds of novel neural 3′ UTR extensions were identified in the Drosophila CNS [25] as well as in mouse and human brains [26]. These collectively exhibited hallmarks of bona fide 3′ UTRs, including enrichment of canonical PAS, DSE, and 3′-seq tags, and included of thousands of conserved miRNA binding sites [25,26]. Other studies of APA isoforms amongst human and zebrafish tissues using 3′-seq technologies similarly observed enhanced expression of distal 3′ UTR isoforms in the nervous system [35,36].
The breadth and magnitude of neural 3′ UTR lengthening are remarkable. A number of validated 3′ UTRs ranged from 10–20 kb, and comprise some of the longest stable 3′ UTRs demonstrated in flies and mammals. Extensive experimental validations (See Box 1 and Figure 2) were performed to confirm that these annotations truly represent continuous mature transcripts, as opposed to unstable RNA fragments or perhaps downstream non-coding RNAs [25,26].
Box 1. Challenges for transcriptome-wide annotation and profiling of 3′ UTRs.
RNA-seq data are excellent for distinguishing alternative transcripts that differ locally (for instance due to alternative promoters, splicing, and/or 3′ UTRs), but using short reads to infer how all these variations relate to whole intact transcripts is challenging. In practice, most algorithms for transcript assembly prefer to generate a concise set of transcript models, as opposed to elaborating every possible alternate transcript that might theoretically be built from a given dataset [94,95]. Accurate transcriptome assembly is also confounded by biased representation of reads, due to library preparation, RNA secondary structure, and inherent properties of polymerases used in library preparation or sequencing.
The existence of widespread neural 3′ UTR extensions, which are necessarily represented by fewer reads than companion shorter isoforms, present a new challenge for transcriptome assembly. Such long transcripts are particularly prone to sequence gaps, whether due to lower coverage, presence of repeat elements, or even shearing during mRNA isolation. In any case, most transcript assemblers are not predicated on the notion that there should exist continuous exons that are tens of kb in length, and attempts to appropriately annotate such 3′ UTR extensions are likely to inappropriately merge many other exons and genes genome-wide. Conversely, the incomplete appreciation of full-length 3′ UTRs comes at a cost of erroneously annotating some 3′ UTR extensions as independent non-coding RNAs [26].
While connectivity of transcribed regions is best experimentally verified using Northern analysis of individual genes (Figure 2), a complementary transcriptome-wide approach is to infer connectivity by size selecting RNAs prior to library cloning and sequencing [96,97]. By segregating multiple size ranges prior to library preparation it is possible to distinguish between mRNAs bearing long 3′ UTR extensions from shorter, potentially non-coding RNAs.
The limitations of RNA-seq to identify 3′ termini with sufficient precision and depth have been partially addressed using 3′-seq methodologies (Figure 2). Various 3′-seq protocols have been developed (e.g. polyA-seq, PAS-seq, 3P-Seq, 3′READS, A-seq, etc.) [8,9], each with purported advantages for specificity and ease of execution. These techniques, however, do not provide direct information about which transcript isoforms utilize each polyA site. In the future, improved technologies for direct sequencing and long-substrate sequencing [98] may obviate all of these shortcomings associated with current fragmented RNA-seq and 3′-seq methods.
Mechanisms to achieve neural 3′ UTR lengthening
What can explain the large extensions of 3′ UTRs observed in the CNS of a broad range of metazoan species? Little is currently known, and molecular insight is lacking for most of the tissue-specific, state-responsive, and stimulus-dependent global APA trends described thus far [9]. The following sections discuss some possibilities for how neural APA might be achieved.
polyA factor abundance may influence 3′ end choice
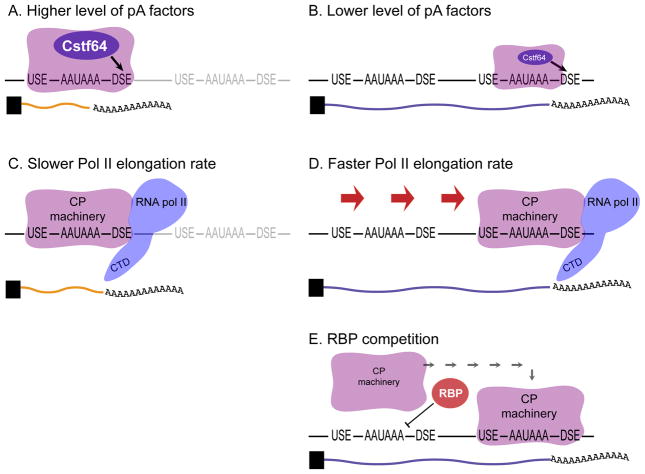
As the proximal polyA site is the first to be encountered by the cleavage machinery during transcription elongation, it is thought that sequence features surrounding this site (such as PAS, DSE) play crucial roles in determining whether cleavage takes place, or alternatively, whether this first polyA site is bypassed. The decision to bypass the first polyA site appears to be influenced by polyA factor abundance. Pioneering studies of IgM heavy chain showed that higher levels of the polyA factor CstF-64 promoted the usage of a “weaker” intronic polyA site in plasma cells, relative to precursor B cells [37,38]. In principle, modulation of polyA factor abundance might represent a more general mechanism for APA (Figure 4A, B).
Figure 4. General mechanisms of APA.
(A) High levels of polyA factors, such as CstF-64, can lead to preferential usage of proximal polyA sites. (B) Lower levels of polyA factors may cause bypass of proximal sites and favor distal polyA site usage. (C) The CP machinery is associated with the carboxy-terminal domain (CTD) of RNA pol II. Slower transcriptional elongation may favor cleavage at more proximal polyA sites, whereas faster elongation rate may promote distal site usage (D). (E) RNA-binding proteins (RBPs) are thought to promote 3′ UTR lengthening by competing with the cleavage and polyadenylation (CP) machinery for cis-elements around proximal polyA sites.
Several studies report that 3′ UTR length is negatively correlated with the abundance of polyA factors in diverse cell types, which could potentially explain the abundance of extended 3′UTRs in the nervous system [22,31,35,39]. However, experimental evidence that polyA factor abundance might globally affect 3′ UTR lengths has been mixed. Reduction of CstF-64 levels did not impart widespread bias toward more proximal APA events [4,40]. Knockdown of another canonical polyA factor, CstF-77, altered tandem 3′ UTR events, but similar frequencies of lengthening and shortening were observed [41]. In contrast, decreasing CFIm68 levels had a broad effect on the transcriptome, in which a majority of significant changes in length biased toward proximal PAS usage [4].
The aforementioned studies performed knockdown of polyA factors in immortalized cell lines, whose APA patterns are not representative of the more extreme 3′ UTR variations observed in tissues such as testis and brain. Therefore, future studies should address if alteration of polyA factor abundance can affect neural APA in bona fide neurons or in animal models.
RNA polymerase II activity may affect APA
The pursuit of APA mechanism might be guided by work in alternative splicing (AS), which is also particularly active in neural tissue. AS can be driven by positive recruitment of splicing machinery or competitive blocking of splice sites by RBPs. However, additional proposed mechanisms take into account that splicing is a dynamic process that is kinetically influenced by transcription elongation rate, chromatin structure and histone modifications [42]. In this regard, an RNA Pol II mutation that reduces elongation rate was linked to preferential usage of proximal polyA sites for several genes in Drosophila [43] (Figure 3C, D). Similarly, in mammalian cells, a positive correlation between transcription levels and proximal polyA site usage has been observed [44]. It remains to be tested whether neurons display altered transcription elongation rates compared to other cell types. Biased nucleosome positioning has also been identified to occur around human PAS [45], suggesting that chromatin status might also influence the usage of alternative polyA sites. Future work should address how these kinetic processes might influence neural specific 3′ UTR lengthening.
Hu proteins regulate neural APA
Perhaps the most well appreciated mechanism of APA regulation involves the competition of RBPs with polyA factors for sequence elements surrounding polyA sites (Figure 4E). Such a model is illustrated by Drosophila Sex-Lethal (SXL), a critical player in sex-determination that regulates polyA site choice in the female germline by competing with CstF-64 for binding to DSE motifs [46]. SXL blocks access to proximal DSEs, thereby promoting bypass of the proximal site and usage of a more distal site (Figure 3). Nuclear polyA binding protein 1 (PABPN1) appears also to regulate APA through such a competition mechanism. PABPN1 binds in the vicinity of proximal polyA sites to suppress proximal PAS usage for >500 genes in cultured human cells [47].
An RBP competition model was recently proposed to explain how the Drosophila neural-specific Hu family protein Embryonic lethal abnormal vision (Elav) promotes neural 3′ UTR lengthening in Drosophila [48]. In situ hybridization and quantitative PCR (qPCR) assays showed that elav mutant embryos lacked CNS-specific 3′ UTR extensions for several genes, while shorter 3′ UTR isoforms remained expressed. Reciprocally, ectopic Elav could induce long neural 3′ UTRs, and direct tethering of Elav in the vicinity of a proximal PAS could promote its bypass. Together, these tests support a model that Elav promotes neural 3′ UTR extension by opposing the usage of proximal PAS [48]. Still, it is worth noting that Elav is expressed pan-neuronally (i.e. equally in both CNS and PNS), but most spatially documented neural 3′ UTR extensions were specific to CNS [25,27]. It is not clear why endogenous Elav does not promote neural 3′ UTR lengthening in the PNS.
Mammalian Hu proteins are well-known to regulate RNA stability in the cytoplasm, but they can also affect 3′ end processing in the nucleus [49]. HuB, HuC and HuD are neuronal whereas HuR is ubiquitously expressed. Interestingly, HuR expresses a longer 3′ UTR preferentially in neurons, while HuB, HuC and HuD can each increase expression of the long HuR 3′ UTR [50]. Furthermore, HuR auto-regulates its own expression via APA in non-neuronal cells [51]. Collectively, these studies suggest that Elav/Hu proteins might promote the accumulation of lengthened 3′ UTRs in neurons through multiple mechanisms, including 3′ end processing in the nucleus and mRNA stabilization in the cytoplasm [10]. A future challenge for studies of Elav/Hu proteins in neurons will be to distinguish the contributions of distinct nuclear and cytoplasmic processing events.
Biological consequences of neural APA
It is clear that APA operates broadly to extend 3′ UTRs across the neural transcriptome, and that neurons have exceptional needs for post-transcriptional control. On these bases, one might assume that failure to implement neural APA should incur substantial phenotypes. However, only in a few cases has this notion been rigorously tested. Nevertheless, these studies provide good reason to believe that broader assessment of the biological impact of neural APA is worth pursuing.
Neural APA can influence miRNA-mediated regulation
At a minimum, many hundreds of genes in Drosophila, mouse and human utilize longer 3′ UTRs specifically in the nervous system. De novo searches for conserved motifs in these 3′ UTR extensions reveal that many of the top hits to correspond to miRNA binding sites [25,26]. Notable enrichment is observed for sites for several neural miRNAs, such as miR-124, miR-137, miR-9, miR-96 and let-7 [26], and inspection of cross-linking and immunoprecipitation (CLIP)-sequencing of Ago complexes from mouse brain [52] shows that many such sites that lay in previously unannotated genomic space are functionally bound [26]. Together, these data suggest that one rationale for neural APA is to bring many broadly-expressed transcripts under distinctive miRNA control in neurons, especially by miRNAs that are abundant in neural tissue.
Only in a handful of cases has the differential impact of miRNAs on neural APA isoforms been studied. One study investigated the requirement of miRNAs in the let-7-Complex (let-7C) for appropriate temporal generation of neural subtypes in the Drosophila mushroom body (MB), an important center for learning and memory. The control of MB lineage development by let-7C was substantially accounted for by derepression of the transcription factor Chinmo [53]. Chinmo is directly repressed by the let-7 and miR-125 members of the let-7C [53]. Enabled by new 3′UTR annotations [25], it was shown that most of the functional target sites for these miRNAs were located in an ~6 kb extension of the chinmo 3′ UTR.
In another example from Drosophila, iab-4/8 miRNAs from the Bithorax-Complex (BX-C) were observed to regulate the homeobox BX-C genes Ultrabithorax and abdominal-A [54–56]. Hox genes are conserved master regulators of segmental identities along the anterior-posterior axis of all animals, and these fly studies provided some of the first evidence that Hox miRNAs could functionally regulate Hox gene expression in vivo. Some of the conserved binding sites for iab-4/8 miRNAs are located in 3′ UTR extensions [54], and it was later shown that 3′ UTRs of BX-C genes are specifically lengthened in the central nervous system as a mechanism for tissue-specific isoform regulation [57].
Misregulation of the mammalian microtubule-associated protein Tau results in insoluble tangles that can underlie Alzheimer’s disease and other tauopathies. miR-34a was recently found to regulate endogenous Tau in neuroblastoma cell lines, and this regulation is mediated by a miR-34 target site located in the extended 3′ UTR [58]. Curiously, reporter studies showed that miR-34 targeting of the extended 3′ UTR of alpha-synuclein increased its activity, in contrast to the more established role of miRNAs as repressors [59]. Similar to Tau, alpha-synuclein also accumulates in insoluble bodies in several neurological disorders, including Parkinson’s disease. These studies expand the biology of miR-34 to neural 3′ UTR extensions, and also provide plausibility of the relevance of neural APA to neurological syndromes. Relevant to this notion, the epigenetic factor methyl CpG-binding protein 2 (MeCP2) is mutated in Rett syndrome and other neurological disorders, and is subject to neural APA. Interestingly, a recent study demonstrated human-specific repression of the long 3′ UTR MeCP2 isoform by miR-483-5p, and that such regulation has impact on neuronal morphology [60].
These precedents likely only scratch the surface of neural 3′ UTR lengthening as a means to bring specific transcript isoforms under distinct miRNA control in neurons. Given the many hundreds of examples of conserved miRNA binding sites in 3′ UTR extensions, and abundant evidence for roles of miRNAs in the developing and adult nervous systems [61,62], it seems clear that this direction is ripe for further investigation.
Extended 3′ UTRs mediate RNA localization to axons and dendrites
Neuronal nuclei are typically far removed from locations of protein function in axons, dendrites or synapses. Given such distances, an energetically favorable strategy might be to transport mRNA rather than protein. There is strong experimental support for local protein synthesis in neuronal dendrites, a process with functional importance for learning and memory [63]. Targeting and translation of mRNAs in growth cones and axons is also widespread and is implicated in axon guidance, regeneration and survival [64].
Several studies have uncovered elements within APA-generated 3′ UTR extensions that confer mRNA localization patterns in neurons (Figure 5). Brain-Derived Neurotrophic Factor (BDNF) is subject to APA in neurons, for which the short 3′ UTR (bdnf-S) confers cell body localization, whereas the long 3′ UTR (bdnf-L) enables localization to dendrites. The functional importance of bdnf-L was shown by a mouse mutant that lacked this isoform [65]. In this model, the insertion of tandem polyadenylation signals downstream of the bdnf-S polyA site was used to prevent expression of bdnf-L while allowing expression bdnf-S to continue. Loss of bdnf-L impaired long-term potentiation (LTP) and altered dendritic spine morphology [65]. A subsequent study showed that the bdnf-L mutant exhibited obesity syndrome, and that the 3′ UTR extension is important for translation of BDNF in response to leptin and insulin [66].
Figure 5. Long 3′ UTR APA isoforms that localize to subcellular regions of neurons.
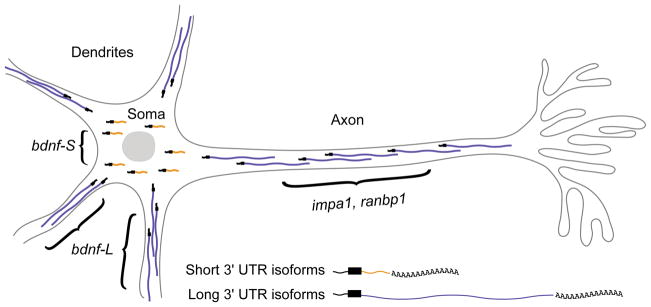
bdnf expresses an extended 3′ UTR isoform that localizes to dendrites, where it undergoes localized translation. In contrast, the short 3′ UTR APA variant of bdnf is restricted to the soma. impa1 and ranbp1 express extended 3′ UTR species that are preferentially localized to axons.
Lengthening of 3′ UTRs can enable conditional targeting of transcripts to axons. Ranbp1 utilizes an extended 3′ UTR variant that provides an axonal localization signal that allows its localized translation [67]. Similarly, an extension of Impa1 confers NGF-dependent axonal localization and axonal translation [68]. A longer 3′ UTR isoform of MKK7 is localized more robustly to growth cones, where it undergoes localized translation and promotes neurite outgrowth [69]. Finally, a long 3′ UTR isoform of importin-β1 harbors localization elements directing expression to axons, where it provides retrograde signals that are utilized in recovery after nerve injury [70].
The trans-acting factors that mediate differential localization of neural APA isoforms mostly remain to be identified, although perhaps hints may be found amongst RBPs that are known to interact with 3′ UTR elements to promote mRNA localization in neurons, such as hnRNP-R [71] and CBF-A [72,73]. G-quadruplex structures in 3′ UTRs have also been documented to drive neurite targeting of PSD95 and camKIIa transcripts [74].
New evidence suggests that extended 3′UTR mRNA isoforms might preferentially accumulate in ribonucleoprotein (RNP) granules- membrane-free cytoplasmic structures composed of RNA and associated RBPs. Translationally silenced mRNAs can be stored in RNP granules and exit to undergo protein synthesis in response to stress [75]. Such granules were recently generated in vitro from cell lysates for the first time [76,77]. Surprisingly, RNA-seq analysis of these granules revealed that they were enriched in transcripts with extended 3′ UTRs. Interestingly, the granule-associated transcripts were enriched in gene ontology categories related to functions/composition of axons, dendrites and synapses. These findings may suggest that extremely long 3′ UTRs could endow mRNAs with distinct physical properties that drive them into neuronal granules.
The relevance of APA to neurological disease
A number of human diseases are associated with disregulation of specific APA isoforms [9]. Since broad neural 3′ UTR lengthening is a property of adult mammalian brains, one may easily hypothesize that defective neural APA might be associated with neurological syndromes. Serotonin function is implicated in anxiety disorder and depression. The serotonin transporter (SERT) has two 3′ UTR APA forms, the longer of which is implicated in anxiolytic effects on behavior [78,79]. Mutation of SNCA, the lpha-synuclein gene, causes a familial form of Parkinson’s disease (PD), and a long 3′ UTR isoform of this gene is associated with PD pathology [59]. A shift to increased usage of the long lpha-synuclein 3′ UTR was also found to occur in response to elevated cytoplasmic dopamine levels and other conditions that invoke oxidative stress.
Disruption in activity-dependent neuronal signaling is thought to occur in many psychiatric disorders. In response to neuronal activity the long (but not short) 3′ UTR of bdnf was found to mediate translational upregulation of BDNF protein [80]. More generally, neuronal activity activates proximal intronic polyA sites across the genome [81]. Further investigation of how neurotransmitter stimulation and activity regulates APA, and the scope of genes affected, seems warranted.
Future directions for elucidating the scope, mechanism, and biology of neural APA
Given the recent appreciation of the breadth of neural APA, much remains to be done to understand all aspects of this phenomenon. In the remainder of this review, we highlight potential areas of future research that are needed to answer fundamental questions regarding the “what”, “how”, and “why” of neural APA? In other words, what is the full catalog of alternative neural 3′ UTRs (see Box 1), how is neural APA achieved, and why is neural APA needed to regulate gene activity?
Cataloging the subcellular distribution of extended 3′ UTRs
As the depth of read coverage afforded with RNA-seq continues to rise, the amount of input RNA required has fallen, even down to single cell levels [82]. The central nervous system (CNS) contains neuronal and non-neuronal populations, including abundant glia. It is unclear how different cell types and subcellular compartments contribute to the 3′ UTR extensions documented in whole brain or dissected brain region samples.
Compartmentalized chambers can be used to purify axons from somata, glial cells and synapses, and microarray analysis of such preparations had identified >300 axonal mRNAs [83]. An alternative method uses microporous filters to fractionate neurites from soma, with which microarray studies identified 80 mRNAs localized to neurites [69]. A recent RNA-seq study estimated 2550 axonal or dendritic localized RNAs [84], and certainly other neural cell compartments could be analyzed. Indeed, it is conceivable that transcriptomes might differ amongst similar compartments of an individual neuron (e.g., between different synapses).
Inter-cell and intra-cell APA trends can be extracted from RNA-seq data, but 3′-seq approaches will be needed to better elucidate and quantify APA-generated transcript isoforms. The combined application of these techniques to purified subcellular regions will uncover the extent to which neurite/axon/dendrite/synaptic localization (Figure 5) is influenced by neural APA.
Does length, per se, matter?
The 3′ UTRs of critical neural genes such as Ntrk3 (encoding the neurotrophin receptor trkC) and Grin2B (encoding the NMDA receptor subunit epsilon-2) have been validated as >15 kb [26]. Although short, conserved regulatory elements are found in these long 3′ UTRs, one wonders if they harbor length-dependent regulatory properties. In this regard, analysis of the conservation in lengths of neural 3′ UTR extensions across species will be informative. Long 3′ extensions might act as scaffolds or molecular sinks that generally recruit RBPs. While such recruitment might be directed in part by specific sequence motifs, many RBPs bind in a more promiscuous fashion. Perhaps RBP sequestration by long 3′ UTRs could regulate protein synthesis at specific subcellular regions (i.e. the extended 3′ UTR of an mRNA exert a non-coding function in trans to influence translation of other nearby mRNAs).
We note that 3′ UTRs tend to harbor A-rich stretches, and longer 3′UTRs will tend to harbor more of these stretches. Perhaps these A-rich regions are utilized endogenously in some fashion, for example by associating with poly-A binding protein (PABP) to stimulate cap-dependent translation, as with normal polyA tail binding. In a similar vein, the recruitment of RBPs could be mediated by repetitive elements, which are highly abundant in vertebrate genomes, including 3′ UTRs. A special class of 3′ UTR repeat occurs particularly with tandem inverted Alu elements – these can promote gene expression when they are bound by Staufen proteins [85] which are known to bind transcripts that are several fold longer than average [86]. Genome-wide approaches to evaluate RNA secondary structure were reported recently [87,88], and it will be interesting to apply such techniques to neurons and their extended 3′ UTR landscape.
Illuminating the mechanism of 3′ UTR extension in neurons
One can imagine perturbations that might affect 3′ UTR choice by indirect or direct means, both of which are relevant to understand. However, factors that directly affect APA are presumably ones that interact with the core CP machinery, or ones that contact 3′ UTRs. General approaches to identify direct APA factors include genetic screening and RNA-affinity chromatography.
A reporter-based cell assay was used to screen hundreds of RNA-binding proteins for modulators of proximal PAS usage [47]. To extend RNAi screening to the neural APA problem, an appropriate model needs to be established. Although many immortalized cell lines have neural character, it remains to be seen whether any broadly execute 3′ UTR lengthening. If not, it may be necessary to utilize primary neurons, or perhaps in vitro differentiated neurons. Another challenge will be to identify appropriate reporters for neural 3′ UTR lengthening. Although episomal reporters linked to 3′ UTRs are convenient, they might not be subject to endogenous features that might impact 3′ end selection, such as chromatin context, transcription rate and splicing events specific to particular gene loci [9]. If so, integrated reporters engineered into large genomic fragments might better recapitulate endogenous APA patterns.
Since neural 3′ UTR lengthening is conserved in Drosophila [25,27] this is an alternative system with potential to identify relevant factors while simultaneously assessing biological relevance. Forward genetic screens can be performed by crossing UAS-RNAi transgene libraries [89,90] to Gal4 drivers. By using neural drivers, one can systematically assay the effects of knockdowns on neural 3′ UTR extensions in embryos or adult heads, either by quantifying endogenous distal 3′ UTR levels or perhaps by utilizing a neural 3′ UTR extension reporter.
A complementary biochemical approach involves RNA-affinity chromatography coupled to mass spectrometry to identify APA regulating factors. This utilizes an RNA “bait” containing a regulatory element(s) from a modulated 3′ UTR extension, which is used to isolate adhering proteins from cell or tissue lysates. This approach was used to identify CP regulating proteins that target a defined upstream sequence element (USE) important for the proximal prothrombin polyA site [91]. Applying this approach to understanding neural APA could be particularly useful if common and conserved elements that enhance “neural” 3′ end formation are identified.
Assessing the biological functions of neural APA
To gain a deeper appreciation of neural 3′ UTR lengthening, it will be imperative to elucidate its impact on the nervous system, with respect to cell specification, differentiation, electrophysiology, and/or behavior. Since neural APA (1) affects at least many hundreds of genes, (2) is a process that exhibits exquisite tissue-specificity and utilizes conserved PAS, and (3) often generates extremely long 3′ UTRs bearing many conserved regulatory sites, including for miRNAs and RBPs, it is easy to infer that this process exists for a reason. On the other hand, it might still be the case that neural APA is an evolutionary conserved phenomenon that occurs, but does not impart substantial impact on neurobiology. These possibilities need to be tested using functional assays and, in particular, genetics.
In vitro systems using cultured neurons may be useful to dissect the molecular function of the transcript isoforms. As shown in studies of the β-actin 3′ UTR, GFP reporters bearing a myristoylation signal can be used to detect local translation at distal cellular sites [92]. Neural APA reporters that utilize extended 3′ UTRs may be useful to assess their impact on localized protein expression, as well as to assess variants with modified PAS or partial 3′ UTR regions.
Perhaps more importantly, it will be necessary to study the function of neural APA in vivo. To date, functional consequences of abrogating usage of an endogenous 3′ UTR extension have only been demonstrated to date for bdnf [65]. An impediment to manipulating endogenous 3′ UTRs in intact animals, such as flies or mice, has traditionally been the considerable requirements of time and effort using conventional gene targeting approaches. Recent advances in CRISPR/Cas9-mediated genome engineering [93] now make these feasible to consider on a large scale. The future should see broader experimentation to assess the consequences of deleting long 3′ UTR neural APA isoforms, or alternatively to force the usage of these extensions in non-neuronal tissues.
Conclusions and Outlook
The tremendous cell diversity, unique architecture and dynamic physiology of neurons have evolved in concert with complex regulatory mechanisms that permit precise and responsive control of gene function and activity. Amongst diverse tissues analyzed, the brain exhibits the greatest degree of alternative splicing and the broadest 3′ UTR real estate, and these figure prominently into post-transcriptional mechanisms utilized in neurons. However, as detailed in this perspective, our appreciation of the breadth, mechanism, and biology of neural APA is still only its infancy.
Still much remains to be learned from descriptive studies of 3′ UTR annotation and their alternative usage. Beyond simply sequencing brain samples more deeply, the analysis of different brain regions and cell types, and especially from within different subcellular compartments, should prove revealing. However, we especially look forward to new directions that elucidate the mechanism and biological relevance of neural APA. By comparison with most studies on molecular strategies of 3′ end formation, understanding how neural APA in implemented cannot rely upon convenient models of immortalized cultured cells. Instead, manipulations will need to be done within intact nervous systems, or perhaps within bona fide differentiated neurons. The same applies to efforts to reveal the functional impact, and biological necessity of neural 3′ UTR extensions. These endeavors will be challenging, but will ultimately contribute fundamental concepts on strategies and usages of post-transcriptional gene regulation. Moreover, they should be of broad relevance to the etiology and – it is to be hoped – amelioration of neurological disease.
Acknowledgments
P.M. was supported by a fellowship from the Canadian Institutes of Health Research. S.S. was supported by the Tri-Institutional Training Program in Computational Biology and Medicine. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund and the National Institute of General Medical Sciences of the National Institutes of Health (R01-NS074037).
Abbreviations
- RBP
RNA-binding protein
- polyA
polyadenylation
- PAS
cleavage and polyadenylation signal
- CP
Cleavage and Polyadenylation
- APA
Alternative Cleavage and Polyadenylation
- 3′ UTR
3′ untranslated region
- DSE
downstream sequence element
- CLIP
Crosslinking and Immunoprecipitation
- CPEB
cytoplasmic polyadenylation element binding
- CNS
central nervous system
- CstF
cleavage stimulation factor
- CPSF
cleavage and polyadenylation specificity factor
- microRNA
miRNA
- RT-PCR
reverse transcription polymerase chain reaction
- qPCR
quantitative PCR
References
- 1.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & development. 1997;11:2755–66. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic acids research. 2010;38:2757–74. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cellular and molecular life sciences: CMLS. 2008;65:1099–122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell reports. 2012;1:753–63. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. The EMBO journal. 2010;29:1523–36. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Molecular cell. 2009;36:178–91. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic acids research. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–17. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews Genetics. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 10.Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA. 2013;19:1019–37. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA. 2013;4:437–61. doi: 10.1002/wrna.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochimica et biophysica acta. 2013;1829:680–8. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briata P, Chen CY, Ramos A, Gherzi R. Functional and molecular insights into KSRP function in mRNA decay. Biochimica et biophysica acta. 2013;1829:689–94. doi: 10.1016/j.bbagrm.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Molecular cell. 2011;41:419–31. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia J, Yao P, Arif A, Fox PL. Regulation and dysregulation of 3′UTR-mediated translational control. Curr Opin Genet Dev. 2013;23:29–34. doi: 10.1016/j.gde.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nature structural & molecular biology. 2007;14:511–8. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 17.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, et al. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature genetics. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome biology. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smibert P, Miura P, Westholm JO, Shenker S, May G, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell reports. 2012;1:277–89. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome research. 2013;23:812–25. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, et al. Neural-specific elongation of 3′ UTRs during Drosophila development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15864–9. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, et al. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic acids research. 2007;35:234–46. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Loomis PA, Zinkowski RP, Binder LI. A novel tau transcript in cultured human neuroblastoma cells expressing nuclear tau. The Journal of cell biology. 1993;121:257–67. doi: 10.1083/jcb.121.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coy JF, Sedlacek Z, Bachner D, Delius H, Poustka A. A complex pattern of evolutionary conservation and alternative polyadenylation within the long 3′-untranslated region of the methyl-CpG-binding protein 2 gene (MeCP2) suggests a regulatory role in gene expression. Human molecular genetics. 1999;8:1253–62. doi: 10.1093/hmg/8.7.1253. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome biology. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, et al. Extensive alternative polyadenylation during zebrafish development. Genome research. 2012;22:2054–66. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes & development. 2013;27:2380–96. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Molecular cell. 1998;2:761–71. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 38.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–52. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 39.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PloS one. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao C, Biesinger J, Wan J, Weng L, Xing Y, et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18773–8. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo W, Ji Z, Pan Z, You B, Hoque M, et al. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS genetics. 2013;9:e1003613. doi: 10.1371/journal.pgen.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends in genetics: TIG. 2013;29:442–8. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. The EMBO journal. 2011;30:2431–44. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji Z, Luo W, Li W, Hoque M, Pan Z, et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Molecular cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gawande B, Robida MD, Rahn A, Singh R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. The EMBO journal. 2006;25:1263–72. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, et al. The poly(a)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–53. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 48.Hilgers V, Lemke SB, Levine M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes & development. 2012;26:2259–64. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. The Journal of biological chemistry. 2007;282:2203–10. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 50.Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic acids research. 2012;40:2734–46. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai W, Zhang G, Makeyev EV. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic acids research. 2012;40:787–800. doi: 10.1093/nar/gkr783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu YC, Chen CH, Mercer A, Sokol NS. let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Developmental cell. 2012;23:202–9. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes & development. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, et al. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes & development. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes & development. 2008;22:14–9. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–60. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- 58.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. Journal of neurochemistry. 2013 doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, et al. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson’s disease pathology. Nature communications. 2012;3:1084. doi: 10.1038/ncomms2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han K, Gennarino VA, Lee Y, Pang K, Hashimoto-Torii K, et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483–5p. Genes & development. 2013;27:485–90. doi: 10.1101/gad.207456.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nature reviews Genetics. 2013;14:535–48. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Current opinion in cell biology. 2013;25:1–7. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 64.Hornberg H, Holt C. RNA-binding proteins and translational regulation in axons and growth cones. Frontiers in neuroscience. 2013;7:81. doi: 10.3389/fnins.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–87. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao GY, An JJ, Gharami K, Waterhouse EG, Vanevski F, et al. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat Med. 2012 doi: 10.1038/nm.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–52. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nature neuroscience. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 69.Feltrin D, Fusco L, Witte H, Moretti F, Martin K, et al. Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol. 2012;10:e1001439. doi: 10.1371/journal.pbio.1001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perry RB, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, et al. Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, et al. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Human molecular genetics. 2010;19:1951–66. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- 72.Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, et al. Transport and localization elements in myelin basic protein mRNA. The Journal of cell biology. 1997;138:1077–87. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raju CS, Goritz C, Nord Y, Hermanson O, Lopez-Iglesias C, et al. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol Biol Cell. 2008;19:3008–19. doi: 10.1091/mbc.E07-10-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO reports. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harbor perspectives in biology. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–79. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Kato M, Han TW, Xie S, Shi K, Du X, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hartley CA, McKenna MC, Salman R, Holmes A, Casey BJ, et al. Serotonin transporter polyadenylation polymorphism modulates the retention of fear extinction memory. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5493–8. doi: 10.1073/pnas.1202044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoon Y, McKenna MC, Rollins DA, Song M, Nuriel T, et al. Anxiety-associated alternative polyadenylation of the serotonin transporter mRNA confers translational regulation by hnRNPK. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1301485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lau AG, Irier HA, Gu J, Tian D, Ku L, et al. Distinct 3′UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15945–50. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–38. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nature methods. 2013 doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, et al. Axonal mRNA in uninjured and regenerating cortical mammalian axons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:4697–707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes & development. 2013;27:1495–510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, et al. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic acids research. 2013;41:9438–60. doi: 10.1093/nar/gkt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–9. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 90.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature methods. 2011 doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. The EMBO journal. 2007;26:2658–69. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, et al. Axonal Localization of transgene mRNA in mature PNS and CNS neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:14481–7. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nature methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28:503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kolle G, Shepherd JL, Gardiner B, Kassahn KS, Cloonan N, et al. Deep-transcriptome and ribonome sequencing redefines the molecular networks of pluripotency and the extracellular space in human embryonic stem cells. Genome research. 2011;21:2014–25. doi: 10.1101/gr.119321.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hurowitz EH, Drori I, Stodden VC, Donoho DL, Brown PO. Virtual Northern analysis of the human genome. PloS one. 2007;2:e460. doi: 10.1371/journal.pone.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nature biotechnology. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]