Abstract
The autoimmune disease systemic lupus erythematosus (SLE) has a complex environmental and multi-factorial genetic basis. Genome wide association studies have recently identified numerous disease-associated polymorphisms, but it remains unclear in which cells and during which step of pathogenesis specific polymorphisms interact to cause disease. Using a mouse model in which the same activating mutation (CD45E613R) causes distinct genetic background-dependent disease phenotypes, we perform a screen for genetic modifiers of autoreactivity between anti-nuclear antibody (ANA)-resistant CD45E613R.B6 and ANA-permissive CD45E613R.BALB/c mice. Within a novel autoreactivity-associated locus on chromosome 9, we identify a putative modifier, TLR9. Validating a role for TLR9 in modifying autoreactivity in the context of the CD45E613R mutation, manipulation of TLR9 gene dosage eliminates ANA in CD45E613R.BALB/c, but confoundingly permits ANA in CD45E613R.B6. We demonstrate that sensitivity to ANA is modulated by strength of TLR9 signal, since stronger TLR9B6 signals, but not weaker TLR9BALB/c signals, negatively regulate CD45E613R B cell development during competitive reconstitution at the central tolerance checkpoint. Our results identify a novel autoreactivity-associated locus and validate Tlr9 as a candidate gene within the locus. We further demonstrate a novel role for TLR9 signal strength in central tolerance, providing insight into the interplay of disease-associated polymorphisms at a discrete step of SLE pathogenesis.
Introduction
Pathogenesis of the clinically heterogeneous autoimmune disease systemic lupus erythematous (SLE) is a multi-step process that is heavily influenced by both genetics and environment (1–3). A hallmark of SLE is the presence of circulating anti-nuclear antibodies (ANA), which can form immune complexes with self nucleic acids and associated proteins (4). These immune complexes can deposit in tissues, trigger inflammation, and cause end organ damage (1). Recent advances have identified numerous candidate genes via genome wide association studies (GWAS) that may contribute to SLE pathogenesis (3). However, it remains incompletely understood how these disease-associated loci cooperate with each other or environmental triggers at various stages of SLE pathogenesis. Furthermore, the variability of clinical presentation has made studying relative contributions of individual loci to the pathogenesis of SLE in patients difficult. Murine models of SLE have been essential for dissecting the multi-step pathogenesis of SLE in a controlled environment (2). These models provide a tractable genetic framework for dissecting the perturbations in signaling networks and cell types responsible for disease.
Regulators and mediators of lymphocyte antigen receptor signaling are commonly dysregulated in SLE (5). However, despite well-documented evidence that perturbations of antigen receptor signaling can alter the developmental tolerance checkpoints and determine cell fate upon activation(5), it remains unclear how genetic context influences whether or not these lymphocytes will break tolerance. The phosphatase CD45 is an essential regulator of antigen receptor signaling, and its absence impairs lymphocyte development, causing a severe combined immunodeficiency (SCID) phenotype in both mice and humans (6). CD45 is expressed on all nucleated hematopoietic cells, and its dysregulation has been associated with increased susceptibility to autoimmune disease. We previously demonstrated that a single amino acid substitution, E613R, in the juxtamembrane wedge domain of CD45 results in a lupus-like phenotype in approximately 40% of mice on a mixed 129/Sv and C57BL/6 (B6) genetic background (7). Mirroring the variable presentation of human SLE, the phenotype of CD45E613R mice is extremely sensitive to genetic context. Despite hyper-responsive antigen receptor signaling, CD45E613R mice fully backcrossed to B6 or 129/Sv genetic backgrounds fail to develop autoantibodies or end organ damage (8–11). However, true B6×129/Sv CD45E613R F1 mice recapitulate the original lupus phenotype with 100% penetrance (12). Further validating this model, the CD45E613R mutation cooperates with established lupus risk alleles to exacerbate disease in the autoimmune resistant B6 genetic background (9, 10). These data indicate that the phenotypic consequences of CD45E613R-induced antigen receptor hyper-responsiveness require additional genetic perturbations to mediate loss of tolerance and systemic autoimmunity.
Here, we further investigate the interplay of alterations in antigen receptor signaling and genetic modifiers on the development of ANA. We demonstrate that the CD45E613R mutation on a BALB/c genetic background results in production of ANA, specifically anti-double stranded DNA (dsDNA) antibodies, without concomitant end organ disease. This provides a tractable system to interrogate a key step in the multi-step pathogenesis of SLE, loss of self-tolerance, without the interference of immune complex-mediated tissue damage. We leverage this phenotype to screen for genetic modifiers of anti-dsDNA IgG production in an unbiased fashion in an F2 cross between ANA-permissive CD45E613R.BALB/c and ANA-resistant CD45E613R.B6 mice. We identify a novel putative modifier locus on chromosomes 9, denoted Wedge Associated Modifier 1 (Wam1), and based on an in silico SNP analysis we identify a putative modifier gene within Wam1, Tlr9. Sequencing and molecular modeling of TLR9BALB/c and TLR9B6 indicate there are five polymorphisms between these alleles, mapping to the ligand-binding ectodomain and immediately adjacent to the intracellular TIR domain. Validating a role for TLR9 in ANA production, we find that genetic ablation of TLR9 eliminates ANA in CD45E613R.BALB/c mice but decreasing gene dosage of TLR9 permits ANA in CD45E613R.B6 mice. We reconcile these opposing phenotypes by demonstrating that sensitivity to ANA is modulated by strength of the TLR9 signal. We demonstrate that the stronger TLR9B6 signal, but not weaker TLR9BALB/c signal, negatively regulates CD45E613R B cell development at the central tolerance checkpoint. Our findings demonstrate that in the context of a single risk allele that causes hyper-responsive BCR signaling, TLR9 signal strength has a previously unappreciated role in setting the threshold for central tolerance against nuclear antigens. With our unbiased approach, we provide new insight into how disease-associated polymorphisms and risk alleles can interact to modulate susceptibility to autoimmune disease.
Materials and Methods
Mice
Mice were obtained from the following sources: C57BL/6.CD45.1 (002014), C57BL/6.CD45.2 (000664), BALB/c.CD45.1 (006584), BALB/c.CD45.2 (001026), BALB/c.IgHb (001107), and BALB/c.Actin.GFP+ (007075) strains (Jackson Laboratory), BALB/c.TCRα−/− and BALB/c.JH−/− (A. Abbas, UCSF), C57BL/6.TLR9−/− mice (J. DeRisi, UCSF), and BALB/c.TLR9−/− (I. Rifkin, Boston University), used with permission from S. Akira (13). CD45E613R mice were generated as previously described (7) and backcrossed at least nine generations to C57BL/6 and BALB/c backgrounds. Fidelity of backcrossing was verified in the UCSF Genomics Core using the Mouse MD Linkage Analysis SNP Array (Illumina). All loci were of the predicted genotype except in CD45E613R.B6, where the two SNPs immediately flanking Ptprc (CD45) were 129/Sv (these SNPs are shared between 129/Sv and BALB/c). Mice were bred and housed in a specific-pathogen free facility and experiments were performed according to UCSF IACUC and NIH guidelines. For aging cohorts, mice were bled monthly beginning at 8 weeks of age to monitor serum autoantibodies. As previously described (7), proteinuria was evaluated using Uristix (Siemens). Briefly, urine applied to a colorimetric dipstick was read on a scale from 0 to ++++, where a score of +++ or ++++ (corresponding to greater than 300mg/dL) was considered positive.
Genetic Modifier Mapping and Statistical Analyses
Genomic DNA was extracted using GenePure Kit according to manufacturer’s instructions. SNP analysis was performed according using the Illumina MD platform by the UCSF Genomics Core. Linkage analysis and quantitative trait loci (QTL) mapping were performed using Mapmaker-QTL and NCBI m36 mouse assembly. Experimental p-values were established by permutation testing (1000 permutations). In silico mapping of potentially interesting genes in candidate loci and SNP analyses were performed using mouse EnsEMBL (http://www.ensembl.org/Mus_musculus/index.html), the UCSC genome browser (http://genome.ucsc.edu/), and the Jackson Laboratory Mouse Genome Informatics database (http://www.informatics.jax.org/).
Autoantibody Assays
ANA were performed as previously described (9). Briefly, HEp-2 slides (Inova Diagnostics) were stained with a 1:40 or 1:100 serum dilution, washed and detected with FITC-conjugated donkey anti-mouse IgG secondary (Jackson Immunoresearch). Images were acquired on an Olympus BX51 epifluorescence microscope with OpenLab software and scored according to staining intensity. Pooled CD45E613R.BALB/c or MRL/lpr serum was used as a positive control.
Anti-dsDNA IgG ELISA was performed as previously described (7). Briefly, 96 well flat bottom plates were coated with Poly-L-lysine (Sigma P2636), then Poly dA:dT (Sigma P0883), and blocked. Serum was diluted 2-fold starting at a dilution of 1:100 (unless otherwise indicated), and antibodies were detected with HRP-Goat-anti Mouse pan IgG (Southern Biotech) and TMB (Sigma). Pooled CD45E613R.BALB/c or MRL/lpr serum was used as a positive control and run in serial dilution on each plate as a quantitation control. Anti-RNP IgG was similarly detected using plates coated with Sm/RNP antigen (Immunovision SRC-3000). Sm/RNP-positive TLR9−/−CD45E613R.B6 serum was used as a positive control.
Antibodies and flow cytometry
The following antibodies were used: CD4-V500, CD11c-PE-Cy7, CD19-V450, CD19-APC-Cy7, CD69-PE-Cy7, IgM-PerCP-Cy5.5, CD45.2-V500, CD45.2-APC (BD Biosciences), IgD-PE, CD45.1-FITC, CD11b-PerCP-Cy5.5, CD21-PB, CD23-PE-Cy7, AA4.1-APC (eBiosciences), and CD86-PB, CD45.1-PB (BioLegend). Following red blood cell lysis, Fc receptor blockade, and staining, cells were processed on a BD FACSVerse or LSR II flow cytometer and analyzed using FlowJo v9.6.4 (Treestar).
Calcium flux
Calcium flux was performed as previously described (5). Briefly, single cell suspensions were prepared from lymph nodes of 6–8 week old mice, loaded with 2µg FastRed /107 cells and 2µg Fluo4/ 107 cells (Molecular Probes), stimulated with goat anti-mouse IgM F(ab’)2 (Jackson Immunoresearch), and collected on a BD LSR II. The intracellular concentration of calcium was determined using the FlowJo kinetic function.
Bone Marrow Chimeras
Donor bone marrow was isolated from femurs of 5–10 week old mice of the indicated genotypes, resuspended at 1.5-2×107 cells/ml in sterile PBS, combined at the indicated ratios, and 100µl injected intravenously into lethally irradiated 6–8 week old hosts. Recipients were treated with antibiotic pellets (Bio-Serv S0443) for two weeks following bone marrow transfer. Beginning 8 weeks following transfer, mice were bled monthly to monitor for engraftment and serum autoantibodies. ANA titers from the terminal time point for each experiment are shown.
B cell signaling flow cytometry
Single cell suspensions were generated from lymph nodes of 8–10 week old mice, resuspended at 2×107 cells/ml, rested in serum free media at 37°C for 1 hour, and stimulated in 96 well V-bottom plates at 37°C with vehicle, CpG 1668 (Invivogen) or F(ab’)2 anti-mouse IgM (Jackson Immunoresearch) for the times indicated in the figure legend. Cells were fixed with pre-warmed 1% paraformaldehyde, washed, permeabilized with ice cold 100% methanol, washed, rehydrated in PBS, stained with antibodies against pERK or IκBα (Cell Signaling Technologies), washed, stained with PE-donkey anti-rabbit IgG (Jackson Immunoresearch) and anti-B220-APC (eBiosciences), and analyzed as described above.
Statistics
Statistical analyses were performed using Prism v5.0a (GraphPad). All p values ≤ 0.05 were considered significant and are shown in the figures. Student’s t test was used for pair-wise comparisons. In experiments comparing more than two groups, a one-way ANOVA test was first performed to evaluate significance, followed by pair-wise t test indicated in the figures.
Results
Genetic background determines the phenotypic consequences of CD45E613R-induced antigen receptor hyper-responsive signaling
Despite hyper-responsive BCR and TCR signaling responses, CD45E613R mice fail to break tolerance and develop ANA or any stigmata of SLE on an F10 B6 genetic background (8–11). However, this mutation can cooperate with other lupus-prone alleles or genetic backgrounds to cause a fulminant autoimmune phenotype on the B6 genetic background, clearly demonstrating a role for genetic modifiers in this model (7, 9, 10, 12). To systematically test the influence of genetic modifiers on the CD45E613R autoimmune phenotype, we backcrossed the CD45E613R for 10 generations to the BALB/c genetic background. A whole genome SNP analysis confirmed F10 CD45E613R.BALB/c mice were >99% homozygous for the BALB/c background (data not shown). Cohorts of B6 and BALB/c CD45WT and E613R mice were monitored monthly for autoantibody production and proteinuria.
Genetic background had a dramatic effect on the phenotype of CD45E613R mice. CD45E613R.B6 and CD45WT.B6 littermate control mice failed to develop detectable ANAs, anti-dsDNA antibodies, or proteinuria through 18 months of age (Fig 1A–C; data not shown). In striking contrast, F10 CD45E613R.BALB/c mice began to develop high-titer ANAs and anti-dsDNA antibodies by 6 weeks of age independent of gender, with complete penetrance by 4 months of age (Fig 1A–C). Only one CD45WT.BALB/c littermate developed low titer ANAs when co-housed under the same conditions. Surprisingly, despite the presence of high titer autoantibodies, CD45E613R.BALB/c mice failed to develop proteinuria or histopathologic evidence of glomerulonephritis or any other end organ disease (data not shown).
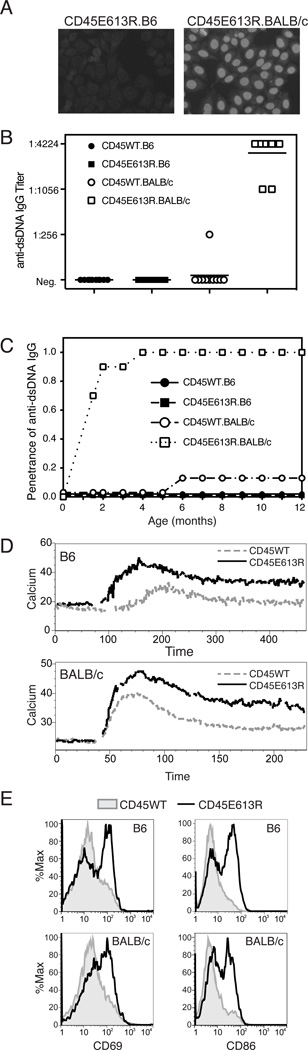
Figure 1. Genetic background influences autoreactivity but not signaling phenotypes of CD45E613R mice.
(A–C) Cohorts of 5 male and 5 female CD45E613R and CD45WT mice were established for each inbred strain and aged for 18 months. Sera were collected monthly to assess for serum autoantibodies and animals were tested for proteinuria and examined for other stigmata of autoimmunity. (A) Representative ANA staining pattern of 1:100 dilutions of sera from 6 month old mice. (B) Relative titer at 12 months and (C) penetrance of anti-dsDNA IgG measured by ELISA. Titer was defined as the last dilution at which a positive result was observed. Reference MRL.lpr sera peaked at 1:1600.
(D) Intracellular calcium levels of Fluo4 and Fast Red loaded B cells upon stimulation with 5µg/ml goat anti-mouse IgM F(ab’)2. Results are representative of 3–5 experiments per genotype, each testing calcium responses in 3 mice/genotype. (E) Representative histograms of CD19+ splenic B cells from 12 month old mice stained with CD69 and CD86. (D–E) Solid black line represents CD45E613R and grey line or filled histogram represents CD45WT of indicated genetic background.
We previously demonstrated that CD45E613R.B6 follicular B cells were hyper-responsive to antigen stimulation (8). To test whether this hyper-responsiveness is sensitive to genetic background and a potential explanation for the different phenotypes, we measured calcium flux in response to antigen receptor stimulation in mature naïve B cells from 6–8 week old mice, prior to disease onset. Calcium flux was similarly augmented in CD45E613R B cells relative to WT controls in both B6 and BALB/c genetic backgrounds (Fig 1D). Consistent with this hyper-responsiveness, the activation markers CD69 and CD86 were similarly upregulated in B cells of aged CD45E613R mice on both genetic backgrounds relative to age-matched CD45WT controls (Fig 1E). These similarities raise the possibility that genetic modifiers may influence the phenotypic consequence of biochemical changes in antigen receptor signaling by either sensitizing cells to or protecting cells from loss of self-tolerance. The data also suggest that additional modifier loci may be necessary for end organ damage in the BALB/c background. Our model also provides further evidence that development of autoreactivity (loss of self-tolerance) and autoimmunity (autoantibody-mediated end organ damage) are distinct steps in SLE pathogenesis(2, 14).
Identification of anti-dsDNA antibody modifier loci in CD45E613R B6 and BALB/c mice
To identify loci that might regulate susceptibility to autoreactivity in the context of the CD45E613R mutation, a genetic modifier screen using an F2 strategy was performed between B6 and BALB/c CD45E613R strains. We generated 43 CD45E613R and 45 CD45WT F1 (B6 × BALB/c) mice and monitored them for SLE phenotypes. In contrast to 100% penetrance of ANA and glomerulonephritis in F1 (B6 × 129/Sv) CD45E613R mice, only 4.6% of F1 (B6 × BALB/c) mice CD45E613R developed positive ANA and no mice developed anti-dsDNA IgG, proteinuria, or other stigmata of autoimmunity by 12 months of age (data not shown). These findings suggest that the loci cooperating with the CD45E613R mutation in causing or preventing autoantibody production operate in a recessive and/or additive fashion in B6 and BALB/c mice.
F1 (B6 × BALB/c) CD45E613R mice were interbred to generate 255 F2 (B6 × BALB/c) progeny. Mice were scored at 4 months of age, a time point when autoantibody production is completely penetrant in CD45E613R.BALB/c mice, for ANAs, anti-dsDNA IgG, and proteinuria. Positive ANA and anti-dsDNA IgG developed in 36.8% of the F2 mice without gender bias (Fig 2A). The frequency of anti-dsDNA antibody positive mice was more than would be expected from a single recessive BALB/c susceptibility locus, suggesting that several combinations of susceptibility genes can result in ANA or that the B6 genome contains susceptibility loci that are neutralized by co-existent resistance alleles. Only 1.5% of the F2 cohort developed proteinuria (data not shown). The small number of F2 mice with proteinuria suggests that this phenotype requires contributions from a larger number of susceptibility genes, some potentially only present in 129/Sv mice, and/or the absence of resistance loci.
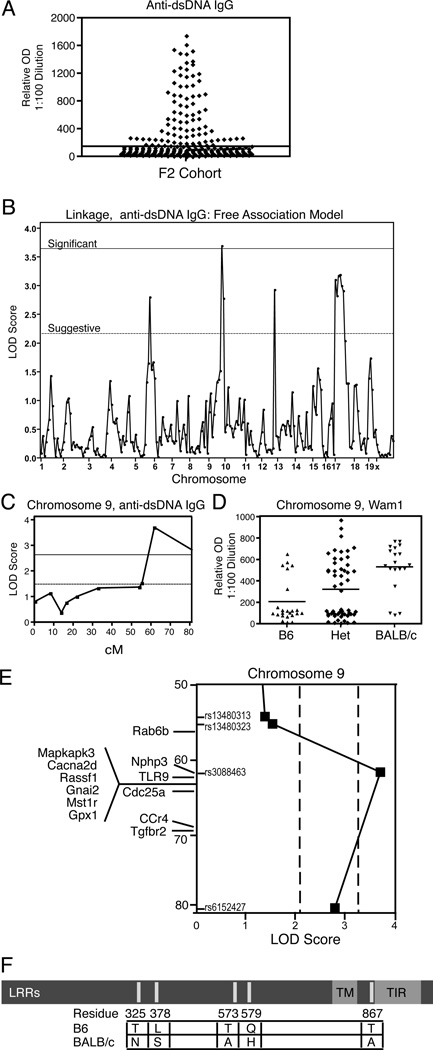
Figure 2. Phenotypic analysis of (B6 × BALB/c) CD45E613R F2 mice and susceptibility loci for ANA.
(A) Relative OD of 1:100 dilutions of sera for anti-dsDNA IgG for all animals in F2 cohort (n = 255). Each symbol represents one mouse. (B) Linkage analysis for anti-dsDNA IgG was performed on 94 F2 (B6 × BALB/c) CD45E613R mice. The solid line indicates the threshold for a significant LOD score and the dashed line indicates the threshold for a suggestive LOD score as based on permutation testing (1000 permutations) for each model. (C) Interval mapping of locus Wam1 on chromosome 9. (D) Anti-dsDNA IgG relative OD of cohort segregated by Wam1 genotype. (E) Genes of immunological interest and relative position within Wam1 are displayed on chromosome 9. (F) TLR9 polymorphsims between strains are displayed. LRR = leucine rich repeat; TM = transmembrane domain; TIR = Toll/interleukin-1 receptor domain.
To identify loci that contribute to ANA susceptibility in the context of altered antigen receptor signaling, a genome wide scan was performed using a medium density Illumina platform on 48 F2 mice with the highest anti-dsDNA IgG ELISA titers and 46 F2 mice with no detectable autoantibodies. Equivalent proportions of male and female mice were in each group. Using established definitions (34) and a free association model, LOD scores for anti-dsDNA antibody production in the suggestive range were identified on chromosomes 6, 12, and 17 and a LOD score in the significant range was found on chromosome 9 (Figure 2B; Table I). Using an additive association model, LOD scores for the loci on chromosomes 9 and 17 became more significant while the association with the chromosome 6 locus remained suggestive and the association on chromosome 12 was no longer apparent (Figure S1A; Table I). Loci with significant LOD scores for autoantibody production were designated Wedge Associated Modifer (Wam1) on chromosome 9 (peak SNP rs3088463) and Wam2 on chromosome 17 (peak SNPs rs6239530 and rs3703275). Interval mapping indicated Wam1 localized to the distal end of chromosome 9 (Fig 2C), an area devoid of known modifier loci in other murine models of SLE (15, 16), and was of BALB/c origin (Figure 2D). Wam2 encompasses the MHC, as well as other genes of immunologic interest, and was interestingly predominantly of B6 origin (Figure S1B,C; Table SII).
Table I.
Loci Associated with Autoimmune Phenotypes by Genome Wide Scan of (B6×BALB/c) CD45E613R F2 Mice.
| Trait | Chrom. | SNP | Position (cM) |
LOD Free |
LOD Dom |
LOD Rec |
LOD Add |
Locus |
|---|---|---|---|---|---|---|---|---|
| Anti-ds DNA | 9 | rs30884 63 |
61.67 | 3.7 | 1.61 | 2.91 | 3.50 | Wam1 |
| 17 | rs62395 30 |
0.24 | 3.11 | 3.11 | 0.33 | 3.50 | Wam2 | |
| rs13459 148 |
6.08 | 2.83 | 2.48 | 1.26 | 2.70 | Wam2 | ||
| CEL- 17_127 36614 |
6.23 | 3.17 | 2.83 | 1.26 | 2.96 | Wam2 | ||
| rs37032 75 |
16.13 | 3.20 | 3.00 | 1.22 | 2.91 | Wam2 | ||
| CEL- 17_374 78448 |
25.17 | 3.00 | 2.91 | 0.89 | 2.59 | Wam2 | ||
| rs42314 94 |
26.83 | 2.91 | 2.89 | 0.57 | 2.37 | Wam2 | ||
| Threshold Significance* |
3.65 | 2.63 | 2.86 | 2.72 | ||||
| Threshold Suggestive* |
2.17 | 1.48 | 1.52 | 1.47 |
Threshold determined by permutation testing (1000 permutations) for each model and trait.
LOD scores above the significance threshold in each model are shown in bold italics.
Because of a potentially novel association with autoimmune disease, we focused our attention on Wam1. Using the Mouse Genome Informatics (MGI) database, we found several genes of immunologic relevance localized to Wam1 (Fig 2E, Table SI). Interestingly, an in silico analysis of SNPs between B6 and BALB/c genetic backgrounds using the MGI database highlighted Toll like receptor 9 (Tlr9) as the only gene in the Wam1 locus with reported SNPs that resulted in non-synonymous coding changes. Consistent with a published report(17), sequencing Tlr9 from our B6 and BALB/c colonies confirmed a total of five non-synonymous coding changes between these strains. Molecular modeling of TLR9B6 and TLR9BALB/c indicates that the amino acid polymorphisms between these alleles map to the leucine rich repeats (LRRs) of the ectodomain and immediately adjacent to the intracellular TIR domain (Fig 2F). Both these domains are important for TLR9 function(18), warranting further studies of TLR9 as a potential genetic modifier.
TLR9 deficiency ablates ANA in CD45E613R.BALB/c mice but permits ANA in CD45E613R.B6 mice
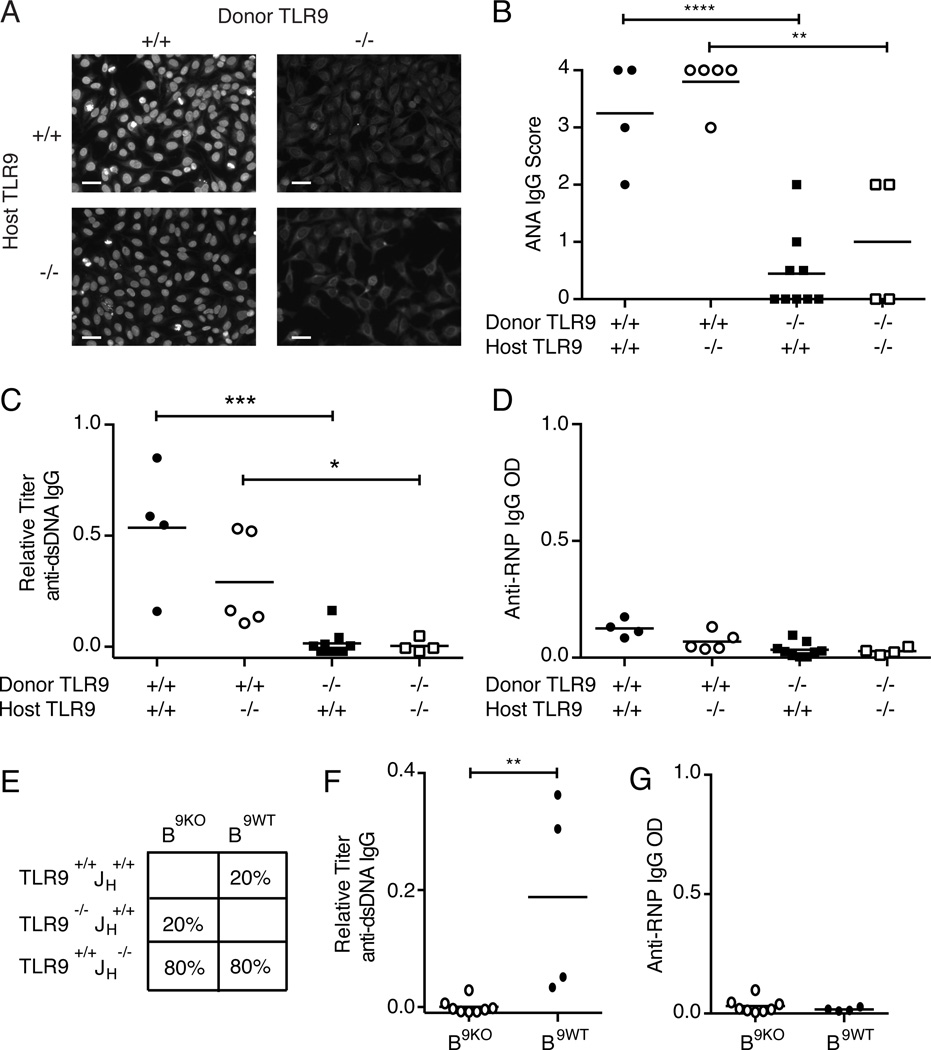
TLR9 is a member of a family of endosomally-restricted nucleic-acid sensing TLRs that autoreactive B cells require to become activated and secrete pathogenic autoantibodies(19, 20). It has been shown to modulate SLE phenotypes in murine models and has been implicated in human SLE (20). To examine whether TLR9 might modify ANA susceptibility between CD45E613R BALB/c and B6 mice, we generated TLR9+/+, TLR9+/−, and TLR9−/− CD45E613R mice on both BALB/c and B6 genetic backgrounds and monitored monthly for serum ANA. While aged TLR9+/+ and TLR9+/− CD45E613R.BALB/c mice developed ANA, TLR9−/− CD45E613R.BALB/c mice did not (Fig 3A,B). TLR9 deficiency also ablated the high titer anti-dsDNA specific IgG with a trend towards a gene dosage effect in heterozygotes, and low titer anti-RNP IgG in CD45E613R.BALB/c sera (Fig 3C,D). These results clearly demonstrate that TLR9 positively regulates autoantibody production in CD45E613R.BALB/c mice.
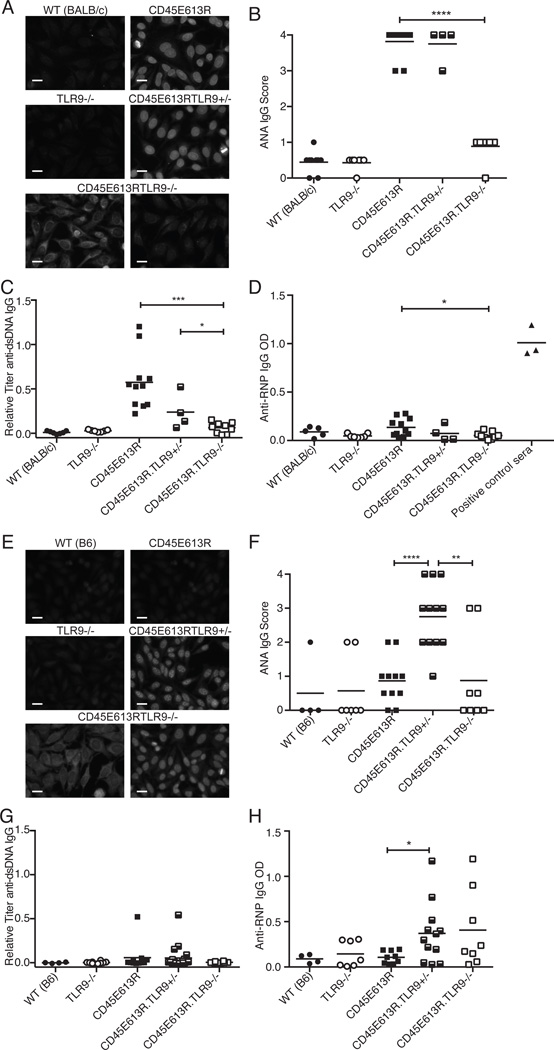
Figure 3. TLR9 regulates ANA in CD45E613R mice.
(A) Representative ANA staining pattern (original magnification 200X, scale bar represents 20µm) of 1:40 diluted sera and (B) score of staining intensity of serum collected from 6 month old mice of the indicated genotypes on the BALB/c background relative to pooled 6–8 month CD45E613R.BALB/c positive control sera. (C) Anti-dsDNA IgG relative to pooled 6–8 month CD45E613R.BALB/c and (D) Anti-RNP IgG titers from the same mice as assessed by ELISA. Data are pooled from three independent aging cohorts.
(E) Representative ANA staining pattern (original magnification 200X, scale bar represents 20µm) of 1:40 diluted sera and (F) score of staining intensity of serum collected from 6 month old mice of the indicated genotypes on the B6 background relative to pooled 6–8 month CD45E613R.BALB/c positive control sera. (G) Anti-dsDNA IgG relative to pooled 6–8 month CD45E613R.BALB/c positive control sera and (H) anti-RNP IgG titers as assessed by ELISA from sera from the same mice. Data are pooled from at least three independent aging cohorts. * p<0.05; ** p<0.01; *** p<0.005; **** p<0.001
In striking contrast, loss of one allele of TLR9 on the ANA resistant CD45E613R.B6 background permitted strong ANA staining in 7 of 12 mice, and complete deficiency for TLR9 permitted ANA production in 2 of 8 mice (Fig 3E,F). Further analyses of ANA specificity revealed that both TLR9+/− and TLR9−/− CD45E613R.B6 mice failed to develop high titer anti-dsDNA antibodies (Fig 3G). However, there was a significant increase in anti-RNP IgG autoantibodies in TLR9+/−CD45E613R.B6 mice. A similar trend was observed in TLR9−/−CD45E613R.B6 mice, but did not reach statistical significance (Fig 3H). We speculate that the phenotype of TLR9−/−CD45E613R.B6 was less pronounced due to TLR9’s known role in activation and expansion of autoreactive B cells. Taken together, these data clearly demonstrate that TLR9 negatively regulates autoantibody production in a gene dosage-dependent manner in CD45E613R.B6 mice.
Interestingly, consistent with observations in the TLR9−/−MRL/lpr model (21, 22), cytoplasmic staining was seen in 3 of 9 TLR9−/−CD45E613R.BALB/c mice and 4 of 8 TLR9−/−CD45E613R.B6 (Fig 3A,E, lower left panels). However, contrasting the TLR9−/−MRL/lpr model, TLR9−/−CD45E613R.BALB/c or B6 mice failed to develop any stigmata of end organ disease (data not shown), consistent with the lack of proteinuria observed in our F2 (B6 × BALB/c) CD45E613R cohort. Furthermore, the lack of end organ damage in TLR9 deficient CD45E613R mice supports the notion that additional genetic modifiers may be required for this step of lupus pathogenesis in the context of this risk allele.
B cell-intrinsic CD45E613R expression is required for ANA in BALB/c mice
Having determined that TLR9 is indeed a modifier of ANA development in CD45E613R mice, we sought to establish which cell lineages require CD45E613R and TLR9 for ANA development in susceptible CD45E613R.BALB/c mice. While CD45 is expressed in all nucleated hematopoietic cells (6) and TLR9 is expressed in both hematopoietic and non-hematopoietic cells (20), it is conceivable that expression either in cis or in trans, in only a subset of cells, is required for regulating tolerance to ANA.
Focusing first on the cellular requirements for the CD45E613R mutation, we generated a series of mixed bone marrow chimeras in CD45WT.BALB/c hosts to examine which immune cell subsets were required for ANA susceptibility. Six months following transfer of equal parts IgHbCD45WT and IgHaCD45E613R marrow, all ANA were IgGa, demonstrating a CD45E613R B cell origin (Fig. 4A). However, this result did not exclude the possibility that other cell lineages were required for development and/or activation of autoreactive CD45E613R B cells. For example, dysregulation of both T and myeloid lineages can promote autoantibody production (23–25).
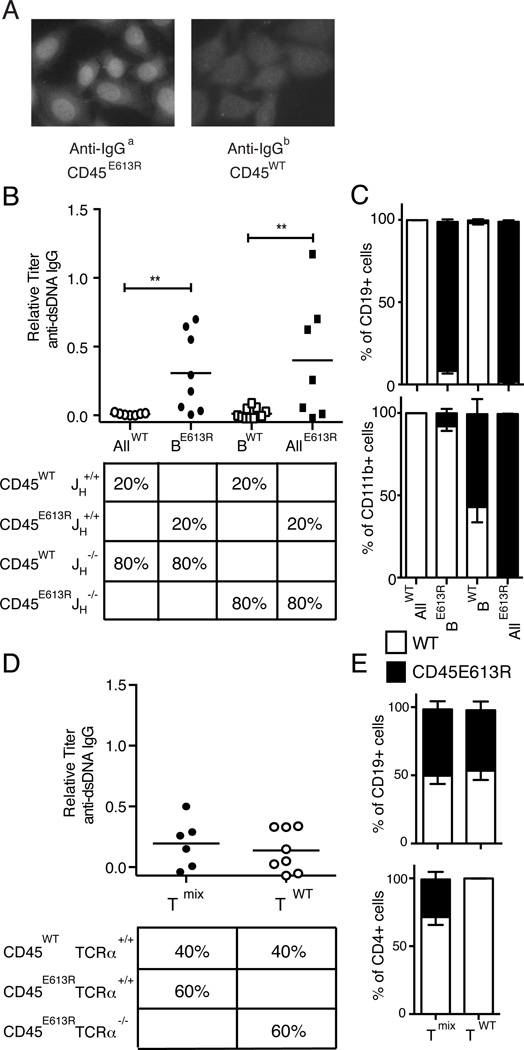
Figure 4. B cell intrinsic CD45E613R is necessary for ANA in CD45E613R.BALB/c mice.
(A) To determine whether ANA are CD45E613R B cell intrinsic, mixed bone marrow chimeras were generated from IgHb CD45WT and IgHa CD45E613R.BALB/c donors. HEp-2 cells were incubated with recipient sera and detected by fluorescent anti-IgGa or anti-IgGb secondary antibodies. Representative ANA staining pattern (original magnification 400X) of 1:40 diluted sera is shown for two independent experiments of 5 mice. (B–C) Mixed bone marrow chimeras were generated using the schema in (B, lower panel) to examine the role of CD45E613R B cells in autoreactivity. (B) Anti-dsDNA IgG serum titers detected by ELISA from chimeras 24–32 weeks after marrow transfer relative to pooled 6–8 month CD45E613R.BALB/c positive control sera. (C) Composition of splenic B cell and monocyte populations from the same chimeras 24–32 weeks after transfer. Data are pooled from two independent experiments of 3–4 mice per condition. (D–E) Bone marrow chimeras were generated using the schema in (D, lower panel) to examine the role of CD45E613R αβ T cells in autoreactivity. (D) Anti-dsDNA IgG serum titers from indicated chimeras at 20–24 weeks after marrow transfer relative to pooled 6–8 month CD45E613R.BALB/c positive control sera. (E) Blood composition of congenically marked CD45WT (CD45.1) and CD45E613R (CD45.2) B and T cells from the chimeras in (D) at 8 weeks after marrow transfer. Data are pooled from three independent experiments of 2–3 mice per condition. ** p<0.02
To more rigorously examine whether B cell-intrinsic CD45E613R is necessary for anti-dsDNA IgG, a 1:4 ratio of B cell sufficient (JH+/+) to B cell deficient (JH−/−) congenically marked CD45WT (CD45.1) or CD45E613R (CD45.2) marrow was transferred into lethally irradiated CD45WT.BALB/c hosts to generate chimeras where all B cells expressed exclusively the CD45WT or CD45E613R allele in the context of an immune compartment primarily derived from the opposite CD45 genotype. Confirming a B cell-intrinsic requirement for CD45E613R, recipients with CD45WT B cells in a predominantly CD45E613R immune compartment (BWT) did not develop anti-dsDNA IgG, similar to negative control chimeras generated from 20% CD45WT and 80% CD45WT JH−/− marrow (AllWT) (Fig. 4B). In contrast, recipients with CD45E613R B cells in a predominantly CD45WT immune compartment (BE613R) produced anti-dsDNA IgG at similar titers and frequencies to positive control chimeras generated from 20% CD45E613R B cell sufficient marrow combined with 80% CD45E613R B cell deficient marrow (AllE613R) (Fig. 4B).
The capacity of CD45E613R B cells to generate anti-dsDNA IgG in the context of a primarily CD45WT immune compartment suggests that B cell-intrinsic CD45E613R is sufficient for ANA. To control for the possibility that CD45E613R myeloid cells could skew these results by preferentially expanding in aged chimeras, we assessed the proportion of CD45E613R CD11b+ cells in the spleen. In both BWT and BE613R chimeras, CD45E613R CD11b+ cells did not preferentially expand and were in fact out-competed by CD45WT cells (Fig. 4C, lower panel). Furthermore, in the context of a common microenvironment, all ANA were of CD45E613R and not CD45WT B cell origin (Fig 4A), arguing that B cell-intrinsic CD45E613R is sufficient for ANA in this model system.
αβ T cells are also implicated in SLE pathogenesis (1) and dysregulated in CD45E613R mice (11). To examine whether CD45E613R αβ T cells were required for autoantibody production, we generated chimeras using congenically marked CD45WT and CD45E613R TCRα+/+ or TCRα−/− marrow at a 2:3 ratio to achieve approximately 50% CD45E613R CD19+ B cells in the peripheral blood 8 weeks post transfer (Fig. 4E, upper panel). Aged chimeras lacking CD45E613R αβ T cells developed anti-dsDNA IgG antibodies at similar frequencies and titers to chimeras containing both CD45E613R and CD45WT αβ T cells (Fig 4D), demonstrating that CD45E613R αβ T cells are dispensable for autoantibody production. Taken together, these data indicate that B cell-intrinsic CD45E613R is necessary and likely sufficient for autoreactivity.
CD45E613R.BALB/c B cells require TLR9 for anti-dsDNA IgG production
Having established a B cell-intrinsic requirement for the CD45E613R mutation, we were curious whether TLR9 was necessary in cis or in trans for ANA production in CD45E613R.BALB/c mice. Unlike CD45, TLR9 expression is not restricted to the hematopoietic compartment, and non-hematopoietic TLR9 expression has been implicated in lupus pathogenesis (26, 27). To test whether hematopoietic TLR9 was required for ANA in CD45E613R.BALB/c mice, we transferred TLR9+/+ or TLR9−/−Actin.GFP+CD45E613R.BALB/c marrow into lethally irradiated TLR9+/+ or TLR9−/−CD45WT recipients. Both lethally irradiated TLR9+/+ and TLR9−/−CD45WT recipients of TLR9+/+CD45E613R marrow developed ANA and anti-dsDNA IgG (Fig 5A–C), indicating that non-hematopoietic TLR9 was dispensable for ANA production. Neither TLR9+/+ nor TLR9−/−CD45WT recipients of TLR9−/− CD45E613R marrow developed ANA, anti-dsDNA, or anti-RNP antibodies, further substantiating a positive regulatory role for TLR9 in BALB/c CD45E613R mice (Fig 5A–D).
Figure 5. CD45E613R.BALB/c B cells require TLR9 for anti-dsDNA IgG production.
(A–D) To test whether non-hematopoietic TLR9 contributes to ANA, TLR9+/+ or TLR9−/−Actin.GFP+CD45E613R marrow was transferred into lethally irradiated TLR9+/+ or TLR9−/−CD45WT.BALB/c hosts as indicated.
(A) Representative ANA staining pattern (original magnification 200X, scale bar represents 40µm) of 1:40 diluted sera and (B) quantification of relative fluorescence intensity of sera from individual chimeras of the indicated genotypes. (C) Relative anti-dsDNA IgG or (D) anti-RNP IgG titers determined by ELISA as in figure 1. Data are pooled from two independent experiments of 2–4 mice per condition. (E–G) To test whether CD45E613R B cells require TLR9 for autoreactivity, bone marrow chimeras were generated with CD45E613R marrow of the indicated TLR9 genotypes transferred into lethally irradiated CD45WT hosts. (E) Scheme of mixed bone marrow chimeras and (F) anti-dsDNA IgG or (G) anti-RNP IgG serum titers from 20–30 weeks following marrow transfer. Data are pooled from three independent experiments. * p<0.05; ** p<0.01; *** p<0.005; **** p<0.001
We next tested whether CD45E613R B cells require TLR9 for ANA production. Chimeras in which all B cells were TLR9−/−CD45E613R (B9KO) or TLR9+/+CD45E613R (B9WT) in the context of a primarily TLR9+/+CD45E613R hematopoietic compartment were monitored monthly for serum ANA (Fig 5E). While half of the positive control chimeras containing TLR9+/+CD45E613R B cells (B9WT) developed anti-dsDNA IgG, none of the chimeras containing TLR9−/−CD45E613R B cells (B9KO) did (Fig 5F). Neither B9KO nor B9WT chimeras had detectable serum anti-RNP IgG (Fig 5G). The absence of anti-dsDNA IgG in chimeras containing TLR9−/−CD45E613R B cells in the context of a primarily TLR9+/+CD45E613R hematopoietic compartment indicates a B cell-intrinsic requirement for TLR9 for ANA in CD45E613R.BALB/c mice. Having identified the cell type in which TLR9 modifies CD45E613R-mediated autoreactivity, we sought to understand how TLR9 modifies this step of SLE pathogenesis by examining B cells between the two genetic backgrounds.
TLR9 signaling differs in B cells depending on genetic background
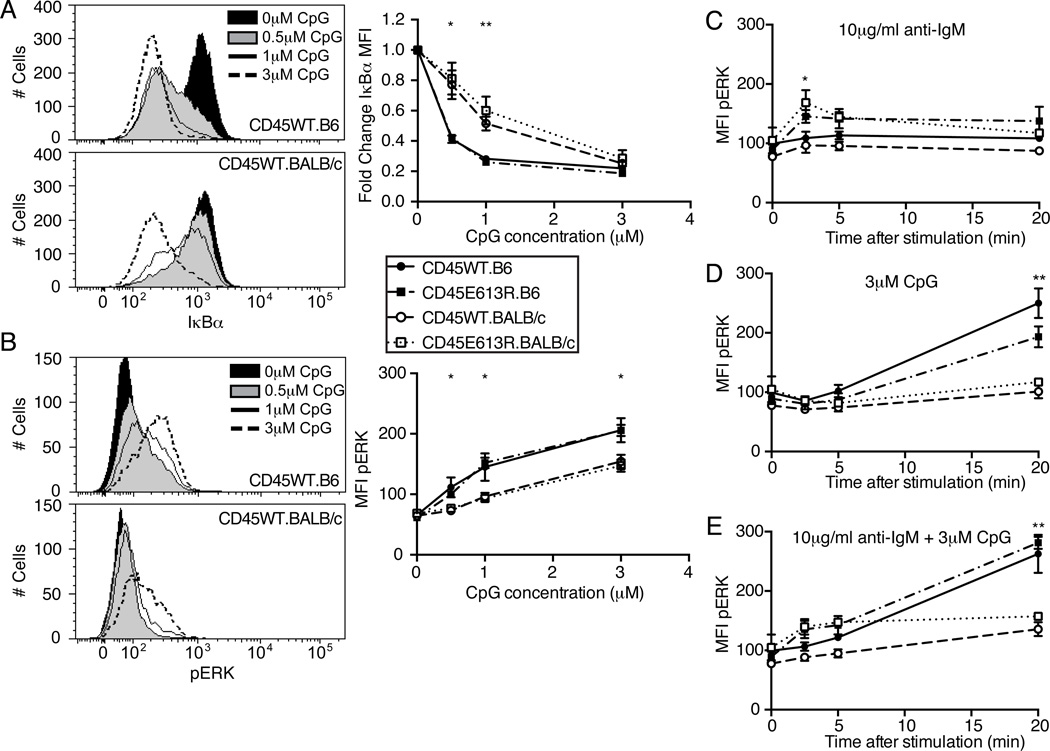
The opposing ANA phenotypes in the absence of TLR9 in B6 versus BALB/c CD45E613R mice were perplexing. The requirement of both CD45E613R and TLR9 for ANA production in BALB/c B cells (Fig 4,Fig 5) is consistent with the established requirement for BCR and TLR signals for activation of autoreactive B cells in the periphery (20). Based upon this model, we would predict that stronger TLR9-mediated signaling should promote increased ANA susceptibility. However, a prior study using a reporter assay to compare responses to CpG stimulation in cells expressing TLR9B6 or TLR9BALB/c in vitro found increased NF-κB signaling in cells expressing TLR9B6 (17).
To address this conundrum, we examined TLR9 signaling in follicular B cells from 8–10 week old mice. Stimulation with low doses of the synthetic TLR9 ligand CpG 1668 resulted in increased IκBα degradation, a measure of NF-κB activity, in B6 relative to BALB/c B cells (Fig 6A). This difference was masked at saturating doses of CpG (Fig 6A, right panel). Similarly, ERK was hyperphosphorylated in CpG-stimulated B6 compared to BALB/c B cells (Fig 6B). However, unlike NF-κB signaling, pERK was elevated in B6 B cells at both low and high CpG doses (Fig 6B, right panel). Interestingly, the response to CpG at either signaling node was not modulated by the CD45E613R mutation.
Figure 6. TLR9 signaling differs in B cells depending on genetic background and is not modulated by CD45E613R.
(A–B) Lymph node B cells from 8–10 week old mice of the indicated genotypes were stimulated for 20 minutes with the indicated concentration of CpG 1668, fixed, permeabilized, and stained for IκBα or pERK. (A) Representative histograms of IκBα expression in B220+ LN B cells following stimulation with indicated dosage of CpG. Right panel, Geometric mean fluorescence intensity (MFI) of IκBα for each CpG dosage was divided by MFI of unstimulated cells from the same mouse and plotted as the mean ± SD of three mice/genotype. (B) Representative histograms of pERK in B220+ LN B cells following stimulation with indicated dosage of CpG. Right panel, MFI for each CpG dosage was plotted as mean ± SD of three mice/genotype. Data are representative of at least two independent experiments. * p<0.05; ** p<0.01; comparing CD45WT.B6 to CD45WT.BALB/c using student’s t test at the indicated time point.
(C–E) Lymph node B cells from 8–10 week old mice of the indicated genotypes were stimulated with 3µM CpG 1668 or 10µg/ml anti-IgM F(ab)2 and stained for pERK. Time course of MFI of pERK in B220+ B cells following 10µg/ml anti-IgM (C), 3µM CpG 1668 (D), or both (E). Data points represent mean ± SD of three mice/genotype and are representative of at least three independent experiments. In (C), * p<0.05 comparing CD45WT and CD45E613R regardless of genetic background. In (D,E) ** p<0.01 comparing B6 to BALB/c regardless of CD45WT or CD45E613R status.
Since autoreactive B cells require signals from both the BCR and a nucleic acid-sensing TLR for activation (20), we hypothesized that the altered BCR signaling mediated by CD45E613R might interact differently with TLR9 signals in these two genetic backgrounds. We explored whether co-stimulation differentially modulated a common signaling node, pERK, in CD45E613R follicular B cells. Following BCR stimulation alone, pERK peaked at 2.5 minutes and was similarly elevated in both B6 and BALB/c CD45E613R B cells relative to CD45WT B cells (Fig 6C). Following CpG stimulation alone, pERK peaked at 20 minutes in both strains and was not influenced by the CD45E613R mutation (Fig. 6D). Surprisingly, regardless of ligand concentration, co-stimulation with anti-IgM and CpG did not have a synergistic effect or modulate the magnitude or timing of the two waves of pERK. (Fig 6E; data not shown). We conclude the CD45E613R mutation dictates augmented ERK phorphorylation in response to BCR stimulation regardless of genetic background. In contrast, the B6 background dictates augmented ERK phorphorylation in response to TLR9 stimulation regardless of the presence or absence of the CD45E613R mutation.
TLR9 negatively regulates CD45E613R.B6 but not CD45E613R.BALB/c B cell development in a competitive microenvironment
While the decreased signaling downstream of TLR9BALB/c compared to TLR9B6 mirrors previous observations (17), it remained inconsistent with our murine results in the context of the current model of TLR9 as an important mediator of autoreactive B cell activation (20). We therefore sought to better understand how stronger TLR9 signaling could promote B cell tolerance against nuclear antigens and protect against ANA. Since genetic elimination of one or more copies of the stronger signaling TLR9B6 allele permits ANA in CD45E613R.B6 mice (Fig 3), we hypothesized that TLR9 was negatively regulating the development of autoreactive B cells in this genetic background.
BCR signal strength regulates tolerance checkpoints in B cell development in several murine models, including CD45E613R (5). We previously observed decreased mature B cell numbers in CD45E613R.B6 mice with an unrestricted BCR repertoire. However, restricting the repertoire with a BCR transgene increased antigen specific B cell numbers in the absence of cognate antigen but decreased antigen specific B cell numbers when exposed to cognate antigen in vivo (8). These data support a model where CD45E613R-mediated increased BCR signal strength results in enhanced deletion of mature B cells by endogenous ligands. Based on the observations TLR9 signaling is elevated in B6 compared to BALB/c B cells and that TLR9 deficiency permits autoreactivity in CD45E613R.B6 mice (Fig 3, Fig 6), we hypothesized that the combined strength of signal from the BCR and TLR9 alters B cell tolerance checkpoints in CD45E613R.B6 but not CD45E613R.BALB/c mice.
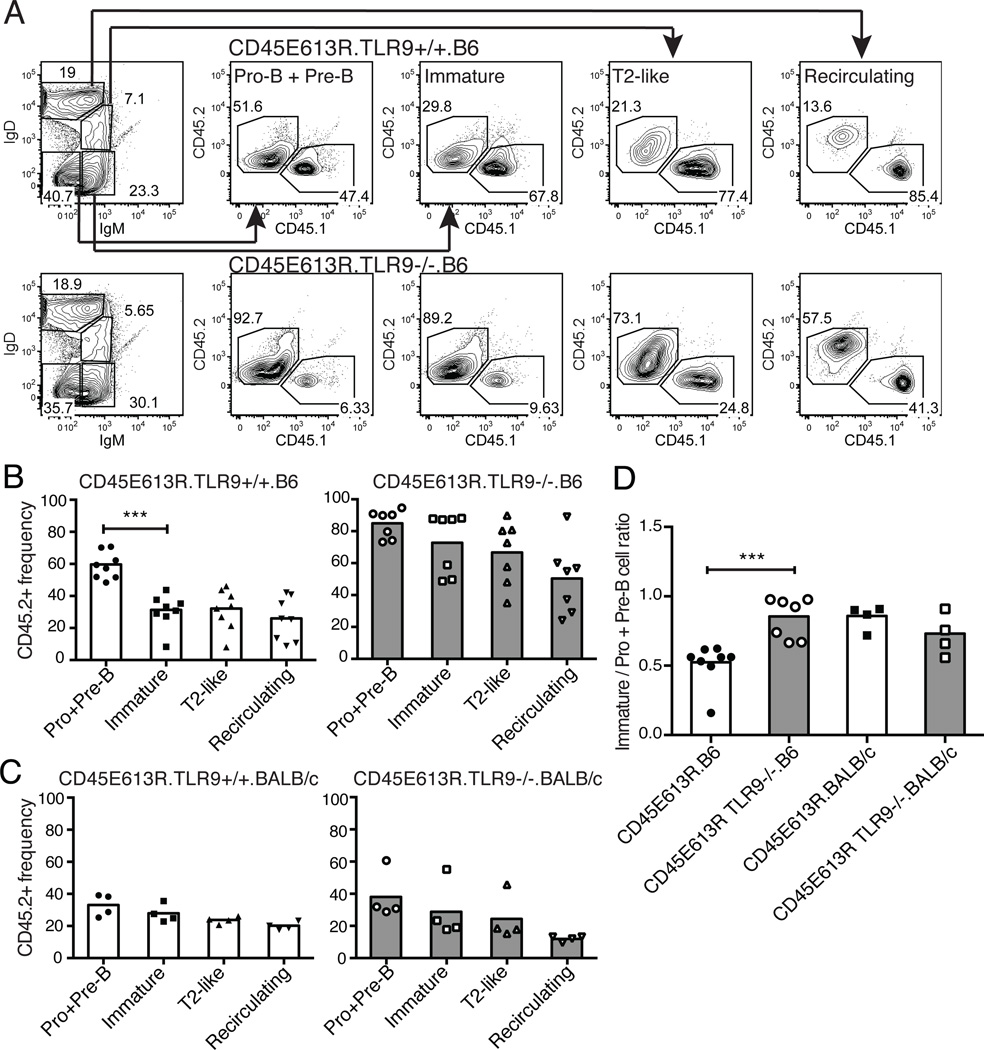
We first analyzed B cell development in the bone marrow and spleen of 8-week-old TLR9+/+, TLR9+/−, or TLR9−/−CD45E613R.B6 mice directly ex vivo, but failed to observe any TLR9-dependent differences (data not shown). To eliminate potential interference of compensatory B cell expansion, we next tested our hypothesis in a more stringent setting: the competitive microenvironment of mixed bone marrow chimeras. We generated chimeras in both the B6 and BALB/c genetic backgrounds using a 2:3 ratio of congenically marked CD45WT (CD45.1) to TLR9+/+ or TLR9−/−CD45E613R (CD45.2) marrow.
An important B cell tolerance checkpoint occurs in the spleen, where immature autoreactive B cells are eliminated or anergized and prevented from developing into follicular mature B cells (28). While there was a CD45E613R-mediated loss of cells at the splenic transitional-mature B cell checkpoint relative to CD45WT in both genetic backgrounds, it was surprisingly not affected by TLR9 (Fig S2D). We next examined the central tolerance checkpoint between pro- and pre-B versus immature B cells in the bone marrow. We noted a significant decrease in the reconstitution efficiency of IgM+IgD−CD19+ immature B cells compared to IgM−IgD−CD19+ pro- and pre-B cells in chimeras generated using TLR9+/+CD45E613R.B6 marrow but not in chimeras containing TLR9−/−CD45E613R.B6 marrow (Fig 7A,B). In contrast, a TLR9-mediated decrease at this checkpoint was not observed in BALB/c chimeras (Fig 7C). These differences were more striking when we calculated the engraftment ratio of CD45E613R immature B cells to pro- plus pre-B cells. This ratio was significantly increased when the CD45E613R cells lacked TLR9 in the B6 but not BALB/c genetic background (Fig 7D). Taken together, these data support a model where the increased signal strength via TLR9 is sufficient to eliminate autoreactive immature CD45E613R B cells in the bone marrow of B6 but not BALB/c mice.
Figure 7. TLR9 negatively regulates CD45E613R.B6 but not CD45E613R.BALB/c B cell development in a competitive microenvironment.
Bone marrow chimeras were generated using 40% CD45WT (CD45.1) and 60% TLR9+/+ or TLR9−/−CD45E613R (CD45.2) marrow transferred to lethally irradiated B6 or BALB/c CD45WT hosts (heterozygous for the CD45.1/2 congenic markers). (A) B cell developmental subsets in the bone marrow were defined using IgM and IgD staining of cells in the CD19+ gate. The frequency of CD45.1+ (CD45WT) and CD45.2+ (CD45E613R) cells in the indicated B cell subset was then assessed. (B) Frequency of CD45.2+ cells in each B cell compartment of chimeras generated using TLR9+/+ (left) or TLR9−/−CD45E613R (right) marrow on the B6 background. (C) Frequency of CD45.2+ cells in each B cell compartment of chimeras generated using TLR9+/+ (left) or TLR9−/−CD45E613R (right) marrow on the BALB/c background. (D) Ratio of the CD45.2+ frequency of immature (IgM+IgD-CD19+) to pro- plus pre-B (IgM-IgD-CD19+) cells for chimeras of the indicated genotype. Data in (B) are compiled from three independent experiments, and in (C) from two independent experiments. *** p<0.005
Discussion
Current data suggest that SLE is the culmination of a multi-step process involving altered lymphocyte homeostasis, breach of tolerance mechanisms, and dysregulated effector functions that result in unchecked inflammation and tissue damage (1, 2). Each point in this process appears to be governed by complex interactions between multiple, and likely distinct, susceptibility loci and the environment (14, 29). Consistent with studies dissecting loci that contribute to autoreactivity and autoimmunity in spontaneous murine lupus models (15), the CD45E613R risk allele is not sufficient for disease in the resistant B6 background. However, with CD45E613R mutant mice, we have a tractable model system that allows us to identify genetic modifiers of each step of disease pathogenesis in the context of the same risk allele. Here we have exploited the CD45E613R model to dissect the genetic parameters of autoreactivity in the absence of the sequellae of autoimmunity. In a screen for QTL conditioning anti-dsDNA IgG production between ANA-resistant CD45E613R.B6 mice and ANA-permissive CD45E613R.BALB/c, we identified a novel locus of BALB/c origin at the distal end of chromosome 9, Wam1. As with any QTL, the position of the initial Wam1 locus is large and further studies will be necessary to identify the causative gene(s). To evaluate the feasibility of utilizing congenic dissection to further characterize Wam1, we performed an in silico analysis that identified a candidate gene, Tlr9. We find that manipulation of TLR9 gene dosage in the context of the CD45E613R risk allele results in completely opposite phenotypes in these distinct genetic backgrounds, validating both our model system and TLR9 as a genetic modifier of autoreactivity.
Although TLR9 deficiency exacerbates end organ damage in several mouse models of SLE, controversy over the role of TLR9 in autoreactivity stems from several groups reporting different alterations to autoantibody specificity in TLR9 deficient SLE mice (21, 30–32). However, in these models ablation of TLR9 did not alter the susceptibility of lupus-prone mice to autoreactivity. Here, we demonstrate that in addition to altering autoantibody specificity, TLR9 plays an important role in modulating loss of tolerance, since loss of TLR9 can shift the permissiveness of mice to autoreactivity. Furthermore, we are able to dissect the contributions of TLR9 to autoreactivity in the absence of end organ damage, providing a better understanding of the contributions of this molecule to a discrete step of the multi-step pathogenesis of SLE.
In CD45E613R.B6 mice, TLR9 negatively regulates ANA, preventing the development of autoantibodies. The increased TLR7-associated anti-RNP IgG observed in TLR9 deficient and heterozygous CD45E613R.B6 mice are consistent with recent evidence suggesting that TLR9 may alter autoantibody specificity in mouse models by negatively regulating TLR7-mediated autoreactive B cell activation (22, 33). An in silico analysis of TLR7 identified no polymorphisms between B6 and BALB/c strains (34). However, in preliminary data (not shown), we observed decreased IκBα degradation in response to the synthetic TLR7 ligand R848 in CD45WT.BALB/c B cells compared to B6. If this difference in TLR7 responsiveness is not modified by CD45E613R, the decreased signaling in BALB/c B cells may account for the absence of TLR7-associated anti-RNP autoantibodies in TLR9−/−CD45E613R.BALB/c mice (Fig 3D). Since previous studies have shown that TLR9 deficiency results in increased TLR7 responses in several models (22, 33), it may be of interest to assess whether CD45E613R modulates this phenotypic difference in the context of ablation of TLR9, particularly in the B6 background. To our surprise, genetic ablation of one copy of Tlr9 in CD45E613R.B6 resulted in a higher frequency of mice developing ANA compared to TLR9−/− CD45E613R.B6 mice. This is likely due to the importance of TLR9 signaling in the activation and expansion of anti-nuclear autoreactive B cells in the periphery(20, 35). In contrast, in CD45E613R.BALB/c mice, TLR9 positively regulates ANA, consistent with the established role of nucleic acid-sensing TLRs in activation of autoreactive B cells (20).
We further exploit our model system by using a chimera approach to dissect the cell types in which TLR9 and CD45E613R act to modulate autoreactivity. Despite the broad expression pattern of CD45 in the hematopoietic compartment, B cell-intrinsic CD45E613R is arguably sufficient for ANA in CD45E613R.BALB/c mice. Furthermore, hematopoietic TLR9 is sufficient for ANA, and TLR9 is required in CD45E613R B cells for autoreactivity. Here we provide mechanistic evidence that strong TLR9 signals negatively regulate B cell autoreactivity. Since TLR9 is known to play an important role in the activation and expansion of autoreactive B cells (20), we initially hypothesized that TLR9 would mediate loss of B cells in the spleen. Surprisingly, CD45E613R-mediated loss of developing B cells at the peripheral tolerance checkpoint was TLR9-independent in both genetic backgrounds, likely due to out-competition by CD45WT B cells in the competitive microenvironment. Instead, we find that TLR9 negatively regulates CD45E613R.B6 B cell development in the bone marrow in a competitive microenvironment and inhibits ANA development. Based on these data, we propose that TLR9 modifies autoreactivity by regulating the central B cell tolerance checkpoint. This is consistent with published studies demonstrating that humans deficient for MYD88, IRAK4, or UNC93B1, important downstream signaling and trafficking adapters for endosomally restricted nucleic-acid sensing TLRs, have defects in purging autoreactive B cells at the central B cell tolerance checkpoint (36). Furthermore, according to expression data from the Immunological Genome Project Consortium, in B6 mice Tlr9 is highly upregulated in immature bone marrow B cells (37), the compartment where we observe loss of CD45E613R.B6 B cell precursors.
We observe that the capacity of TLR9 to promote B cell tolerance depends on strength of TLR9 signal, where TLR9B6 is sufficient but TLR9BALB/c is not for tolerance against anti-nuclear antigens in the context of hyper-responsive BCR signaling. However, TLR9BALB/c is required in B cells for ANA production in permissive CD45E613R.BALB/c mice and TLR9+/−CD45E613R.B6 develop ANA at higher penetrance than TLR9−/−CD45E613R.B6 mice, demonstrating that some level of TLR9 signaling promotes ANA, likely upon activation of autoreactive B cells. Our results highlight the importance of understanding the genetic context in which risk alleles exist for their function. In human populations, polymorphisms in the promoter region of TLR9 have been shown to be risk alleles for SLE in some populations, but meta-analyses of these studies have generated conflicting results (38–40). We propose that our results provide a framework for understanding how polymorphisms in a putative disease-associated gene may have opposing effects in the context of other risk alleles.
Our data highlights the importance of understanding the functional consequences of activating or inhibiting TLR9 in humans. Currently, many SLE patients are treated with hydroxychloroquine, an inhibitor of endosomal acidification that prevents activation of several endosomal nucleic acid-sensing TLRs, including TLR9 and TLR7 (41). It has also been demonstrated that activation of endosomal nucleic acid-sensing TLRs contributes to glucocorticoid resistance in SLE patients via increased type I interferon production (42). For these reasons, there has been increased interest in more specific TLR7 and TLR9 antagonists as possible therapeutics (43). Additionally, TLR9 agonists are also being considered as possible vaccine agonists (44, 45). Here, we demonstrate an important tolerogenic role for TLR9 in B cell development in a murine lupus model that depends upon genetic context. Our results indicate that distinct alleles of TLR9 can have opposing effects on the development of ANA, an important mediator of tissue damage in SLE. Therefore, it will be of further interest to explore the dual nature of this molecule and whether it similarly modulates human B cell tolerance in the context of other established risk alleles.
Supplementary Material
Acknowledgements
We thank the Hermiston and Weiss laboratories for helpful discussions and Drs. Anthony DeFranco and Jason Cyster for critical reading of the manuscript.
Footnotes
This work was supported by NSF GRFP1144247 and NIH T32AI007334 (RM), NIH K08AR059723 and Rosalind Russell Medical Research Foundation Bechtel Award (JZ), NIH PO1AI355297 (AW), and the Campini, Pepp Family, and St. Baldrick’s Foundations and NIH RO1AI089831 (MH). The authors declare no conflicts of interest.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Kanta H, Mohan C. Three checkpoints in lupus development: central tolerance in adaptive immunity, peripheral amplification by innate immunity and end-organ inflammation. Genes Immun. 2009;10:390–396. doi: 10.1038/gene.2009.6. [DOI] [PubMed] [Google Scholar]
- 3.Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann. Rheum. Dis. 2013;72(Suppl 2):ii56–ii61. doi: 10.1136/annrheumdis-2012-202351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ippolito A, Wallace DJ, Gladman D, Fortin PR, Urowitz M, Werth V, Costner M, Gordon C, Alarcon GS, Ramsey-Goldman R, Maddison P, Clarke A, Bernatsky S, Manzi S, Bae SC, Merrill JT, Ginzler E, Hanly JG, Nived O, Sturfelt G, Sanchez-Guerrero J, Bruce I, Aranow C, Isenberg D, Zoma A, Magder LS, Buyon J, Kalunian K, Dooley MA, Steinsson K, van Vollenhoven RF, Stoll T, Weisman M, Petri M. Autoantibodies in systemic lupus erythematosus: comparison of historical and current assessment of seropositivity. Lupus. 2011;20:250–255. doi: 10.1177/0961203310385738. [DOI] [PubMed] [Google Scholar]
- 5.Zikherman J, Weiss A. Antigen receptor signaling in the rheumatic diseases. Arthritis Res. Ther. 2009;11:202. doi: 10.1186/ar2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermiston ML, Zikherman J, Zhu JW. CD45, CD148, and Lyp/Pep: critical phosphatases regulating Src family kinase signaling networks in immune cells. Immunol. Rev. 2009;228:288–311. doi: 10.1111/j.1600-065X.2008.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majeti R, Xu Z, Parslow TG, Olson JL, Daikh DI, Killeen N, Weiss A. An inactivating point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 8.Hermiston ML, Tan AL, Gupta VA, Majeti R, Weiss A. The juxtamembrane wedge negatively regulates CD45 function in B cells. Immunity. 2005;23:635–647. doi: 10.1016/j.immuni.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Gupta VA, Hermiston ML, Cassafer G, Daikh DI, Weiss A. B cells drive lymphocyte activation and expansion in mice with the CD45 wedge mutation and Fas deficiency. J. Exp. Med. 2008;205:2755–2761. doi: 10.1084/jem.20081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zikherman J, Hermiston M, Steiner D, Hasegawa K, Chan A, Weiss A. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J. Immunol. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermiston ML, Zikherman J, Tan AL, Lam VC, Cresalia NM, Oksenberg N, Goren N, Brassat D, Oksenberg JR, Weiss A. Differential impact of the CD45 juxtamembrane wedge on central and peripheral T cell receptor responses. Proc. Natl. Acad. Sci. U. S. A. 2009;106:546–551. doi: 10.1073/pnas.0811647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zikherman J, Parameswaran R, Hermiston M, Weiss A. The structural wedge domain of the receptor-like tyrosine phosphatase CD45 enforces B cell tolerance by regulating substrate specificity. J. Immunol. 2013;190:2527–2535. doi: 10.4049/jimmunol.1202928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 14.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N. Engl. J. Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 15.Wakeland EK, Liu K, Graham RR, Behrens TW. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397–408. doi: 10.1016/s1074-7613(01)00201-1. [DOI] [PubMed] [Google Scholar]
- 16.Kono DH, Theofilopoulos AN. Genetics of SLE in mice. Springer Semin. Immunopathol. 2006;28:83–96. doi: 10.1007/s00281-006-0030-7. [DOI] [PubMed] [Google Scholar]
- 17.Anderson AE, Worku ML, Khamri W, Bamford KB, Walker MM, Thursz MR. TLR9 polymorphisms determine murine lymphocyte responses to Helicobacter: results from a genome-wide scan. Eur. J. Immunol. 2007;37:1548–1561. doi: 10.1002/eji.200636562. [DOI] [PubMed] [Google Scholar]
- 18.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 20.Green NM, Marshak-Rothstein A. Toll-like receptor driven B cell activation in the induction of systemic autoimmunity. Semin. Immunol. 2011;23:106–112. doi: 10.1016/j.smim.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Nickerson KM, Christensen SR, Shupe J, Kashgarian M, Kim D, Elkon K, Shlomchik MJ. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J. Immunol. 2010;184:1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groom JR, Fletcher CA, Walters SN, Grey ST, Watt SV, Sweet MJ, Smyth MJ, Mackay CR, Mackay F. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J. Exp. Med. 2007;204:1959–1971. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothlin CV, Lemke G. TAM receptor signaling and autoimmune disease. Curr. Opin. Immunol. 2010;22:740–746. doi: 10.1016/j.coi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scapini P, Hu Y, Chu CL, Migone TS, Defranco AL, Cassatella MA, Lowell CA. Myeloid cells, BAFF, and IFN-gamma establish an inflammatory loop that exacerbates autoimmunity in Lyn-deficient mice. J. Exp. Med. 2010;207:1757–1773. doi: 10.1084/jem.20100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summers SA, Steinmetz OM, Ooi JD, Gan PY, O'Sullivan KM, Visvanathan K, Akira S, Kitching AR, Holdsworth SR. Toll-like receptor 9 enhances nephritogenic immunity and glomerular leukocyte recruitment, exacerbating experimental crescentic glomerulonephritis. Am. J. Pathol. 2010;177:2234–2244. doi: 10.2353/ajpath.2010.100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frieri M, Samih MA, Dzhindzhikhashvili M, Liu H, Balsam L, Rubinstein S. Toll-like receptor 9 and vascular endothelial growth factor levels in human kidneys from lupus nephritis patients. J. Nephrol. 2012;25:1041–1046. doi: 10.5301/jn.5000091. [DOI] [PubMed] [Google Scholar]
- 28.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat. Immunol. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 29.Chung SA, Brown EE, Williams AH, Ramos PS, Berthier CC, Bhangale T, Alarcon-Riquelme ME, Behrens TW, Criswell LA, Graham DC, Demirci FY, Edberg JC, Gaffney PM, Harley JB, Jacob CO, Kamboh MI, Kelly JA, Manzi S, Moser-Sivils KL, Russell LP, Petri M, Tsao BP, Vyse TJ, Zidovetzki R, Kretzler M, Kimberly RP, Freedman BI, Graham RR, D C Langefeld, and for the International Consortium for Systemic Lupus Erythematosus Genetics. Lupus Nephritis Susceptibility Loci in Women with Systemic Lupus Erythematosus. J. Am. Soc. Nephrol. 2014 doi: 10.1681/ASN.2013050446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Peng SL. Toll-like receptor 9 signaling protects against murine lupus. Arthritis Rheum. 2006;54:336–342. doi: 10.1002/art.21553. [DOI] [PubMed] [Google Scholar]
- 31.Yu P, Wellmann U, Kunder S, Quintanilla-Martinez L, Jennen L, Dear N, Amann K, Bauer S, Winkler TH, Wagner H. Toll-like receptor 9-independent aggravation of glomerulonephritis in a novel model of SLE. Int. Immunol. 2006;18:1211–1219. doi: 10.1093/intimm/dxl067. [DOI] [PubMed] [Google Scholar]
- 32.Lartigue A, Courville P, Auquit I, Francois A, Arnoult C, Tron F, Gilbert D, Musette P. Role of TLR9 in anti-nucleosome and anti-DNA antibody production in lpr mutation-induced murine lupus. J. Immunol. 2006;177:1349–1354. doi: 10.4049/jimmunol.177.2.1349. [DOI] [PubMed] [Google Scholar]
- 33.Desnues B, Macedo AB, Roussel-Queval A, Bonnardel J, Henri S, Demaria O, Alexopoulou L. TLR8 on dendritic cells and TLR9 on B cells restrain TLR7-mediated spontaneous autoimmunity in C57BL/6 mice. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1497–1502. doi: 10.1073/pnas.1314121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickerson KM, Christensen SR, Cullen JL, Meng W, Luning Prak ET, Shlomchik MJ. TLR9 promotes tolerance by restricting survival of anergic anti-DNA B cells, yet is also required for their activation. J. Immunol. 2013;190:1447–1456. doi: 10.4049/jimmunol.1202115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isnardi I, Ng YS, Srdanovic I, Motaghedi R, Rudchenko S, von Bernuth H, Zhang SY, Puel A, Jouanguy E, Picard C, Garty BZ, Camcioglu Y, Doffinger R, Kumararatne D, Davies G, Gallin JI, Haraguchi S, Day NK, Casanova JL, Meffre E. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heng TS, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 38.Lee YH, Lee HS, Choi SJ, Ji JD, Song GG. Associations between TLR polymorphisms and systemic lupus erythematosus: a systematic review and meta-analysis. Clin. Exp. Rheumatol. 2012;30:262–265. [PubMed] [Google Scholar]
- 39.Li J, Tao JH, Gao W, Fan Y, Lu MM, Li R, Li XP, Ye DQ. Lack of association of Toll-like receptor 9 polymorphisms with susceptibility to systemic lupus erythematosus in an Asian population: a meta-analysis. Mod. Rheumatol. 2012;22:550–556. doi: 10.1007/s10165-011-0573-x. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Liang Y, Qin B, Li C, Zhong R. TLR9 polymorphisms and systemic lupus erythematosus risk in Asians: a meta-analysis study. Cytokine. 2012;57:282–289. doi: 10.1016/j.cyto.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Tang C, Godfrey T, Stawell R, Nikpour M. Hydroxychloroquine in lupus: emerging evidence supporting multiple beneficial effects. Intern. Med. J. 2012;42:968–978. doi: 10.1111/j.1445-5994.2012.02886.x. [DOI] [PubMed] [Google Scholar]
- 42.Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, Meeker T, Chan JH, Wright T, Punaro M, Bolland S, Soumelis V, Banchereau J, Coffman RL, Pascual V, Barrat FJ. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrat FJ, Meeker T, Chan JH, Guiducci C, Coffman RL. Treatment of lupus-prone mice with a dual inhibitor of TLR7 and TLR9 leads to reduction of autoantibody production and amelioration of disease symptoms. Eur. J. Immunol. 2007;37:3582–3586. doi: 10.1002/eji.200737815. [DOI] [PubMed] [Google Scholar]
- 44.Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nat. Rev. Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 45.Galluzzi L, Vacchelli E, Eggermont A, Fridman WH, Galon J, Sautes-Fridman C, Tartour E, Zitvogel L, Kroemer G. Trial Watch: Experimental Toll-like receptor agonists for cancer therapy. Oncoimmunology. 2012;1:699–716. doi: 10.4161/onci.20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.